Abstract
The Rad50, Mre11 and Nbs1 complex is involved in many essential chromosomal organization processes dealing with DNA ends, including two major pathways of DNA double-strand break repair, homologous recombination and non-homologous end joining. Previous data on the structure of the human Rad50 and Mre11 (R/M) complex suggest that a common role for the protein complex in these processes is to provide a physical link between DNA ends such that they can be processed in an organized and coordinated manner. Here we describe the DNA binding properties of the R/M complex. The complex bound to both single-stranded and double-stranded DNA. Scanning force microscopy analysis of DNA binding by R/M showed the requirement for an end to form oligomeric R/M complexes, which could then migrate or transfer away from the end. The R/M complex had a lower preference for DNA substrates with 3′-overhangs compared with blunt ends or 5′-overhangs. Interestingly, ATP binding, but not hydrolysis, increased the preference of R/M binding to DNA substrates with 3′-overhangs relative to substrates with blunt ends and 5′-overhangs.
INTRODUCTION
DNA ends are a common intermediate in genome metabolism (1). Ends are always present at the termini of linear chromosomes in specialized structures known as telomeres. A DNA end also arises during replication, when a replication fork encounters a single-strand gap or when the fork reverses due to a DNA lesion that blocks its progress. Pairs of ends occur at DNA double-strand breaks (DSBs), which can be caused by endogenous and exogenous DNA-damaging agents. DNA ends are also intermediates in genome rearrangements. For example, DSBs are necessary intermediates during meiotic recombination that creates genetic diversity and during specific mitotic recombination events that create diversity in antibody and T-cell receptor genes.
Despite being frequent intermediates, DNA ends are extremely potent inhibitors of normal cellular function. This is the reason for capture of the natural chromosomal ends into T-loop structures, where the DNA end is not exposed but folded back into a structure, resembling a recombination intermediate (2). Promiscuous recombination of DNA ends causes deleterious chromosomal rearrangements and genomic instability. These events are often the precursors to mutations, uncontrolled cell growth and carcinogenesis. To prevent these catastrophic events it is essential that DNA ends are sequestered or rapidly repaired. Eukaryotic cells primarily utilize two, mechanistically unrelated, pathways to repair DNA ends: non-homologous end joining (NHEJ) and homologous recombination. Repair through NHEJ directly joins two DNA ends at regions of very limited or no sequence homology. Because DSB repair through NHEJ is untemplated, it is error prone. This pathway requires a pair of DNA ends and thus can only repair a true DSB. Homologous recombination, on the other hand, uses the information on a homologous template DNA, such as the sister chromatid, to heal DNA ends. Any DNA sequences missing due to possible degradation at an end can be recovered, making this pathway error free (3). In addition, single DNA ends, resulting from, for instance, replication of a gapped template, can be repaired by homologous recombination (4–7). Although the NHEJ and recombination pathways for DSB repair are mechanistically unrelated, they share the requirement to keep the potential repair partner DNA molecules in close proximity.
An evolutionarily conserved protein complex, containing Rad50, Mre11 and Nbs1 (R/M/N), is involved in diverse aspects of genome metabolism that involve DNA end processing (8). The importance of R/M/N is underscored by the fact that all components are essential in mammalian cells (9–12). In addition, hypomorphic mutations in the human MRE11 and NBS1 genes cause the cancer predisposition syndromes ataxia telangiectasia-like disorder (ATLD) and Nijmegen breakage syndrome (NBS), respectively (13,14). Furthermore, hypomorphic mutations in the murine Rad50 and Nbs1 genes also result in cancer predisposition (12,15,16). Apart from direct involvement in DNA end processing, the R/M/N complex has also been implicated in DNA damage-induced cell cycle regulation via Nbs1 (17).
Structural studies of R/M and its components have revealed an interesting architecture and provided clues to understanding aspects of its function. Both the Pyrococcus furiosus Mre11 and Rad50 globular domains have been crystallized and models for their atomic level structures have been proposed (18,19). The R/M complex contains two Rad50 molecules and two Mre11 molecules. Dimerization of Mre11 presumably contributes to the stability of the R/M complex (19). Contrary to previous notions, scanning force microscopy (SFM) studies have demonstrated that Rad50 forms an intramolecular coiled-coil by folding back onto itself (20). The R/M complex contains two of these coiled-coil arms, emanating from a globular domain. DNA is bound by the globular domain while the flexible arms protrude away. The complex can tether DNA molecules, presumably through multiple interactions of the zinc hooks located at the very tip of the protruding flexible arms (20,21). These observations suggest that a common role of the R/M complex in different aspects of chromosome metabolism is to hold DNA molecules in close proximity.
The R/M/N complex and its components display a variety of biochemical activities relevant to DNA end processing. Mre11, by itself and in the complex, has 3′→5′ exonuclease activity on double-stranded (ds)DNA, as well as endonuclease activity on single-stranded (ss)DNA (22,23). Small regions of DNA homology in the substrate molecules inhibit the nuclease activity (24), perhaps in coordination with the annealing activity of Mre11 (25). The Rad50 amino acid sequence predicts that the N- and C-termini form globular structures with characteristics of Walker type A and B ATPase domains, respectively. For one Rad50 homolog, these domains have been shown to associate and form a functional ATPase (18). However, no influence of ATP on the biochemical activities of human R/M has been detected to date (23,26). Only for Saccharomyces cerevisiae R/M does the presence of ATP result in additional activity to degrade 3′-overhangs (27). The human complex of R/M including Nbs1, however, does have activities that are influenced by ATP. In this case, the presence of ATP promotes opening of fully paired DNA hairpins, induces a relatively weak DNA unwinding activity and alters endonuclease specificity (26). Crystallography revealed that ATP binding causes a change in the relative orientation of two ATPase domains of P.furiosus Rad50, which has been suggested to affect DNA binding (19). We therefore investigated the influence of ATP on the interaction of human R/M with DNA. The complex bound to both ssDNA and dsDNA and had a preference for binding to DNA substrates containing blunt ends or ends with 5′-overhangs relative to DNA molecules ending in 3′-overhangs. Interestingly, ATP binding, but not hydrolysis, changed the preference for R/M association with the different DNA substrates.
MATERIALS AND METHODS
Protein expression and purification
Human R/M was produced by co-infection of Sf21 cells with baculoviruses expressing C-terminally His6-tagged human Rad50 and untagged human Mre11 (generous gifts of T. Paull and M. Gellert) at multiplicities of infection of ∼8 and 5, respectively. Cells were harvested after 72 h. The purification was performed as described previously (25). Equilibrium density centrifugation experiments with the truncated versions of the archaeal structural homologs of Rad50 and Mre11 have shown that this complex is a heterotetramer (R2/M2) (19). Therefore, we assumed a molecular mass of 470 kDa for the human R/M complex. The functionality of the R/M preparation was confirmed by testing the nuclease activity (23). As described previously for human Mre11 by itself, the R/M complex was also able to stimulate annealing of complementary oligonucleotides, although at a reduced efficiency (25 and data not shown).
Antibodies
The hRad50 antibodies (affinity purified, no. 551) used for immunoblotting were a generous gift of C. Heyting. The hMre11 antibodies (serum, no. 2244) used for immunoblotting were generated as described previously (25).
Size fractionation
Fifty micrograms of purified R/M were loaded on a Superose 6 PC 3.2/30 size fractionation column (Amersham Biosciences). Protein buffer (150 mM KCl, 25 mM Tris–HCl pH 7.8 and 10% glycerol) was used for equilibration and fractionation. Fifty microliter fractions were collected, of which 10 µl was analyzed by immuno-spotblotting with Rad50 and Mre11 antibodies.
DNA substrates
The 160 bp dsDNA fragment used in gel retardation experiments was obtained by a PCR reaction with M13 ssDNA as template, using 22 nt primers that hybridized to positions 6425 and 6584. The 160 nt ssDNA fragment used in gel retardation experiments was obtained by a PCR reaction as described above with one biotinylated primer. After the PCR reaction, DNA was bound to magnetic streptavidin beads (Dynal) following the manufacturer’s protocols. The non-biotinylated DNA strand was eluted by incubating the DNA-bound beads in 0.1 M NaOH for 3 min. The ssDNA-containing supernatant was removed and neutralized with an equal volume of 0.1 M HCl. Both the ssDNA and dsDNA fragments were subsequently purified using a QiaQuick PCR purification kit (Qiagen).
The DNA substrates used for SFM were produced by digestion of pcDNA3.1(+) (Invitrogen). A 5.4 kb blunt-ended fragment was created by StuI (New England Biolabs) digestion, after which the DNA was phenol extracted. To create a 4.8 kb fragment with non-complementary 4 and 3 nt 3′-overhangs, plasmid DNA was digested with KpnI and DraIII (NEB), respectively. To create a 4.5 kb fragment with non-complementary 4 nt 5′-overhangs, plasmid DNA was digested with BglII and XhoI (NEB). After double digestion, DNA fragments were separated by electrophoresis and the desired fragments were isolated from the gel with the use of a QiaQuick gel extraction kit (Qiagen). A singly nicked, circular 3 kb DNA substrate was generated by DNase I treatment of pBlueScript (Stratagene) as described (28).
Gel retardation experiments
Binding of R/M to DNA was analyzed using gel mobility shift assays as described (25). Briefly, a 32P-end-labeled 160 nt or bp DNA fragment was incubated at 2.5 nM with R/M at the indicated concentrations in binding buffer (50 mM HEPES– KOH pH 8.0, 10 mM Tris–HCl, 100 mM KCl, 4.2% glycerol and 0.5 mg/ml BSA). Binding reactions were separated on a 0.3% agarose gel in 0.5× TBE.
ATPase assays
To assay ATPase activity, radiolabeled [γ-32P]ATP at 5 nM, supplemented with non-labeled ATP to the indicated concentrations, was incubated with 825 nM R/M in 100 mM KCl, 10 mM Tris–HCl pH 7.8, 4% glycerol, 10 mM HEPES–KOH pH 8.0, 5 mM MgCl2, 0.2 mg/ml BSA for 0, 5, 10 and 20 min at 30°C. Reactions were terminated by the addition of EDTA to 167 mM. Samples were analyzed by thin layer chromatography (Merck TLC plates), run in 0.75 M KH2PO4 pH 4.5, and subsequently quantified by phosphorimaging.
The maximal rate of R/M-mediated ATP hydrolysis was determined from a plot of the hydrolysis rate (V) during the first 20 min of the reaction at different substrate concentrations ([S]). These data are plotted in a double reciprocal plot (see Fig. 4A).
Figure 4.
R/M possesses a low ATPase activity that influences the affinity for different DNA ends. (A) Analysis of the ATPase activity of the human R/M complex. The rate of ATP hydrolysis (V) was determined by analysis of the first 20 min of the reaction at different ATP concentrations ([S]). Shown is a double reciprocal plot of 1/V against 1/[S] from which the Km and Vmax values were determined. Error bars indicate SEM of independent triplicate experiments. (B) Quantification of R/M affinity for different DNA substrates in the presence of ATP. Blunt-ended DNA or DNA with either 3′- or 5′-overhangs was incubated at 3 nM with 56 nM R/M in the presence of ATP or AMP-PNP. Shown is the percentage of DNA molecules bound by R/M oligomeric complexes for different substrates either in the absence of ATP (white bars), the presence of ATP (black bars) or the presence of AMP-PNP (hatched bars). Error bars indicate SEM of independent triplicate experiments with at least 100 DNA molecules analyzed for each data point.
Scanning force microscopy
DNA–protein complexes for SFM were formed in 20 µl reactions as described for the gel mobility shift experiments, with the exception of the inclusion of 2 mM MgCl2 and the omission of BSA. ATP, when present, was at a concentration of 2 mM. DNA and protein concentrations were as described in the figure legends. Reactions were diluted 10-fold in deposition buffer (10 mM HEPES–KOH pH 7.5 and 10 mM MgCl2) and deposited on freshly cleaved mica. After ∼1 min the mica was washed with water (glass distilled; Sigma) and exposed to a stream of filtered air. Samples were imaged in air at room temperature and humidity, using a NanoScope IIIa (Digital Instruments), operating in tapping mode with a type E scanner. Silicon tips (Nanoprobes) were from Digital Instruments or Nanofactory.
The binding of the R/M complex to DNA was quantified by counting different types of R/M complex bound to the different DNA substrates. Specifically, monomeric R/M complexes or small multimers with less than 10 protein arms were distinguished from oligomeric complexes. The protein–DNA complexes were further categorized according to whether they were located at the end or internally on the DNA molecule or whether they tethered multiple DNA molecules.
RESULTS
Production and purification of the human Rad50/Mre11 complex
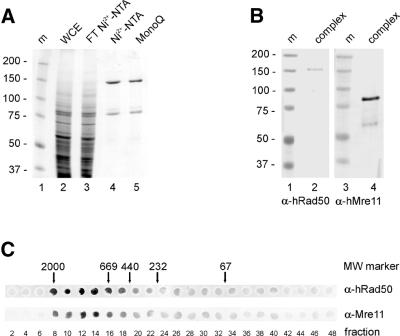
The human R/M complex was purified from Sf21 cells co-infected with baculoviruses expressing histidine-tagged Rad50 and untagged Mre11. A protein profile of fractions from the purification is shown in Figure 1A. The final fraction (lane 5) was estimated to contain Rad50 and Mre11 in approximately stoichiometric amounts. The identity of the proteins in this fraction was confirmed by immunoblot analysis (Fig. 1B) using Rad50 and Mre11 antibodies (lanes 2 and 4, respectively). Size fractionation showed that the purified Rad50 and Mre11 were present in a complex, because the separate Rad50 and Mre11 proteins could not be detected at their respective positions of 150 and 85 kDa (Fig. 1C). The complex eluted at a position corresponding to a molecular mass of ∼1.5 MDa, as compared with the elution pattern of molecular weight standards in a parallel analysis. Because the complex does not have a globular shape (20) its elution profile cannot be used to estimate its molecular mass or the stoichiometry of its components. However, previous biophysical analyses of an archaeal R/M complex have shown that it consists of a heterotetramer containing two Rad50 and two Mre11 molecules (19), which would yield a molecular mass of ∼470 kDa.
Figure 1.
Purification of the human R/M complex. (A) SDS–PAGE analysis of human R/M complex purification, stained with Coomassie Brilliant Blue. Lane 1, m, molecular size markers (molecular mass indicated in kDa); lane 2, WCE, soluble fraction of extracted infected Sf21 cells; lane 3, flow-through of a Ni2+–NTA column; lane 4, pool of eluted fractions of the Ni2+–NTA column; lane 5, R/M preparation eluted from a MonoQ column. (B) Immunoblot analysis of the purified complex. Lanes 1 and 3, m, molecular size markers (molecular mass indicated in kDa); lanes 2 and 4, 500 ng of protein from the MonoQ fractions. Lane 2 was probed with anti-Rad50 antibodies and lane 4 with anti-Mre11 antibodies. (C) Size fractionation analysis of the protein complex. Fractions from a Superose 6 column were analyzed by immuno-spotblotting with anti-Rad50 and anti-Mre11 antibodies as indicated. Positions of molecular size markers relative to eluted fraction numbers are indicated (molecular mass indicated in kDa).
Rad50/Mre11 binds ssDNA and dsDNA
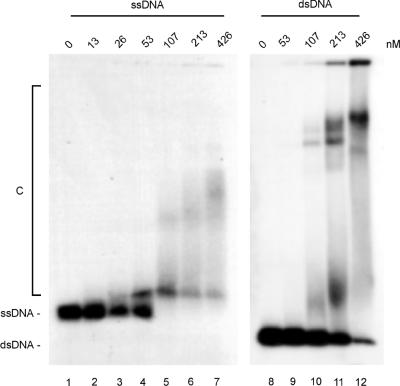
The R/M complex was tested in DNA binding reactions including either a radiolabeled 160 nt ssDNA or 160 bp dsDNA fragment (see Materials and Methods). Distinct DNA–protein complexes were formed with both ssDNA and dsDNA (Fig. 2). Binding to ssDNA resulted in at least two complexes that differed in mobility, while binding to dsDNA resulted in at least three distinct complexes. Stable complexes formed at lower protein concentrations on ssDNA than on dsDNA (compare lanes 4 and 5 with 9 and 10). To confirm that the binding to the 160 nt ssDNA fragment was not due to secondary structures in the DNA substrate, binding to a 90 nt dA oligonucleotide substrate was tested as well. The complex did bind to this substrate with a similar affinity, confirming that the complex had the capabilty to bind to ssDNA (data not shown). Binding to short (50 nt or bp) ssDNA or dsDNA fragments did not result in the formation of DNA–protein complexes that were stable enough to be detected as discrete bands in gel mobility shift experiments (data not shown). The presence or absence of divalent cations and/or ATP did not influence R/M binding to DNA as assayed by this method.
Figure 2.
DNA binding by the R/M complex. Radiolabeled 160 nt ssDNA or 160 bp dsDNA fragments were incubated at a concentration of 2.5 nM DNA fragments with the indicated concentrations of R/M for 20 min at 25°C. Samples were analyzed by agarose gel electrophoresis, followed by autoradiography. Positions of unbound ssDNA, dsDNA and DNA–protein complexes (C) are indicated.
Formation of Rad50/Mre11 oligomers at different DNA end structures
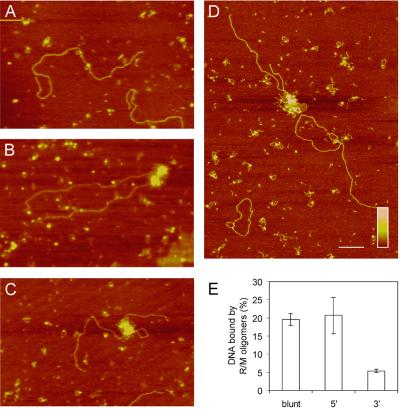
The biochemical activities of the R/M complex that have been described to date differ for various DNA end structures and therefore we investigated the influence of the type of DNA ends on R/M binding. To analyze the DNA binding properties of the human R/M complex in more detail, we used SFM. Protein–DNA complexes can be detected by SFM at much lower, presumably more physiologically relevant, protein to DNA ratios than are needed for gel assays (20,29). Furthermore, by directly observing protein–DNA complexes, different architectures can be distinguished. DNA substrates were prepared from the same plasmid DNA digested with restriction endonucleases to produce substrates with either blunt ends or single-strand overhangs with either 3′ or 5′ polarity. As shown in Figure 3A–C, different distinct classes of R/M–DNA complexes were observed. The first class consisted of single R/M complexes bound to linear DNA via their globular domains with their two coiled-coil arms protruding away from the DNA (Fig. 3A). In addition, we classified small multimeric R/M complexes, with up to 10 coiled-coil arms, as belonging to this class. The second class consisted of much larger oligomeric R/M complexes bound to DNA (Fig. 3B and C). Previous experiments indicated that a DNA end is required for the formation of oligomeric R/M complexes (20). Therefore, we quantitated this behavior of the R/M complex by performing competition experiments with a mixture of linear 5 kb DNA substrates and nicked circular 3 kb DNA substrates (Fig. 3D). While 14% of the linear DNA molecules were bound by oligomeric R/M complexes, only 2% of the circular DNA substrates were bound by this form of R/M. In contrast, for monomeric R/M complexes an equal binding efficiency, of ∼35%, was observed for both linear and circular DNA substrates. The oligomeric R/M complexes were not only observed at the ends of these 5 kb DNA substrates, but a majority (up to >80%) could be found at internal locations on DNA molecules. Both end-bound and internally bound R/M oligomers could tether distant DNA sites, inter- as well as intramolecularly (Fig. 3B and C, respectively). In accordance with previous data, tethering of DNA by single or small multimeric R/M complexes was never observed (20). The percentage of DNA molecules bound by R/M oligomers was determined for each DNA substrate (Fig. 3E). Approximately 20% of the substrate molecules with blunt ends and 5′-overhangs were bound by R/M oligomers. In contrast, only 5% of the DNA substrates with 3′-overhangs were bound by R/M oligomers. Of the DNA molecules bound by R/M oligomers, the relative contribution of end-bound, internally bound and tethered DNA molecules did not differ significantly among the different DNA substrates.
Figure 3.
Influence of DNA end structure on R/M binding. Examples of R/M binding to 5 kb linear DNA molecules. DNA molecules were incubated at 3 nM with 56 nM R/M in the absence of ATP. (A) Monomeric R/M protein complexes bound to linear DNA. (B) Binding of oligomeric complexes to linear DNA substrates. In the example shown, oligomeric complexes are located at DNA ends and tether two DNA molecules. (C) An example of internally bound oligomeric complexes, resulting in intramolecular tethering. (D) Scanning force micrograph of a reaction mixture containing 5 kb linear DNA substrates and 3 kb nicked circular DNA substrates at a 1:1 ratio. R/M forms oligomeric complexes only on linear DNA substrates. The scale bar is 200 nm and color represents height from 0 to 1 nm (dark to light) as shown by the key insert. (E) Quantification of R/M binding to different DNA substrates. Blunt-ended DNA or DNA with either 3′- or 5′-overhangs was incubated at ∼3 nM with 56 nM R/M in the absence of ATP. Shown is the percentage of DNA molecules bound by R/M oligomeric complexes for different DNA substrates. Error bars indicate SEM of independent triplicate experiments with at least 100 DNA molecules analyzed for each data point.
ATP hydrolysis by the Rad50/Mre11 complex
The human Rad50 amino acid sequence, as well as that of other Rad50 homologs, includes an N-terminal Walker A motif and a C-terminal Walker B motif (30,31), indicating potential ATPase activity. ATP-dependent DNA binding activities in the absence of the Mre11 subunits have been demonstrated for S.cerevisiae Rad50 and the catalytic domains of the P.furiosus structural Rad50 homolog (32,33). ATP hydrolysis has only been directly demonstrated for P.furiosus Rad50 (18). Furthermore, for the structural R/M homolog of Escherichia coli, SbcCD, ATP-dependent DNA binding and nuclease activities have been revealed (34). We measured the ATPase activity of the purified human R/M complex by determining the rate of ATP hydrolysis at different substrate concentrations (Fig. 4A). The Km for ATP of the R/M complex was 33 nM and its Vmax was 222 fmol min–1. Thus, the R/M complex has a relatively weak ATPase activity with a turnover rate of 0.026 min–1 complex–1. The ATPase activity of a number of other proteins involved in DSB repair, such as human Rad51 and Rad54, dramatically increases in the presence of ssDNA and dsDNA, respectively (35,36). However, the kinetics of the R/M complex ATPase activity did not change dramatically upon addition of either ssDNA or dsDNA (data not shown).
Influence of ATP on R/M oligomer formation at different DNA end structures
To investigate whether ATP affected DNA binding by R/M, we analyzed binding of R/M to the different DNA substrates in the presence of ATP. The efficiency of formation of large R/M oligomers on the different DNA substrates, in the presence (black bars) and absence of ATP (white bars), is graphically represented in Figure 4B. Binding of DNA substrates containing either blunt ends or 5′-overhangs was slightly reduced in the presence of ATP, while binding to substrates ending in 3′-overhangs was increased significantly. The addition of ATP increased binding to DNA substrates with 3′-overhangs to ∼20%, just as was observed for DNA substrates with blunt ends and 5′-overhangs in the absence of ATP.
Subsequently, we tested whether ATP hydrolysis was required to influence the formation of R/M oligomers on the different DNA substrates. ATP was replaced with the non-hydrolyzable ATP analog AMP-PNP in the DNA binding reactions. As shown in Figure 4B (hatched bars), the presence of AMP-PNP enhanced the effect observed with ATP. The binding to DNA substrates with blunt ends or 5′-single-stranded overhangs was further decreased to about half of the binding in the absence of ATP. In contrast, R/M oligomer formation on 3′-overhangs was increased 1.5-fold compared with binding in the presence of ATP and 6-fold compared with binding in the absence of ATP.
DISCUSSION
The Rad50/Mre11 complex is essential for many aspects of chromosomal metabolism. Here, we have characterized the DNA binding properties of the human R/M complex by SFM. R/M preferentially binds to particular DNA end structures and this preference can be modified by addition of ATP. R/M requires a DNA end to form oligomers which can subsequently migrate or transfer on the DNA.
The crystal structure of P.furiosus Mre11 has revealed a likely DNA-binding pocket with two manganese ions at the nuclease active site. Modeling of DNA substrates into this pocket suggests that dsDNA substrates need to be deformed to get the phosphodiester backbone of the DNA close enough to the active site metal ions for catalysis to occur. In contrast, the inherently more flexible ssDNA can easily be positioned in close proximity to the metal ions (19). The analysis of DNA binding by gel retardation experiments showed that R/M binds to both ssDNA and dsDNA substrates (Fig. 2). The geometry of the DNA-binding pocket has implications for the specificity of the Mre11 nuclease activity. Mre11 and the R/M complex display a manganese-dependent 3′→5′ exonuclease activity on DNA substrates with blunt ends or 5′-overhangs, while substrates with 3′-overhangs are not degraded (23,27). The results of the experiments we describe here suggest that the nuclease activity of the R/M complex might be linked to its DNA binding properties. In the absence of ATP, the R/M complex prefers binding to DNA substrates with blunt ends or 5′-overhangs compared with substrates with 3′-overhangs (Fig. 3E).
Biochemical experiments have shown that addition of ATP stimulates the degradation of substrates with 3′-overhangs by S.cerevisiae R/M complex (27). For the human R/M/N complex, ATP has also been shown to stimulate degradation of DNA substrates with 3′-overhangs (26). This ATP-induced effect on Mre11 is likely to be mediated through Rad50, although a direct influence of ATP on the catalytic domain of Mre11 cannot be excluded. The crystal structure of P.furiosus Rad50 catalytic domains provides some insight into how ATP binding by Rad50 might influence DNA binding by Mre11. The P.furiosus Rad50 catalytic domain dimer forms two ATPase sites. The presence of AMP-PNP in the Rad50 crystals induces a 30° rotation of the N-terminal globular domain with respect to the C-terminal globular domain. Furthermore, the crystal structure has revealed that the Rad50 dimer contains a positively charged groove, which could be a potential DNA-binding site (18). From these observations it has been suggested that the structural change in Rad50 induced by ATP binding could alter the conformation of DNA bound to the R/M complex, thereby influencing the position of the DNA in the Mre11 DNA-binding pocket and active site. Alternatively, the structural change in Rad50 could directly modify the conformation of the Mre11 DNA-binding pocket. Our results show that the presence of ATP stimulates the binding of the R/M complex to DNA substrates with 3′-overhangs, while the binding to substrates with blunt ends and 5′-overhangs decreases slightly. A similar, but more pronounced, effect is observed in the presence of AMP-PNP (Fig. 4B). Because AMP-PNP cannot be hydrolyzed to ADP, the presence of AMP-PNP results in a R/M complex that is presumably continuously in its ‘ATP-bound’ conformation, which apparently has the most dramatic effect on preference for binding to different ends. These data show that the ATP binding by Rad50 affects the DNA binding specificity of the complex and could, therefore, by extension, influence the nuclease activities of Mre11.
In addition to ATP influencing human R/M end-binding preference (Fig. 4B), DNA ends themselves influence the interaction among R/M complexes. Human R/M binds both to circular and linear DNA molecules (20). However, R/M forms larger oligomeric complexes on linear DNA molecules and not on circular DNA molecules (Fig. 3D). Interestingly, although R/M requires a DNA end to oligomerize, the position of the R/M oligomers is not restricted to the DNA end of the 5 kb DNA molecules used in our study (Fig. 3B–D). The R/M complexes formed on circular and linear DNA are functionally different; the smaller R/M complexes found on circular DNA do not interact with each other even if they are present on the same DNA molecule. In contrast, tethering of different DNA molecules via interactions of larger R/M oligomers, most likely through the coiled-coil arms, is observed for ∼30% of linear DNA molecules bound by R/M oligomers (Fig. 3B). We also observe intramolecular DNA tethering via two R/M oligomers bound at internal positions on a DNA molecule (Fig. 3C). R/M oligomers observed at internal positions on linear DNA are possibly a consequence of migration or transfer of the complex along the DNA after end binding, since in direct competition experiments large R/M oligomers are almost never observed on circular DNA (20) (Fig. 3D).
Interestingly, a similar mode of DNA binding with respect to DNA ends and inward migration is observed for another end-binding protein complex, the Ku70/Ku86 heterodimer. The ring structure of Ku70/Ku86 requires the complex to initially interact with a DNA end, after which it can migrate along the DNA (37–39). In vitro and in vivo experiments have demonstrated that the R/M and the Ku70/86 protein complexes are involved in DNA end metabolism. They are key components of DSB repair pathways, but are also implicated in V(D)J recombination and telomere length maintenance (8,40–45). Based on their structures, the R/M complex and the Ku70/86 heterodimer have been proposed to recognize and structurally organize DNA ends (20,38,46,47). The inward migration or transfer of the R/M and Ku70/86 complexes is probably a key aspect of their function because it allows a molecular hand-off between proteins required for structural organization of DNA ends and end-processing factors. Due to chromatin structure or other interacting factors, the inward migration is likely to be limited, which has two advantages. It will allow access of factors with specific functions, such as (further) nucleolytic processing, nucleotide addition or strand ligation, to the very DNA ends, while it still enables protein complexes such as R/M to function in proximity of the DNA ends to keep them organized.
Acknowledgments
ACKNOWLEDGEMENTS
We thank K. P. Hopfner and M. Modesti for helpful discussions. This work was supported by grants from the Dutch Cancer Society (KWF) and the Chemical Sciences section (CW) of the Netherlands Organization for Scientific Research (NWO).
REFERENCES
- 1.Cromie G.A., Connelly,J.C. and Leach,D.R. (2001) Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell, 8, 1163–1174. [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. (2002) Protection of mammalian telomeres. Oncogene, 21, 532–540. [DOI] [PubMed] [Google Scholar]
- 3.Kanaar R., Hoeijmakers,J.H. and van Gent,D.C. (1998) Molecular mechanisms of DNA double strand break repair. Trends Cell Biol., 8, 483–489. [DOI] [PubMed] [Google Scholar]
- 4.Rothstein R., Michel,B. and Gangloff,S. (2000) Replication fork pausing and recombination or “gimme a break”. Genes Dev., 14, 1–10. [PubMed] [Google Scholar]
- 5.Cox M.M., Goodman,M.F., Kreuzer,K.N., Sherratt,D.J., Sandler,S.J. and Marians,K.J. (2000) The importance of repairing stalled replication forks. Nature, 404, 37–41. [DOI] [PubMed] [Google Scholar]
- 6.Kowalczykowski S.C. (2000) Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci., 25, 156–165. [DOI] [PubMed] [Google Scholar]
- 7.Michel B., Flores,M.J., Viguera,E., Grompone,G., Seigneur,M. and Bidnenko,V. (2001) Rescue of arrested replication forks by homologous recombination. Proc. Natl Acad. Sci. USA, 98, 8181–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haber J.E. (1998) The many interfaces of Mre11. Cell, 95, 583–586. [DOI] [PubMed] [Google Scholar]
- 9.Xiao Y. and Weaver,D.T. (1997) Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res., 25, 2985–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo G., Yao,M.S., Bender,C.F., Mills,M., Bladl,A.R., Bradley,A. and Petrini,J.H. (1999) Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl Acad. Sci. USA, 96, 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J., Petersen,S., Tessarollo,L. and Nussenzweig,A. (2001) Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol., 11, 105–109. [DOI] [PubMed] [Google Scholar]
- 12.Kang J., Bronson,R.T. and Xu,Y. (2002) Targeted disruption of NBS1 reveals its roles in mouse development and DNA repair. EMBO J., 21, 1447–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart G.S., Maser,R.S., Stankovic,T., Bressan,D.A., Kaplan,M.I., Jaspers,N.G., Raams,A., Byrd,P.J., Petrini,J.H. and Taylor,A.M. (1999) The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell, 99, 577–587. [DOI] [PubMed] [Google Scholar]
- 14.Varon R., Vissinga,C., Platzer,M., Cerosaletti,K.M., Chrzanowska,K.H., Saar,K., Beckmann,G., Seemanova,E., Cooper,P.R., Nowak,N.J., Stumm,M., Weemaes,C.M., Gatti,R.A., Wilson,R.K., Digweed,M., Rosenthal,A., Sperling,K., Concannon,P. and Reis,A. (1998) Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell, 93, 467–476. [DOI] [PubMed] [Google Scholar]
- 15.Bender C.F., Sikes,M.L., Sullivan,R., Erskine Huye,L., Le Beau,M.M., Roth,D.B., Mirzoeva,O.K., Oltz,E.M. and Petrini,J.H.J. (2002) Cancer predisposition and hematopoietic failure in Rad50S/S mice. Genes Dev., 16, 2237–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Jager M. and Kanaar,R. (2002) Genome instability and Rad50S: subtle yet severe. Genes Dev., 16, 2173–2178. [DOI] [PubMed] [Google Scholar]
- 17.Carney J.P., Maser,R.S., Olivares,H., Davis,E.M., Le Beau,M., Yates,J.R.,III, Hays,L., Morgan,W.F. and Petrini,J.H. (1998) The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell, 93, 477–486. [DOI] [PubMed] [Google Scholar]
- 18.Hopfner K.P., Karcher,A., Shin,D.S., Craig,L., Arthur,L.M., Carney,J.P. and Tainer,J.A. (2000) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell, 101, 789–800. [DOI] [PubMed] [Google Scholar]
- 19.Hopfner K.P., Karcher,A., Craig,L., Woo,T.T., Carney,J.P. and Tainer,J.A. (2001) Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell, 105, 473–485. [DOI] [PubMed] [Google Scholar]
- 20.de Jager M., van Noort,J., van Gent,D.C., Dekker,C., Kanaar,R. and Wyman,C. (2001) Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell, 8, 1129–1135. [DOI] [PubMed] [Google Scholar]
- 21.Hopfner K.P., Craig,L., Moncalian,G., Zinkel,R.A., Usui,T., Owen,B.A., Karcher,A., Henderson,B., Bodmer,J.L., McMurray,C.T., Carney,J.P., Petrini,J.H. and Tainer,J.A. (2002) The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature, 418, 562–566. [DOI] [PubMed] [Google Scholar]
- 22.Trujillo K.M., Yuan,S.S., Lee,E.Y. and Sung,P. (1998) Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem., 273, 21447–21450. [DOI] [PubMed] [Google Scholar]
- 23.Paull T.T. and Gellert,M. (1998) The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell, 1, 969–979. [DOI] [PubMed] [Google Scholar]
- 24.Paull T.T. and Gellert,M. (2000) A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl Acad. Sci. USA, 97, 6409–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Jager M., Dronkert,M.L., Modesti,M., Beerens,C.E., Kanaar,R. and van Gent,D.C. (2001) DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res., 29, 1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paull T.T. and Gellert,M. (1999) Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev., 13, 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trujillo K.M. and Sung,P. (2001) DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50.Mre11 complex. J. Biol. Chem., 276, 35458–35464. [DOI] [PubMed] [Google Scholar]
- 28.Ristic D., Wyman,C., Paulusma,C. and Kanaar,R. (2001) The architecture of the human Rad54-DNA complex provides evidence for protein translocation along DNA. Proc. Natl Acad. Sci. USA, 98, 8454–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyman C., Rombel,I., North,A.K., Bustamante,C. and Kustu,S. (1997) Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science, 275, 1658–1661. [DOI] [PubMed] [Google Scholar]
- 30.Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aravind L., Walker,D.R. and Koonin,E.V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raymond W.E. and Kleckner,N. (1993) RAD50 protein of S.cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res., 21, 3851–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hopfner K.P., Karcher,A., Shin,D., Fairley,C., Tainer,J.A. and Carney,J.P. (2000) Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J. Bacteriol., 182, 6036–6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Connelly J.C., de Leau,E.S., Okely,E.A. and Leach,D.R. (1997) Overexpression, purification, and characterization of the SbcCD protein from Escherichia coli. J. Biol. Chem., 272, 19819–19826. [DOI] [PubMed] [Google Scholar]
- 35.Benson F.E., Stasiak,A. and West,S.C. (1994) Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J., 13, 5764–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swagemakers S.M., Essers,J., de Wit,J., Hoeijmakers,J.H. and Kanaar,R. (1998) The human RAD54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J. Biol. Chem., 273, 28292–28297. [DOI] [PubMed] [Google Scholar]
- 37.Paillard S. and Strauss,F. (1991) Analysis of the mechanism of interaction of simian Ku protein with DNA. Nucleic Acids Res., 19, 5619–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker J.R., Corpina,R.A. and Goldberg,J. (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature, 412, 607–614. [DOI] [PubMed] [Google Scholar]
- 39.de Vries E., van Driel,W., Bergsma,W.G., Arnberg,A.C. and van der Vliet,P.C. (1989) HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA-multimeric protein complex. J. Mol. Biol., 208, 65–78. [DOI] [PubMed] [Google Scholar]
- 40.Featherstone C. and Jackson,S.P. (1999) Ku, a DNA repair protein with multiple cellular functions? Mutat. Res., 434, 3–15. [DOI] [PubMed] [Google Scholar]
- 41.Manis J.P., Gu,Y., Lansford,R., Sonoda,E., Ferrini,R., Davidson,L., Rajewsky,K. and Alt,F.W. (1998) Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J. Exp. Med., 187, 2081–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casellas R., Nussenzweig,A., Wuerffel,R., Pelanda,R., Reichlin,A., Suh,H., Qin,X.F., Besmer,E., Kenter,A., Rajewsky,K. and Nussenzweig,M.C. (1998) Ku80 is required for immunoglobulin isotype switching. EMBO J., 17, 2404–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H.T., Bhandoola,A., Difilippantonio,M.J., Zhu,J., Brown,M.J., Tai,X., Rogakou,E.P., Brotz,T.M., Bonner,W.M., Ried,T. and Nussenzweig,A. (2000) Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science, 290, 1962–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu H.L., Gilley,D., Blackburn,E.H. and Chen,D.J. (1999) Ku is associated with the telomere in mammals. Proc. Natl Acad. Sci. USA, 96, 12454–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu X.D., Kuster,B., Mann,M., Petrini,J.H. and Lange,T. (2000) Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nature Genet., 25, 347–352. [DOI] [PubMed] [Google Scholar]
- 46.Hopfner K.P., Putnam,C.D. and Tainer,J.A. (2002) DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol., 12, 115–122. [DOI] [PubMed] [Google Scholar]
- 47.Jones J.M., Gellert,M. and Yang,W. (2001) A Ku bridge over broken DNA. Structure, 9, 881–884. [DOI] [PubMed] [Google Scholar]