Abstract
We showed previously that bacterially expressed full-length human wild-type p53b(1–393) binds selectively to supercoiled (sc)DNA in sc/linear DNA competition experiments, a process we termed supercoil-selective (SCS) binding. Using p53 deletion mutants and pBluescript scDNA (lacking the p53 recognition sequence) at native superhelix density we demonstrate here that the p53 C-terminal domain (amino acids 347–382) and a p53 oligomeric state are important for SCS binding. Monomeric p53(361–393) protein (lacking the p53 tetramerization domain, amino acids 325–356) did not exhibit SCS binding while both dimeric mutant p53(319– 393)L344A and fusion protein GCN4–p53(347–393) were effective in SCS binding. Supershifting of p53(320–393)–scDNA complexes with monoclonal antibodies revealed that the amino acid region 375–378, constituting the epitope of the Bp53-10.1 antibody, plays a role in binding of the p53(320–393) protein to scDNA. Using electron microscopy we observed p53–scDNA nucleoprotein filaments produced by all the C-terminal proteins that displayed SCS binding in the gel electrophoresis experiments; no filaments formed with the monomeric p53(361– 393) protein. We propose a model according to which two DNA duplexes are compacted into p53–scDNA filaments and discuss a role for filament formation in recombination.
INTRODUCTION
The p53 tumor suppressor protein protects cells from malignant transformation by regulating the responses of cell growth and death to genotoxic agents (reviewed in 1–3). Stress-induced active p53 protein triggers growth arrest or cell death by apoptosis at least in part via transcriptional activation of a set of genes containing p53 recognition sites. In an unstressed cell, p53 participates in various processes of DNA repair and DNA recombination by virtue of its ability to interact with protein components of the repair and recombination machineries and via various DNA-binding activities (4).
p53 protein contains 393 amino acids that comprise several functional domains (5,6). The N-terminal domain (amino acids 1– to ∼100) encompasses a transactivation region (amino acids 1–42) and a proline-rich region with five copies of the sequence PXXP (amino acids 61–94). The evolutionarily highly conserved central (core) domain (amino acids ∼100 to ∼300) is involved in sequence-specific binding to promoters of p53-regulated genes (7). This domain also interacts with internal regions of single-stranded (ss)DNA (8), three-stranded DNA substrates mimicking early recombination intermediates (9), insertion/deletion mismatches (10) and cruciforms (11) and also manifests a 3′→5′ exonuclease activity (12). Point mutations in this domain are the most frequent alterations in p53 found in human cancers (13). The C-terminal region of the protein contains a flexible linker (amino acids ∼300 to ∼325), a tetramerization domain (amino acids ∼325–356) and a basic C-terminal DNA-binding domain (amino acids 363–382). The ability of the C-terminus to bind to single-stranded gaps in double-stranded (ds)DNA (14), γ-irradiated DNA in vitro (15) and ssDNA ends and to catalyze DNA strand transfer (8,16) presumably accounts for the role of p53 in DNA repair and recombination.
Recently, it was shown that p53 protein binds strongly to negatively as well as to positively supercoiled (sc) plasmid DNA (17,18). It has been suggested that both the core domain and the C-terminal domain regulate the binding of p53 to scDNA (17–19). Competition between scDNAs and their linearized (lin) forms revealed a strong preference for scDNAs by wild-type p53, suggesting a new p53–DNA binding mode denoted supercoil-selective (SCS) binding (20). The p53 core domain exhibited only weak preferential binding to scDNA in the sc/lin competition assay, supporting the previous notion that some other domain (probably C-terminal) is involved in selective binding of the full-length p53 to scDNA (20,21).
Several proteins bind preferentially to scDNA. For example, prokaryotic topoisomerase I (ω protein) distinguishes DNA topology by binding to local regions with single-stranded character stabilized by the underwinding of the double helix (22). It has been proposed that the preference of eukaryotic topoisomerases and HMG box-containing proteins (e.g. chromosomal HMG1 and xUBF transcription factor) for crossovers in scDNA accounts for their ability to distinguish the topological state of both negatively and positively scDNA (23,24).
In this study, we have analyzed the contributions of the p53 domains to preferential binding to scDNA. Using bacterially expressed p53 deletion mutants and chimeric proteins we demonstrate that dimerization of the C-terminal segment is a prerequisite for the strong SCS binding of p53. Visualization of p53–scDNA complexes by electron microscopy reveal the ability of p53 dimers to stabilize two DNA duplexes in close vicinity, leading ultimately to the formation of nucleoprotein filaments.
MATERIALS AND METHODS
Recombinant plasmids
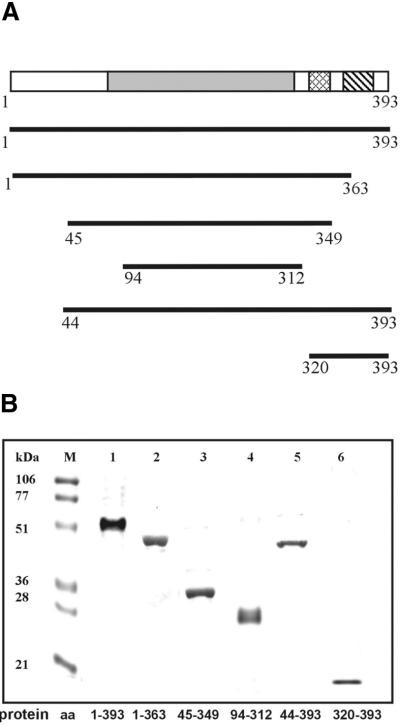
Plasmids encoding human wild-type p53(1–393), p53(1–363), p53(45–349) and p53(44–393) were kindly provided by C. Midgley (25). Core domain-containing recombinant plasmid p53(94–312) was from S. Gorina (26). C-terminal p53 fragment p53(320–393) inserted in pGEX-2T vector was from M. Protopopova (8,14). Figure 1A schematically shows the p53 fragments expressed from these plasmids.
Figure 1.
Purity of p53 deletion mutants isolated from bacterial extracts. (A) p53 fragments expressed in bacteria, shown as lines below the map of p53 domains. The evolutionarily conserved domains are indicated: core DNA-binding domain (shaded; amino acids ∼100–300), tetramerization domain (cross-hatched; amino acids 325–356) and basic C-terminal DNA-binding domain (hatched; amino acids 363–382). Numbers of first and last p53 amino acid residues are indicated for each construct. (B) Gel electrophoresis of full-length p53 and its fragments after their purification by FPLC (see Materials and Methods). Approximately 1 µg of peak fraction of each protein was analyzed by 10–15% gradient SDS–PAGE. Standard protein molecular weight marker (M) was used to compare molecular masses of the proteins.
PCR and standard cloning procedures were used (27) to prepare plasmids for expression of p53 C-terminal proteins. To clone p53(319–393)L344A mutant, p53 cDNA (kindly provided by M. G. Luciani) containing a substitution of CTG by GCG (Leu→Ala at amino acid position 344) was used as a template for PCR amplification of the p53 region coding for amino acid residues 319–393. The dimerization domain (amino acids 252–280) of transcription factor GCN4 (plasmid pLZ335 kindly provided by T. Halazonetis; 28) was PCR amplified and inserted into plasmid pGEX containing p53(347–393) to create chimeric GCN4–p53(347–393) protein. The p53 insert in plasmid pGEX-p53(363–393) started from the AGG codon for K363; the artificial G361 and S362 codons originated from the pGEX vector. Since thrombin cleavage (see below) leaves both amino acids G and S fused to p53 protein, the final product contained amino acid residues identical to p53(361–393). DNA sequencing of each plasmid confirmed that no additional mutations were introduced into the p53 coding regions during PCR.
Supercoiled pBluescript SK II plasmid DNA (17) was isolated from bacterial strain TOP10 as described in the Qiagen protocol (Qiagen, Germany). SmaI restriction enzyme (Takara, Japan) was used for linearization of pBluescript SK II (Stratagene).
p53 recombinant proteins purification
Each of the p53 constructs was expressed in Escherichia coli strain BL21 using a two-step induction at 18°C to limit protein aggregation (29). The proteins p53b(1–393), p53(1–363), p53(44–393) and p53(45–349) were purified according to a protocol described previously (30) with some modifications. Bacterial lysate was loaded at 4°C and 1 ml/min onto a 5 ml HiTrap–heparin column (Pharmacia, Sweden) that had been pre-equilibrated in buffer A (10% glycerol, 25 mM Na HEPES, pH 7.6, 5 mM DTT, 1 mM benzamidine) with 50 mM KCl. After washing the column with 8 column vol of buffer A containing 50 mM KCl, an 8 column vol linear gradient (from 0.05 to 1 M KCl in buffer A) was initiated at 0.5 ml/min. The p53 forms eluted at 0.5 M KCl [p53b(1–393)] or 0.4 M KCl [p53(1–363), p53(44–393) and p53(45–349)]. p53-containing fractions were concentrated using ammonium sulfate precipitation and MICROSEP 10K centrifugation (Pall Filtron, Germany). Samples were then applied to a Superdex 200 column (HR 10/30; Pharmacia, Sweden) equilibrated in buffer A with 200 mM KCl. The peak fractions of p53b(1–393), p53(1–363) and/or p53(44–393) were eluted after 15, 22 and/or 27 min (flow rate 0.5 ml/min), respectively. p53(94–312) was isolated as described previously (29).
Proteins p53(320–393), p53(319–393)L344A, p53(361–393) and GCN4–p53(347–393) were expressed as fusions with GST. Lysis of bacteria was performed in 1× PBS (150 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, 2 mM NaH2PO4, pH 7.3), 0.1% Triton X-100, 1 mM benzamidine and 0.1 mM PMSF. The lysate was loaded onto a 5 ml HiTrap-GST column (Pharmacia, Sweden). After washing with 1× PBS (10 column vol), the fusion protein was eluted with 10–20 mM reduced glutathione. Protein-containing fractions were loaded onto MonoS columns (HR 5/5; Pharmacia, Sweden) and eluted with a 40 ml linear gradient (from 0 to 1 M KCl in buffer A). The GST tag was cleaved with 1 U thrombin (Calbiochem, USA) per 5 mg fusion protein. Except for the p53(361–393) protein (separated on MICROSEP 10K), each p53 protein was separated from GST protein on MonoS columns using a 40 ml linear gradient (0–1 M NaCl in 50 mM Tris–HCl, pH 8.8, or 25 mM Na HEPES, pH 7.6). Most of the pure p53 proteins eluted at 0.5 M NaCl (31). The purity and appropriate size of each protein was analyzed with Coomassie blue staining of SDS–PAGE gels (Fig. 1B) and western blotting (not shown). Mouse monoclonal antibodies (mAb) DO-1 (recognizing amino acids 20–25), DO-11 (amino acids 176–185), DO-12 (amino acids 256–270), Bp53-6.1 (amino acids 381–390), Bp53-10.1 (amino acids 375–379) and ICA-9 (amino acids 388–393) were applied to detect epitopes present within the p53 fragments.
Competition assay
Competition experiments with a mix of supercoiled (native superhelix density) and SmaI linearized pBluescript SK II DNAs (in equimolar concentrations) were performed as described previously (20). The total amount of DNA was in the range 0.3–1.6 µg (higher amounts of DNA and p53 were used in samples with low p53/DNA ratios to keep the p53 level detectable for monoclonal antibodies). The lin/scDNA mix was incubated with p53 fragment (p53 tetramer/DNA molar ratio = 1–20) in binding buffer (5 mM Tris–HCl, pH 7.6, 0.5 mM EDTA, 50 mM KCl and 0.01% Triton X-100) for 30 min at 0°C. Samples were loaded onto a 1.3% agarose gel containing 0.33× Tris–borate–EDTA (TBE) buffer; in this system linDNA migrates faster than scDNA. After 8–12 h electrophoresis (at 4–6 V/cm), gels were blotted onto a nitrocellulose transfer membrane Protran R (Schleicher and Schuell, Germany) and each p53 fragment was detected with the corresponding mAb using the ECL detection system (20).
Supershifting experiment
p53(320–393) protein was incubated with scDNA (0.3 µg) for 15 min on ice (p53 tetramer/DNA molar ratio = 5). Then, affinity purified mouse mAb was added (mAb/p53 tetramer molar ratio = 1.5 or 3) and incubation continued for a further 15 min at room temperature (32). Samples were run on a 1% agarose gel in 0.33× TBE and stained with ethidium bromide. The gel was blotted and p53 protein detected using rabbit anti-p53 polyclonal CM1 antibody.
Electron microscopy
Complexes of p53 with DNA were formed in a buffer containing 10 mM Na HEPES, pH 7.5, 50 mM KCl, 0.01% Triton X-100. p53 constructs (final concentration 100 or 500 nM where indicated for a given stable oligomeric state in solution) were added to a reaction mixture containing equimolar concentrations of linear and native supercoiled pBluescript DNA (5 nM each). Samples were incubated on ice for 30–60 min and an aliquot was withdrawn and diluted 20-fold into 10 mM Na HEPES, pH 7.5, 10 mM KCl. Electron microscopic samples were prepared as described (29). Samples were analyzed with a Philips CM12 electron microscope operated in a tilted dark field mode. Images were scanned using Agfa DuoScan T2500 (Agfa, Germany). For printing, images were flattened with a high-pass filter (radius 250 pixels) and subsequently adjusted for contrast/brightness using Adobe Photoshop.
RESULTS
The C-terminal segment of p53 is important for p53 SCS binding
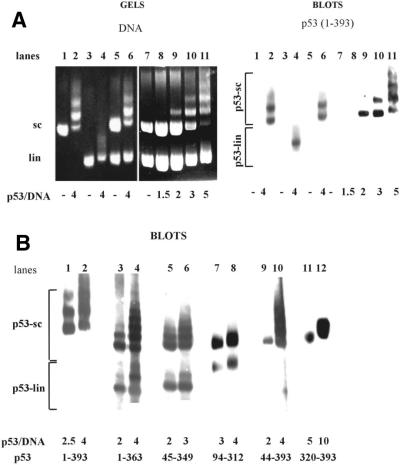
We studied the interaction of bacterially expressed truncated forms of human p53 (Fig. 1) with linDNA and scDNA in order to elucidate the relative contributions of different domains to the preferential binding of p53 to scDNA (17,20). An equimolar mixture of supercoiled and linear (SmaI linearized) forms of pBluescript DNA was incubated with various p53 fragments and the formation of both p53–scDNA and p53–linDNA complexes was analyzed by agarose gel electrophoresis (Fig. 2A, left) and immunoblotting assay of the agarose gel (Fig. 2A, right, and B) as described (20). Note that under the conditions used (1.3% agarose, 0.33× TBE buffer) linDNA migrates faster than scDNA (Fig. 2A, compare lanes 1 and 3). Incubation of either scDNA (lane 2) or linDNA (lane 4) with p53b(1–393) protein led to the appearance of a new band(s) located above the position of the corresponding free DNA. Superposition of the left and right panels clearly shows that p53 was located exclusively within retarded bands, indicating that these bands represent p53–DNA complexes. When the equimolar mixture of sc/linDNA was incubated with an increasing amount of p53b(1–393) protein (at either 0 or 37°C) only p53–scDNA complexes were observed (Fig. 2, lanes 6 and 9–11; data for 37°C not shown).
Figure 2.
p53 deletion mutants bind scDNA with different selectivities in competition experiments. (A) scDNA (0.3 µg; lanes 1 and 2) or linDNA (0.3 µg; lanes 3 and 4) alone was incubated with p53b(1–393) protein (lanes 2 and 4) at a p53 tetramer/DNA molar ratio of 4. A mixture of linear and supercoiled (sc/lin ratio 1:1) forms of pBluescript DNAs containing in total 0.6 µg (lanes 5 and 6), 0.8 µg (lanes 7, 10, 11) or 1.6 µg (lanes 8 and 9) was incubated with p53b(1–393) protein in the molar ratio range 1.5–5.0 protein (tetramer)/DNA (lanes 6 and 8–11). Samples were run on 1.3% agarose gels in 0.33× TBE and stained with ethidium bromide (left). Note that under the conditions used linDNA migrates faster than scDNA. The gels were blotted (right) and p53–scDNA (p53-sc) and/or p53–linDNA (p53-lin) complexes were detected using DO-1 antibody. (B) Immunoblots of two representative lanes showing results of the competition assays with the following proteins: lanes 1 and 2, p53(1–393); lanes 3 and 4, p53(1–363); lanes 5 and 6, p53(45–349); lanes 7 and 8, p53(94–312); lanes 9 and 10, p53(44–393); lanes 11 and 12, p53(320–393). Mouse monoclonal antibodies DO-1 (recognizing amino acids 20–25), DO-11 (amino acids 176–185), DO-12 (amino acids 256–270), Bp53-6.1 (amino acids 381–390), Bp53-10.1 (amino acids 375–379) and ICA-9 (amino acids 388–393) were applied to detect epitopes present within the p53 proteins.
It is evident that p53b(1–393) displays very high selectivity for scDNA in a sc/linDNA competition assay (Fig. 2A, lanes 6 and 9–11, and B, lanes 1 and 2). This selective binding of p53 to scDNA was termed SCS binding (20). The selectivity for scDNA binding was due to topological constraints not dependent on the presence of DNA ends, since p53 bound similarly to relaxed circular DNA as to linear templates (data not shown). In contrast, p53(1–363), lacking the basic C-terminal DNA-binding domain (amino acids 363–382), formed both p53–scDNA and p53–linDNA complexes (Fig. 2B, lanes 3 and 4) with a weak preference for scDNA. Similarly, both the p53(45–349) and p53(94–312) proteins, containing the p53 core domain, showed only a weak preference for scDNA (Fig. 2B, lanes 5–8). Taken together, these data imply an essential role of the p53 C-terminal region in SCS binding.
To further analyze the role of the p53 C-terminal region we purified two N-terminally truncated deletion mutants p53(44–393) and p53(320–393). In the sc/lin competition experiments, both proteins bound scDNA with high selectivity (Fig. 2B, lanes 9–12), suggesting that the last ∼70 amino acid residues (amino acids 320–393) of the C-terminal part are sufficient for SCS binding.
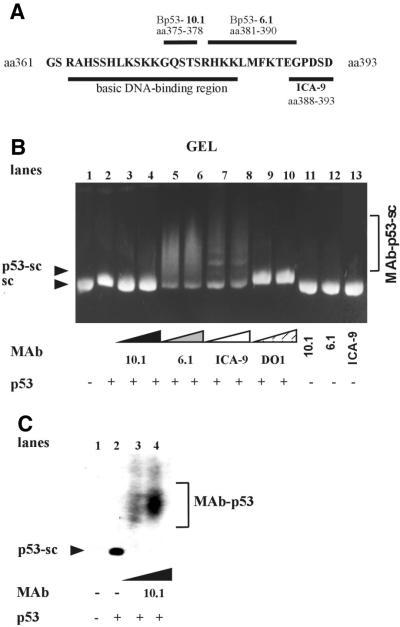
Amino acid residues 375–378 of the basic C-terminal region (amino acids 363–382) participate in binding to scDNA
We mapped the DNA-binding site (Fig. 3A) of the C-terminal protein p53(320–393) using specific mAbs. scDNA (Fig 3B, lane 1) was incubated with the p53(320–393) protein forming a p53(320–393)–scDNA complex that migrated with slightly lower mobility than free scDNA in agarose gels (Fig. 3B, lane 2). After addition of antibody ICA-9 (recognizing amino acids 388–393) (33) the p53(320–393)–scDNA complex was retarded further (supershifted), suggesting the formation of large (ICA-9)–p53(320–393)–scDNA complexes (Fig. 3B, lanes 7 and 8). The Bp53-6.1 epitope (amino acids 381–390) (34) in the p53(320–393)–scDNA complex was also accessible to the mAb, as indicated by smearing of the band (Fig. 3B, lanes 5 and 6). In agreement with these results, the p53(319–382) protein (lacking the last 11 amino acid residues of the C-terminus) selectively bound to scDNA (data not shown), suggesting that the extreme C-terminal region is not essential for SCS binding.
Figure 3.
Amino acid residues 375–378 participate in p53(320–393) binding to scDNA. (A) Schematic representation of amino acid residues within the C-terminal region (amino acids 361–393) of human p53. The basic DNA-binding domain (amino acids 363–382) and epitopes of each monoclonal antibody used in the supershifting experiments are indicated by lines above and/or below the corresponding sequence. N-terminal specific antibody DO-1 (recognizing amino acids 20–25) was used as a negative control. The supershifting experiment (B) was performed as follows. First, pBluescript scDNA (0.3 µg) was incubated with p53(320–393) protein (lanes 2–10) at a p53 tetramer/DNA molar ratio = 5. Then, increasing amounts of monoclonal antibodies were added as indicated below the lanes. Samples were run on 1% agarose gels in 0.33× TBE. The gel was blotted (C) and developed with rabbit anti-p53 antibody (see Materials and Methods); only the left four lanes are shown. The presence of p53(320–393) protein in the reaction mixture is indicated by + and/or –, respectively. The positions of p53–scDNA (p53-sc), immune mAb–p53–scDNA (MAb-p53-sc) and mAb–p53 (MAb-p53) complexes are indicated.
Addition of mouse antibody Bp53-10.1 (recognizing amino acids 375–378) (34) to the p53(320–393)–scDNA complex did not result in supershifting (Fig. 3B, lanes 2–4). To check p53 binding to scDNA the gel was blotted and analyzed for p53 protein using the rabbit antibody CM1 (Fig. 3C). The p53(320–393) protein was dissociated from the scDNA (lanes 3 and 4) and fully titrated with Bp53-10.1 antibodies (lane 4). These results suggest that the Bp53-10.1 antibody competed with scDNA for binding to identical p53 amino acid residues. Similar results have been obtained with antibody pAb421 recognizing amino acids 371–380 (data not shown). We infer that amino acid residues 375–378 participate in binding of the p53(320–393) protein to scDNA.
The oligomeric state of the p53 C-terminal segment is critical for its SCS binding
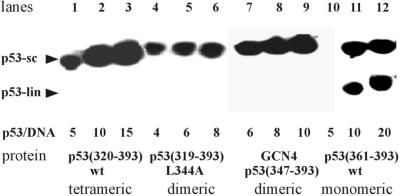
To analyze the role of the tetramerization domain (amino acids 325–356) we used the C-terminal protein p53(319–393) L344A (containing an Ala substitution at amino acid residue L344) and the GCN4–p53(347–393) fusion protein (containing the dimerization domain of yeast transcription factor GCN4 fused to the p53 C-terminal segment). The composition of both proteins limited their oligomeric state in solution to dimers (data not shown) (28,35). In the sc/lin competition assay, mutant p53(319–393)L344A as well as the GCN4– p53(347–393) fusion protein selectively bound scDNA (Fig. 4, lanes 4–6 and 7–9, respectively), showing for the first time that not only tetramers but also p53 dimers display SCS binding and that the p53 tetramerization domain is replaceable by a dimerization domain. In agreement with this result, glutathione S-transferase (GST)–p53(347–393) fusion protein (GST is a homodimer in solution; 36) bound selectively to scDNA (data not shown). On the other hand, neither p53(361–393) nor p53(347–393) monomeric proteins showed any preference for scDNA, forming protein–DNA complexes with both linDNA and scDNA in the sc/lin competition assay (Fig. 4, lanes 10–12 and data not shown). These results imply that the oligomeric state is critical for SCS binding.
Figure 4.
p53 oligomeric state of the C-terminal protein is critical for strong SCS binding. Tetrameric p53(320–393) (lanes 1–3), monomeric p53(361– 393) (lanes 10–12), dimeric mutant p53(319–393)L344A (lanes 4–6) and dimeric fusion GCN4–p53(347–393) (lanes 7–9) were analyzed for selective binding to scDNA in sc/lin (sc/lin ratio 1:1) competition assays (see Materials and Methods). The total amount of DNA was 0.8 µg; protein/DNA molar ratios are indicated. Bound p53 proteins were detected using monoclonal antibodies Bp53-10.1 and Bp53-6.1. Three representative lanes for each p53 construct tested are shown.
Strong SCS binding of p53 C-terminal proteins correlates with their ability to form DNA–protein filaments
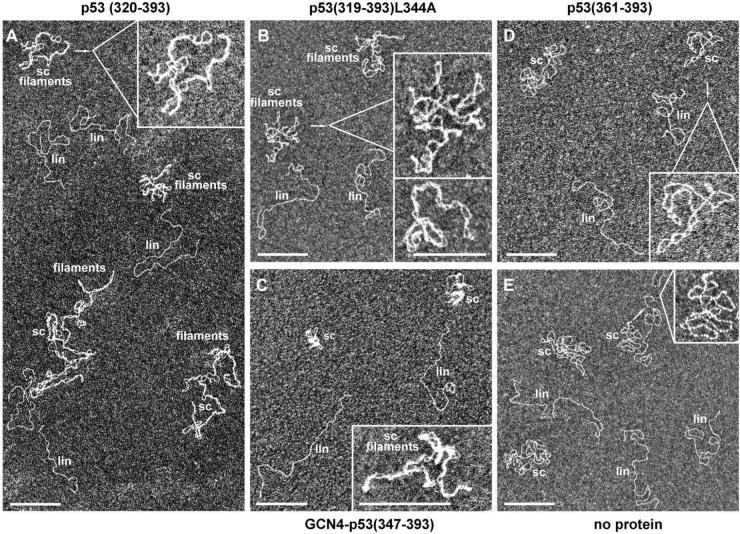
We employed electron microscopy to analyze p53 selective binding to scDNA in the equimolar mixture of scDNA and linDNA (5 nM each). For better discrimination between free and protein-bound scDNA, the samples were deposited from 10 mM KCl since under these conditions scDNA does not display a clear plectonemic form but rather a partially open or irregular configuration (Fig. 5E) (37). The p53(320–393) protein (100 nM, i.e. protein/DNA molar ratio = 10) showed a strong selective binding to scDNA, forming long DNA– protein filaments, whereas practically no interaction was detected with linDNA (Fig. 5A). Both p53(319–393) L344A and GCN4–p53(347–393), which are dimers in solution (28,35), also exhibited selective binding to scDNA, with complexes resembling those formed by p53(320–393), although with minor differences (Fig. 5B and C). At protein/DNA molar ratio = 10, both dimeric proteins formed complexes with less clear filamentous structure (Fig. 5B, upper inset, and C). The regular filamentous structures were observed at higher protein/DNA molar ratios (Fig. 5B, lower inset, and C, inset). In contrast, p53(361–393) neither generated filaments with various DNA targets nor did it perturb the scDNA configuration (Fig. 5D). Other possible complexes were not detected, probably due to their low molecular mass. In view of the above data, we infer that there is a strong correlation between the ability of p53 proteins to selectively bind scDNA in the sc/linDNA competition experiments (Fig. 4) and to form DNA–protein filaments (Fig. 5).
Figure 5.
Electron microscopy of the protein–DNA complexes formed by different p53 C-terminal proteins with sc/lin DNA mixture. The panel represents images of the complexes obtained after the incubation of a mixture of supercoiled pBluescript and pBluescript/EcoRI DNAs (5 nM each) with p53(320–393) (A), p53(319–393)L344A (B), GCN4–p53(347–393) (C) and p53(361–393) (D). Protein concentration was 100 nM for a given oligomeric state of the protein in solution (protein/DNA molar ratio = 10). Insets in (A), (B) (upper) and (D) represent magnified views of supercoiled molecules in the presence of proteins. The lower inset in (B) and the inset in (C) represent images of the complexes obtained at a 500 nM concentration of GCN4–p53(347–393) and p53(319–393)L344A proteins, respectively (protein/DNA molar ratio = 50). (E) The sc/lin DNA mixture in the absence of proteins; inset, magnified view of supercoiled DNA. Note that under the conditions used for mounting DNA for electron microscopic imaging (10 mM Na HEPES, 10 mM KCl) native supercoiled DNA has a loosely interwound appearance, in accordance with Cherny and Jovin (37). sc and lin indicate supercoiled and linear DNA, respectively; filaments indicates DNA–protein filaments. For further details see Materials and Methods. The scale bar represents 200 nm.
DISCUSSION
Role of p53 domains in SCS binding
In this work we have analyzed the preferential binding of different p53 domains to scDNA. Our previous results suggested that both the core domain and the C-terminal part of p53 participate in the preferential binding of full-length p53 to scDNA (17,19,38). Recently, we showed that the isolated core domain displays a preference for binding scDNA in sc/lin competition experiments, although this bias is much weaker than that observed with the bacterially expressed full-length p53b(1–393) (20). Here we demonstrate that the C-terminal region (amino acids 320–393) exhibits SCS binding to scDNA comparable to that of the p53b(1–393) protein (Fig. 2), suggesting that participation of the C-terminal region is of critical importance for scDNA selectivity. Considering the role of the core domain in SCS binding by p53b(1–393), it is possible that either the core domain modulates the C-terminus SCS binding in p53b(1–393) or that the mechanism of p53b(1–393) binding to scDNA differs fundamentally from that of the C-terminal segment, i.e. involving direct core domain interactions as well. The hypothesis that other domains may modulate binding of the C-terminal segment is supported by our observation that fusion of a GST tag to the p53(320–393) protein diminishes binding to scDNA (M.Brázdová, J.Paleček and D.I.Cherny, unpublished results).
Neither the p53(361–393) nor p53(347–393) protein bound selectively to scDNA (Figs 4 and 5). We conclude from these findings that the p53 tetramerization domain (amino acids 325–356), responsible for the oligomerization state of the protein, is critical for SCS binding by the p53(320–393) protein. This conclusion is in agreement with Mazur et al. (18), who demonstrated that monomeric p53(319–393)AAA protein (containing Ala substitutions at positions L323, Y327 and L330) did not exhibit preferential binding to scDNA. We have shown that the p53 C-terminal segment (amino acids 347–393) acquires the ability to selectively bind to scDNA upon fusion to the yeast GCN4 dimerization domain (Figs 4 and 5). These results suggest that (i) the p53 tetramerization domain is replaceable with a dimerization domain (e.g. GCN4) and (ii) the oligomeric state of the p53 C-terminal segment plays a dominant role in SCS binding relative to amino acid composition of the tetramerization domain (amino acids 325–356). However, the amino acid segment 347–356 of this region appears necessary for SCS binding since the GCN4–p53(356–393) fusion protein showed no selectivity for scDNA in the sc/lin competition assay (J.Paleček, unpublished results) while the GCN4–p53(347–393) construct exhibited SCS binding (Fig. 4). Thus, amino acid residues 347–356 contribute either to a proper conformation of the GCN4–p53(347–393) fusion protein or participate directly in the p53–scDNA interactions.
From the successful functional replacement of the p53 tetramerization domain with the GCN4 dimerization domain, it appears that dimers of the C-terminal segment (amino acids 347–393) are sufficient for SCS binding. Thus, dimerization modulates the DNA binding selectivity of the p53 C-terminal region (see below; Fig. 6).
Figure 6.
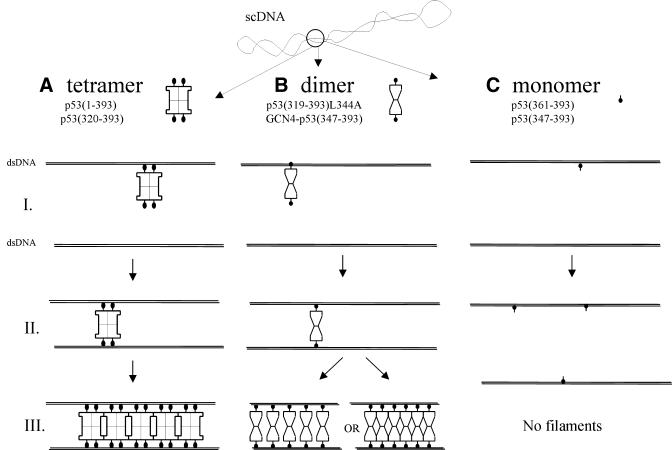
A model of p53 C-terminus interaction with two DNA helices of scDNA. The C-terminal fragment of p53, consisting of a DNA-binding domain (black circles) and an oligomerization domain (empty formations, shown for predominant tetrameric or dimeric protein forms, respectively) binds single DNA duplexes (stage I). Unbound DNA-binding domains of p53 tetramers or dimers are capable of interacting further with an opposing DNA duplex, located close to the first one, due to the plectonemic configuration of scDNA [stage II in (A) and (B)], thus stabilizing DNA strands in close proximity. Then, the next p53 molecule binds opposing DNA duplexes, leading to the synapsing of two non-contiguous duplexes, imaged as a long DNA–protein filament [stage III in (A) and (B)]. Bound proteins can be positioned either in juxtaposed registers due to protein–protein interactions (cooperative binding, the most probable mode for protein tetramers) or separately (the most probable mode for protein dimers). Both modes of positioning of bound proteins may occur concurrently within a single filament. Monomeric p53 binds DNA duplex but does not manifest protein–protein interactions either in solution or in the bound state. Hence, no filaments can be produced (C).
p53 dimers bind two dsDNA duplexes forming filamentous structures
Our recent studies suggested several potential p53-binding motifs within scDNA. It was proposed (20) that the weak preference of p53(94–312) for scDNA may reflect binding to the internal single-stranded regions (8), possibly occurring in native negatively scDNA but absent in linDNA (39). Jett et al. (11) reported binding of the p53(94–312) protein to a cruciform arising in scDNA containing an (AT)34 insert. Using electron microscopy we found only a small fraction of globular complexes formed by p53(94–312), p53(1–363) and insect cell-expressed full-length p53i(1–393) at scDNA crossovers (D.I.Cherny, M.Brázdová, J.Paleček, E.Paleček and T.M.Jovin, submitted for publication). From these data we hypothesize that the interaction of p53(94–312) core domain (and the other proteins lacking the C-terminus and/or containing inactive C-terminus) with any of the above motifs may contribute to the weak preferential binding to scDNA.
The ability of the p53 C-terminal proteins to form filaments correlates well with their strong selective binding to scDNA observed in gel shift sc/lin competition experiments (Figs 4 and 5). Analysis of the p53–scDNA filaments showed that two DNA segments from opposing (distant) duplex strands are linked via p53 proteins (Figs 5 and 6). The high probability of two DNA duplexes coming together in scDNA could account for the ability of p53 to bind scDNA with high selectivity in the sc/lin competition experiments (37). The most attractive explanation for SCS binding of the C-terminal proteins is that the protein dimers (and tetramers) bind, most probably in a cooperative fashion, DNA regions with close lateral contacts inherent to the plectonemic form of natural negatively scDNA. In this regard it is worth noting that filaments formed with scDNA containing only a small number of supercoils, e.g. about five or six (D.I.Cherny, M.Brázdová, J.Paleček, E.Paleček and T.M.Jovin, submitted for publication).
All incubations of the p53 proteins with DNA were carried out under conditions (50 mM KCl) in which scDNA has the plectonemic form (37,40). The conditions for electron microscopic sample preparation (10 mM KCl) were chosen such that scDNA adopted a loosely interwound configuration of irregular shape (Fig. 5E). The clear appearance of DNA synapses under these conditions strongly indicates the presence of nucleoprotein filaments. The electrostatic repulsion between DNA duplexes was probably decreased by binding of the positively charged basic p53 C-terminal domain (Fig. 3A) stabilizing close contacts between DNA duplexes under the low salt conditions (10 mM KCl) (41).
The above considerations led us to propose the model of p53 C-terminus binding to scDNA presented in Figure 6. We suggest that each dimer of the tetramer binds one DNA duplex (Fig. 6A). A similar mechanism is proposed for dimeric peptides (Fig. 6B); one p53 DNA-binding region in the dimer binds one DNA duplex (Fig. 6B, I) while the second p53 DNA-binding region in the dimer interacts with the opposing duplex (Fig. 6B, II). Since dimeric proteins formed filaments while monomeric proteins did not (Fig. 6C) we assume that binding of the first p53 molecule (dimer or tetramer) stabilizes two duplexes in close proximity, forming an initial complex (Fig. 6A and B, II). Further binding of proteins results in formation of higher order structures in which duplex segments are synapsed within long filaments (Fig. 6A and B, III). Since multiples of tetramers of wild-type p53 are found in solution (42,43) it is possible that cooperative DNA–protein and protein–protein interactions contribute to filament formation. In the case of predominantly dimeric forms of p53, filaments could also arise via merely DNA–protein contacts. It is clear that the binding to opposing duplexes is facilitated by DNA supercoiling to an extent increasing with superhelix density, inasmuch as the duplex-to-duplex distance varies inversely with superhelix density (reviewed in 37).
Since the bacterially expressed full-length p53b(1–393) also displays filament formation with scDNA of low superhelix density (D.I.Cherny, M.Brázdová, J.Paleček, E.Paleček and T.M.Jovin, submitted for publication) and of native superhelix density (D.I.Cherny, unpublished results), we suggest that its scDNA binding mode may be similar to that of the C-terminal segments described here.
Biological significance of p53–DNA filament formation
Processes such as DNA recombination and repair, control of supercoiling by topoisomerases and/or gene regulation through DNA looping are accompanied by formation of close contacts between DNA duplexes (40 and references therein). Binding of proteins to such intermediate structures is of critical importance. For instance, it was proposed that vaccinia topoisomerase-mediated DNA synapsis plays a role in viral recombination and in packaging of the 200 kb vaccinia genome during virus assembly (44). We speculate that segments of two aligned chromosomes (exhibiting close contacts between DNA duplexes) could serve as a substrate for p53–DNA filament formation. p53 core domain binding to early recombination intermediates like three-way junctions (45,46) could catalyze formation of an initial nucleoprotein complex. The propagation of filaments (through the C-terminus) may then block the progression of recombination. Recently, Prabhu et al. (47) demonstrated that p53 blocks branch migration promoted by proteins such as RuvAB (as well as spontaneous branch migration) and modulates the cleavage by Holliday junction resolution proteins such as RuvC. In addition, inasmuch as p53 C-terminus can catalyze strand transfer (8,16) and interacts with Rad51 protein (48), it is possible that the propagation of p53 filaments could regulate Rad51-mediated strand transfer. Thus, we propose that the highly efficient formation of nucleoprotein filaments by p53 protein at sites of close contacts between DNA duplexes may play a significant role in fundamental genetic processes.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to C. Midgley, M. G. Luciani, T. Halazonetis, S. Gorina, G. Selivanova, M. Protopopova and K. G. Wiman for the supply of plasmids. Technical assistance by L. Karlovska and K. O. Koutska is acknowledged. This work was supported by grants of the Grant Agency of the Czech Republic, no. 301/02/0831 to B.V. and E.P. and no. 301/99/D078 to J.P., by the Grant Agency of the Academy of Sciences of the Czech Republic, A4004110 to E.P., by the Volkswagen Stiftung, I/72942 to E.P. and T.M.J., by grant NC/5343-3 of the IGA MH CR to M.F. The Academy of Sciences of the Czech Republic supported this work by grant nos S 5004009 and Z 5004920.
REFERENCES
- 1.Oren M. and Rotter,V. (1999) Introduction: p53—the first twenty years. Cell. Mol. Life Sci., 55, 9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogelstein B., Lane,D. and Levine,A.J. (2000) Surfing the p53 network. Nature, 408, 307–310. [DOI] [PubMed] [Google Scholar]
- 3.Appella E. and Anderson,C.W. (2001) Post-translational modifications and activation of p53 by stresses. Eur. J. Biochem., 268, 2764–2772. [DOI] [PubMed] [Google Scholar]
- 4.Albrechtsen N., Dornreiter,I., Grosse,F., Kim,E., Wiesmuller,L. and Deppert,W. (1999) Maintenance of genomic integrity by p53: complementary roles for activated and non-activated p53. Oncogene, 18, 7706–7717. [DOI] [PubMed] [Google Scholar]
- 5.Hupp T.R. (1999) Regulation of p53 protein function through alterations in protein-folding pathways. Cell. Mol. Life Sci., 55, 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jayaraman L. and Prives,C. (1999) Covalent and noncovalent modifiers of the p53 protein. Cell. Mol. Life Sci., 55, 76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Deiry W.S., Kern,S.E., Pietenpol,J.A., Kinzler,K.W. and Vogelstein,B. (1992) Definition of a consensus binding site for p53. Nature Genet., 1, 45–49. [DOI] [PubMed] [Google Scholar]
- 8.Bakalkin G., Selivanova,G., Yakovleva,T., Kiseleva,E., Kashuba,E., Magnusson,K.P., Szekely,L., Klein,G., Terenius,L. and Wiman,K.G. (1995) p53 binds single-stranded DNA ends through the C-terminal domain and internal DNA segments via the middle domain. Nucleic Acids Res., 23, 362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudenhoffer C., Kurth,M., Janus,F., Deppert,W. and Wiesmuller,L. (1999) Dissociation of the recombination control and the sequence-specific transactivation function of p53. Oncogene, 18, 5773–5784. [DOI] [PubMed] [Google Scholar]
- 10.Szak S.T. and Pietenpol,J.A. (1999) High affinity insertion/deletion lesion binding by p53. Evidence for a role of the p53 central domain. J. Biol. Chem., 274, 3904–3909. [DOI] [PubMed] [Google Scholar]
- 11.Jett S.D., Cherny,D.I., Subramaniam,V. and Jovin,T.M. (2000) Scanning force microscopy of the complexes of p53 core domain with supercoiled DNA. J. Mol. Biol., 299, 585–592. [DOI] [PubMed] [Google Scholar]
- 12.Janus F., Albrechtsen,N., Knippschild,U., Wiesmuller,L., Grosse,F. and Deppert,W. (1999) Different regulation of the p53 core domain activities 3′-to-5′ exonuclease and sequence-specific DNA binding. Mol. Cell. Biol., 19, 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hainaut P., Hernandez,T., Robinson,A., Rodriguez-Tome,P., Flores,T., Hollstein,M., Harris,C.C. and Montesano,R. (1998) IARC Database of p53 gene mutations in human tumors and cell lines: updated compilation, revised formats and new visualisation tools. Nucleic Acids Res., 26, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zotchev S.B., Protopopova,M. and Selivanova,G. (2000) p53 C-terminal interaction with DNA ends and gaps has opposing effect on specific DNA binding by the core. Nucleic Acids Res., 28, 4005–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reed M., Woelker,B., Wang,P., Wang,Y., Anderson,M.E. and Tegtmeyer,P. (1995) The C-terminal domain of p53 recognizes DNA damaged by ionizing radiation. Proc. Natl Acad. Sci. USA, 92, 9455–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakalkin G., Yakovleva,T., Selivanova,G., Magnusson,K.P., Szekely,L., Kiseleva,E., Klein,G., Terenius,L. and Wiman,K.G. (1994) p53 binds single-stranded DNA ends and catalyzes DNA renaturation and strand transfer. Proc. Natl Acad. Sci. USA, 91, 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paleček E., Vlk,D., Stankova,V., Brazda,V., Vojtěšek,B., Hupp,T.R., Schaper,A. and Jovin,T.M. (1997) Tumor suppresor protein p53 binds preferentially to supercoiled DNA. Oncogene, 15, 2201–2209. [DOI] [PubMed] [Google Scholar]
- 18.Mazur S.J., Sakaguchi,K., Appella,E., Wang,X.W., Harris,C.C. and Bohr,V.A. (1999) Preferential binding of tumor suppressor p53 to positively or negatively supercoiled DNA involves the C-terminal domain. J. Mol. Biol., 292, 241–249. [DOI] [PubMed] [Google Scholar]
- 19.Fojta M., Kubicarova,T., Vojtěšek,B. and Paleček,E. (1999) Effect of p53 protein redox states on binding to supercoiled DNA and linear DNA. J. Biol. Chem., 274, 25749–25755. [DOI] [PubMed] [Google Scholar]
- 20.Paleček E., Brázdová,M., Brazda,V., Paleček,J., Billová,S., Subramaniam,V. and Jovin,T.M. (2001) Binding of p53 and its core domain to supercoiled DNA. Eur. J. Biochem., 268, 573–581. [DOI] [PubMed] [Google Scholar]
- 21.Fojta M., Brázdová,M., Cernocka,H., Pečinka,P., Brazda,V., Paleček,J., Jagelska,E., Vojtěšek,B., Pospisilova,S., Subramaniam,V. et al. (2000) Effects of oxidation agents and metal ions on binding of p53 to supercoiled DNA. J. Biomol. Struct. Dyn., Sp. Iss. S1, 177–184. [DOI] [PubMed] [Google Scholar]
- 22.Kirkegaard K. and Wang,J.C. (1985) Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J. Mol. Biol., 185, 625–637. [DOI] [PubMed] [Google Scholar]
- 23.Zechiedrich E.L. and Osheroff,N. (1990) Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J., 9, 4555–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stros M. and Reich,J. (1998) Formation of large nucleoprotein complexes upon binding of the high-mobility-group (HMG) box B-domain of HMG1 protein to supercoiled DNA. Eur. J. Biochem., 251, 427–434. [DOI] [PubMed] [Google Scholar]
- 25.Midgley C.A., Fisher,C.J., Bartek,J., Vojtěšek,B., Lane,D. and Barnes,D.M. (1992) Analysis of p53 expression in human tumours: an antibody raised against human p53 expressed in Escherichia coli. J. Cell Sci., 101, 183–189. [DOI] [PubMed] [Google Scholar]
- 26.Cho Y., Gorina,S., Jeffrey,P.D. and Pavletich,N.P. (1994) Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science, 265, 346–355. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Waterman J.L., Shenk,J.L. and Halazonetis,T.D. (1995) The dihedral symmetry of the p53 tetramerization domain mandates a conformational switch upon DNA binding. EMBO J., 14, 512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherny D.I., Striker,G., Subramaniam,V., Jett,S.D., Paleček,E. and Jovin,T.M. (1999) DNA bending due to specific p53 and p53 core domain-DNA interactions visualized by electron microscopy. J. Mol. Biol., 294, 1015–1026. [DOI] [PubMed] [Google Scholar]
- 30.Hupp T.R., Meek,D.W., Midgley,C.A. and Lane,D.P. (1992) Regulation of the specific DNA binding function of p53. Cell, 71, 875–886. [DOI] [PubMed] [Google Scholar]
- 31.Brázdová M., Kizek,R., Havran,L. and Paleček,E. (2002) Determination of glutathione-S-transferase traces in preparations of p53 C-terminal domain (aa320–393). Bioelectrochemistry, 55, 115–118. [DOI] [PubMed] [Google Scholar]
- 32.Brazda V., Paleček,J., Pospisilova,S., Vojtěšek,B. and Paleček,E. (2000) Specific modulation of p53 binding to consensus sequence within supercoiled DNA by monoclonal antibodies. Biochem. Biophys. Res. Commun., 267, 934–939. [DOI] [PubMed] [Google Scholar]
- 33.Hupp T.R. and Lane,D.P. (1995) Two distinct signaling pathways activate the latent DNA binding function of p53 in a casein kinase II-independent manner. J. Biol. Chem., 270, 18165–18174. [DOI] [PubMed] [Google Scholar]
- 34.Pospisilova S., Brazda,V., Amrichova,J., Kamermeierova,R., Paleček,E. and Vojtěšek,B. (2000) Precise characterisation of monoclonal antibodies to the C-terminal region of p53 protein using the PEPSCAN ELISA technique and a new non-radioactive gel shift assay. J. Immunol. Methods, 237, 51–64. [DOI] [PubMed] [Google Scholar]
- 35.Ellenberger T.E., Brandl,C.J., Struhl,K. and Harrison,S.C. (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell, 71, 1223–1237. [DOI] [PubMed] [Google Scholar]
- 36.Maru Y., Afar,D.E., Witte,O.N. and Shibuya,M. (1996) The dimerization property of glutathione S-transferase partially reactivates Bcr-Abl lacking the oligomerization domain. J. Biol. Chem., 271, 15353–15357. [DOI] [PubMed] [Google Scholar]
- 37.Cherny D.I. and Jovin,T.M. (2001) Electron and scanning force microscopy studies of alterations in supercoiled DNA tertiary structure. J. Mol. Biol., 313, 295–307. [DOI] [PubMed] [Google Scholar]
- 38.Paleček E., Brázdová,M., Cernocka,H., Vlk,D., Brazda,V. and Vojtěšek,B. (1999) Effect of transition metals on binding of p53 protein to supercoiled DNA and to consensus sequence in DNA fragments. Oncogene, 18, 3617–3625. [DOI] [PubMed] [Google Scholar]
- 39.Paleček E. (1991) Local supercoil-stabilized DNA structures. Crit. Rev. Biochem. Mol. Biol., 26, 151–226. [DOI] [PubMed] [Google Scholar]
- 40.Lyubchenko Y.L. and Shlyakhtenko,L.S. (1997) Visualization of supercoiled DNA with atomic force microscopy in situ. Proc. Natl Acad. Sci. USA, 94, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yakovleva T., Pramanik,A., Kawasaki,T., Tan-No,K., Gileva,I., Lindegren,H., Langel,U., Ekstrom,T.J., Rigler,R., Terenius,L. et al. (2001) p53 latency. C-terminal domain prevents binding of p53 core to target but not to nonspecific DNA sequences. J. Biol. Chem., 276, 15650–15658. [DOI] [PubMed] [Google Scholar]
- 42.Stenger J.E., Mayr,G.A., Mann,K. and Tegtmeyer,P. (1992) Formation of stable p53 homotetramers and multiples of tetramers. Mol. Carcinog., 5, 102–106. [DOI] [PubMed] [Google Scholar]
- 43.Pavletich N.P., Chambers,K.A. and Pabo,C.O. (1993) The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev., 7, 2556–2564. [DOI] [PubMed] [Google Scholar]
- 44.Shuman S., Bear,D.G. and Sekiguchi,J. (1997) Intramolecular synapsis of duplex DNA by vaccinia topoisomerase. EMBO J., 16, 6584–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S., Cavallo,L. and Griffith,J. (1997) Human p53 binds Holliday junctions strongly and facilitates their cleavage. J. Biol. Chem., 272, 7532–7539. [DOI] [PubMed] [Google Scholar]
- 46.Sturzbecher H.W., Donzelmann,B., Henning,W., Knippschild,U. and Buchhop,S. (1996) p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. EMBO J., 15, 1992–2002. [PMC free article] [PubMed] [Google Scholar]
- 47.Prabhu V.P., Simons,A.M., Iwasaki,H., Gai,D., Simmons,D.T. and Chen,J. (2002) p53 blocks RuvAB promoted branch migration and modulates resolution of Holliday junctions by RuvC. J. Mol. Biol., 316, 1023–1032. [DOI] [PubMed] [Google Scholar]
- 48.Susse S., Janz,C., Janus,F., Deppert,W. and Wiesmuller,L. (2000) Role of heteroduplex joints in the functional interactions between human rad51 and wild-type p53. Oncogene, 19, 4500–4512. [DOI] [PubMed] [Google Scholar]