Abstract
DNA methylation is now seen as a primary signal in the cell for mediating transcriptional repression through chromatin formation. The construction and evaluation of enzymes capable of influencing this process in vivo is therefore of significant interest. We have fused the C5-cytosine DNA methyltransferases, M.HhaI and M.HpaII, which both methylate 4 bp sequences containing a CpG dinucleotide, to a three zinc finger protein recognising a 9 bp DNA sequence. DNA methylation analyses demonstrate specific DNA methylation by both enzymes at target sites comprising adjacent methyltransferase and zinc finger subsites, targeted M.HpaII being the most specific. Binding analysis of the targeted M.HpaII enzyme reveals an 8-fold preference for binding to its target site, compared to binding to a zinc finger site alone, and an 18-fold preference over binding to a methyltransferase site alone, thereby demonstrating enhanced binding by the fusion protein, compared to its component proteins. Both DNA binding and methylation are specific for the target site up to separations of ∼40 bp between the zinc finger and methyltransferase subsites. Ex vivo plasmid methylation experiments are also described that demonstrate targeted methylation. These targeted enzymes, however, are shown to be not fully mono-functional, retaining a significant non-targeted activity most evident at elevated protein concentrations.
INTRODUCTION
Members of the prokaryotic cytosine-5 DNA methyltransferase (C5-Mtase) family of enzymes share a high degree of amino acid sequence homology, most pronounced in 10 highly conserved regions (1,2). This homology to some extent reflects the shared catalytic function of all these enzymes, that of transferring a methyl group from the cofactor, S-adenosyl-l-methionine (AdoMet), to the 5 position of the target cytosine base in DNA. M.HhaI and M.HpaII are type II C5-Mtases from Haemophilus haemolyticus and Haemophilus parainfluenzae, respectively, and recognise the specific DNA sequences 5′-GCGC-3′ and 5′-CCGG-3′, respectively, methylating the underlined cytosine in each instance. Crystal structures for one of these enzymes, M.HhaI, as well as for M.HaeIII, complexed with DNA and cofactor have been described (3,4). Whilst demonstrating subtle differences in DNA–protein contact signatures between the two proteins, both these studies revealed that in each case the target cytosine is flipped out of the DNA helix into the nucleotide-binding pocket of the enzyme, where the methyl transfer reaction can proceed. The functional characteristics of these enzymes, together with a wealth of information relating both amino acid sequence and structure to function, suggests to us that C5-Mtases may have potential as tools both in general research and in the treatment of disease, if their catalytic activity could be delivered to predetermined regions of the genome. The construction and evaluation of a three zinc finger fusion with a prokaryotic methylase was initially reported by Xu and Bestor (5). While they were able to demonstrate a degree of targeted binding and methylation, they also suggested that the protein was not fully mono-functional, each component protein exhibiting a significant independent activity. However, although members of the C5-Mtase family are highly homologous in terms of primary amino acid sequence, the functional characteristics of these proteins can vary significantly. For example, M.SssI, the methyltransferase used in this early work, contains both a processive methyltransferase activity (6) and a topoisomerase activity (7), both of which would be expected to modulate gene targeting by a zinc finger protein, to varying extents. Therefore, in an effort to evaluate the properties of other C5-Mtases in a gene-targeted scenario, we have constructed fusions between a three zinc finger DNA-binding protein and the C5-Mtases M.HhaI and M.HpaII.
A primary application that we envisage for targeted C5-Mtases is as a tool for the study of the spread of methylation patterns within eukaryotes. While there is evidence that DNA methylation does spread from an initial focus (8–10), no clear trends or rules have been established. Recent observations, such as the existence of a novel family of mammalian C5-Mtases (11) and the role of histone deacetylases in mediating methylation-induced packaging of DNA (see below), suggest that a series of complex factors might exist that contribute to DNA methylation spreading in mammalian cells. Previous experimental procedures for studying methylation spread in vivo generally involved in vitro methylation and subsequent stable transfection of plasmid vectors (see for example 12), with the integration site and flanking sequences varying between each experiment. This makes interpretation of data extremely difficult, due to large variations in flanking sequence effects. Furthermore, it has recently been demonstrated that the integration event itself can lead to marked alterations in the DNA methylation patterns of cellular genes and DNA segments (13). Development of the enzymes described in this study will allow an alternative approach to the study of DNA methylation spread in eukaryotes that closely approaches the in vivo state. These enzymes, when expressed sequentially in a clonal cell line whose genome harbours a targetable DNA insert containing multiple C5-Mtase recognition sites, will enable a range of methylation densities to be generated at a defined locus, with a knowledge that both flanking sequences and initial methylation status are identical in each instance.
A second potential application of targeted C5-Mtase enzymes stems from observations that mutation of the invariant catalytic cysteine of C5-Mtases to a glycine residue can generate an enzyme that binds with extremely high affinity to its DNA recognition site (see for example 14,15). These proteins are cytotoxic when expressed in Escherichia coli and the ability to direct such toxic enzymes to specific and unique gene sequences within the mammalian cell may ultimately provide both greater insight into cellular repair processes and lead to a potential strategy for targeted cell death.
Lastly, the correlation between DNA methylation of eukaryotic gene promoters and transcriptional repression is widely accepted (for a review see 16,17) and some of the mechanisms by which this relationship is implemented have been elucidated. These include reduced or abolished binding of transcription factors when their DNA binding site is methylated, as well as proteins that specifically bind to methylated DNA, thus occluding transcription factor binding directly. The observation that methylcytosine-binding proteins accelerate compaction of methylated DNA into a transcriptionally inactive chromatin form prompted studies that have revealed extensive interactions between methylcytosine-binding proteins and histone deacetylase proteins, mediated by a host of adaptor molecules (18–20). With these mechanisms in place within the cell, targeted promoter DNA methylation, accomplished using derivatives of the enzymes described in this study, may represent a novel gene therapy approach for the inheritable silencing of genes associated with disease. This would be of particular relevance in the case of viral genomes, which are known to be sensitive to promoter methylation.
We now report the construction and evaluation of first generation targeted HhaI and HpaII C5-Mtases. These enzymes are assessed for their ability to specifically methylate targeted versus non-targeted DNA sequences. The most promising of these current enzymes, targeted HpaII, has been studied in more detail, both in terms of binding specificity and affinity for different DNA substrates as well as methyltransferase specificity. The results of experiments designed to assess the cooperative interaction between the zinc finger and methyltransferase components of the fusion enzyme are also reported. The results of ex vivo studies are presented that point the way towards successful function of these enzymes under in vivo conditions in the mammalian cell.
MATERIALS AND METHODS
General reagents and chemicals
General reagents were supplied by Sigma unless otherwise stated. Restriction enzymes, Vent DNA polymerase, T4 DNA ligase and T4 polynucleotide kinase were purchased from New England Biolabs. Ultra pure deoxyribonucleotide solutions and poly(dI)·poly(dC) were purchased from Pharmacia Biotech. [γ-32P]ATP (>111 TBq/mmol) was purchased from ICN and [3H-methyl]AdoMet (>1.85 TBq/mmol) from Amersham International. All oligodeoxynucleotides used in this study were synthesised by MWG BioTech (Germany), by the standard CE-phosphoramidite method.
Bacterial strains and plasmids
Escherichia coli ER1647 [F– fhuA2 Δ(lacZ)r1 supE44 trp31 mcrA1272::Tn10(Tetr) his-1 rpsL104(Strr) xyl-7 mtl-2 metB1 Δ(mcrC-mrr)102::Tn10(Tetr) recD1014] was used throughout this study and was obtained from New England Biolabs. Bacteria were grown in liquid culture in Luria–Bertani medium (1% w/v Tryptone, 1% w/v NaCl and 0.5% w/v yeast extract) and on LA medium (1% w/v Tryptone, 1% w/v NaCl, 0.5% w/v yeast extract and 1.5% w/v agar) supplemented, if appropriate, with antibiotics as described before (21). The gene encoding the three zinc finger protein recognising the 9 bp sequence 5′-GCAGAAGCC-3′ (22) was a generous gift from Dr Yen Choo (LMB, University of Cambridge, UK). pUCHhaI and pUCHpaII are pUC18-based plasmids harbouring the entire M.HhaI and M.HpaII ORFs and were a kind gift from Dr Geoff Wilson (New England Biolabs). pGEX5X-3, pET22b and pLsyE (pACYC derivative) vectors were purchased from Pharmacia Biotech and Novagen.
Construction of targeted and non-targeted C5-Mtases
The gene encoding M.HhaI (Genbank accession no. J02677, bases 437–1420) was amplified by PCR using pUCHhaI (10 ng) as template and primers complementary to the 5′ end (5′-GGCGCGGATCCTCGACTAGTGAATTCATGATTG AAATAAAAGATAAACAGCTCACAGGATTACGCTTT-3′) and 3′ end (5′-GCGCGGTACCTTACTCGAG ATATGGTTTGAAATTTAATGA-3′) of the M.HhaI gene. The M.HhaI PCR product was restricted with BamHI and XhoI (underlined in the above primer sequences) and ligated into these same sites in pET22b to produce pHhaI. The PCR product was also ligated into the same sites in pGEX5X-3 in order to produce a non-targeted, GST–HhaI construct (pGHhaI).
Amplification of the three zinc finger peptide described by Choo et al. (22) was performed using primers complementary to the 5′ end (5′-GCGCAAGCTTCGCATATGGCAGA AGAGAAGCCTTTTCAGTGTCGAA-3′) and 3′ end (5′- GCGCGGATCCCTTCTCGCCTGTGTGGGTCTTTAGGT GTCTCTGAAGAGTAGC-3′) of the three zinc finger gene. The start methionine codon of the first zinc finger and end lysine codon of finger three are in bold in the respective primer sequence. The PCR product was restricted with NdeI and BamHI (underlined in the primer sequences above) and subcloned into these same sites in pHhaI, immediately 5′ to the M.HhaI cDNA, to produce pzfHhaI. Double-stranded oligodeoxynucleotide coding for the flexible (Gly4Ser)3 peptide linker was subcloned between the BamHI and EcoRI restriction sites which separate the two cDNAs in pzfHhaI to produce pzfLHhaI. The resulting peptide sequence between the zinc finger and Mtase proteins was therefore TGEKGS(G4S)3SGEFM. Zinc finger and Mtase residues are underlined. The BamHI and EcoRI restriction sites are incorporated into the sequences coding for GS and EF dipeptides.
The zinc finger–linker–M.HhaI gene was excised from pzfLHhaI, as a NdeI–XhoI fragment, and ligated into a pGEX5X-3 vector containing a modified multiple cloning site (P.J.Hurd, unpublished data), cut with the same enzymes to produce pGZfHhaI.
M.HpaII (Genbank accession no. L17342, bases 1594–2670) was amplified by PCR using pUCHpaII (10 ng) as template and the following primers, complementary to the 5′ end (5′-GCGCGCGAATTCATGAAAGATGTGTTAG ATGATAACTTGTTA-3′) and 3′ end (5′-GCGCGCGTC GACGTCATATAAATTTCCTAATTTTTCTAA-3′). After restriction of the PCR product with EcoRI and SalI (sites underlined in the primer sequences above), the fragment was ligated into pGZfHhaI, pre-restricted with EcoRI and XhoI, resulting in direct replacement of the M.HhaI gene with that of M.HpaII (pGZfHpaII). Construction of a vector coding for M.HpaII as a fusion with GST was by direct ligation of the C5-Mtase as a BamHI–SalI fragment into the modified pGEX5X-3 vector, to give pGHpaII. The integrity of constructs was confirmed by sequencing.
Expression from all vectors allowed purification of the recombinant protein via glutathione–agarose affinity chromatography.
Protein expression and purification
Recombinant plasmids were used to transform E.coli ER1647 cells and single colonies were used to induce high level expression of the targeted enzyme. Induction of protein expression was carried out as follows. A single colony was inoculated into 50 ml of LB medium containing 0.5% glucose, grown overnight at 37°C and cells were harvested by centrifugation. The cell pellet was resuspended in 1 ml of LB medium and then used to inoculate 500 ml of LB medium supplemented with 0.5% glucose and 100 µM ZnSO4. Growth was allowed to continue at 37°C until the OD600 reached 0.8. Protein expression was induced by the addition of isopropyl-β,d-thiogalactopyranoside to a final concentration of 1.0 mM and incubation was continued for a further 4 h at 30°C. Induced cells were harvested and lysed by sonication on ice; solubilisation of the recombinant enzymes was aided by the addition of Triton X-100 to 1% (v/v). Soluble recombinant enzymes were purified to homogeneity (as judged by 10% SDS–PAGE and Coomassie blue staining; 21) by glutathione affinity chromatography using glutathione–Sepharose (Sigma), essentially as described before (23). All protein concentrations were determined by quantitative analysis of an SDS polyacrylamide gel containing sample protein and a protein standard of known concentration. Imaging was performed using a Las 1000 CCD camera (Fujifilm) and Aida software (Raytest; Isotopenmessgeräte, GmbH).
Gel retardation analysis
All oligodeoxynucleotides were annealed in 10 mM Tris–HCl, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol (pH 7.9 at 25°C) in a PCR machine. Successful annealing was analysed by 3% agarose gel electrophoresis and ethidium bromide staining. Annealed oligodeoxynucleotide duplexes were labelled at the 5′ end with [γ-32P]ATP using phage T4 polynucleotide kinase. Unincorporated label was removed by gel filtration using Bio-Spin-6 columns (Bio-Rad Ltd). Typically, 0.05 pmol targeted or non-targeted enzyme was incubated in C5-Mtase buffer (20 mM HEPES, pH 7.5, 5 mM dithiothreitol, 20 µM ZnSO4, 5 mM MgCl2, 50 mM KCl, 10% glycerol, 0.1% Nonidet P-40, 0.1 mg/ml BSA) with 0.03 pmol labelled double-stranded oligodeoxynucleotide and 300 ng poly(dI)·poly(dC), in a final reaction volume of 30 µl. S-adenosyl-l-homocysteine (AdoHcy) was added to a final concentration of 100 µM. Incubations were carried out for 35 min at room temperature. The reaction mixture was subsequently loaded onto a pre-run, 4% (w/v) non-denaturing polyacrylamide (acrylamide/bisacrylamide 19:1) gel in 1× TBE buffer and electrophoresed for 1 h at 100 V at room temperature. Electrophoresis was carried out as described previously. All protein–DNA complexes were visualised after autoradiography with intensifying screens at –70°C overnight. (The high levels of protein relative to probe concentrations that were used in gel retardation assays were dictated by the conditions that yielded significant retardation of the probe, and therefore reflect the percentage activities of these protein preparations after the solubilisation and purification steps described above.)
For dissociation constant determination, competitor oligonucleotide was added at time 0, over a final concentration range 15.4–1078 nM (i.e. DNA always in excess). The reaction products were electrophoresed as described above. Dried down gels were imaged using an FLA-2000 Phosphorimager (Fuji). All experiments were performed at least in triplicate, unless stated otherwise, and used the same purified protein preparation. The proportion of bound competitor DNA was determined by subtraction of the retarded probe signal in the presence of competitor from that of a control shift in the absence of competitor, which was assigned an arbitrary value of 1.0 (i.e. 100%), using the imaging software Aida. This ensured that each gel shift assay could be normalised relative to its own internal control. Data was plotted as amount of competitor bound (normalised against zero competitor level shift intensities) against total amount of competitor added in each binding reaction. Data was fitted to the binding isotherm described by the equation [DNA:P] = ([Ptot][DNA]/[DNA] + Kd using Prizm (v.3.0) software (Graph Pad Inc., San Diego, CA), where [DNA:P] is the complex concentration, [Ptot] is the total protein concentration and [DNA] is the free DNA concentration. Under experimental conditions where the protein concentration is kept constant and the DNA levels are increased, the apparent dissociation constant (Kd) is always estimated as the concentration of free DNA when half the protein has been bound. [As an additional test of the data fit, Kd values derived from the non-linear regression approach described above were also compared with those calculated from data that were transformed and subjected to Scatchard analysis. In all cases, plots were clearly linear, suggesting single binding site kinetics, and Kd values were found to be corroborative (data not shown). Attempts to fit data to double binding site models failed in all cases.]
Oligodeoxynucleotide methylation assays
Unless otherwise stated, 1 pmol enzyme was incubated in the relevant C5-Mtase buffer (see previous section) with 3 pmol double-stranded oligodeoxynucleotide, 200 ng poly(dI)·poly(dC) and 1.65 µM [3H-methyl]AdoMet in a final reaction volume of 30 µl. Reactions were allowed to proceed for 30 min at 37°C before being stopped by the addition of SDS to 1% or AdoHcy (100 µM), followed by heat treatment for 5 min at 72°C in formamide gel loading buffer (21). The reaction mixture was subsequently loaded onto a pre-run 6% (w/v) denaturing (7 M urea) polyacrylamide (acrylamide/bisacrylamide 19:1) gel in 1× TBE buffer and electrophoresed for 1 h at 100 V at room temperature. Following electrophoresis, gels were fixed in 10% methanol, 10% acetic acid and either treated with the flurographic reagent Amplify (Amersham International) or dried down directly. Duplexes that had been methylated were identified by autoradiography with intensifying screens after 5 days at –70°C. Titrational methylation assays were performed similarly to above, with protein and oligonucleotide concentrations as indicated in the relevant figures. Time course methylation assays contained 3 pmol duplex oligonucleotide and 20 fmol enzyme. For both these experiments, reactions were stopped by direct addition of formamide loading buffer and subsequent incubation at 85°C for 10 min. After electrophoresis, gels were treated and dried down as described previously and exposed overnight to a tritium imaging plate, which was subsequently scanned using an FLA 2000 Phosphorimager and quantified using Aida software.
Ex vivo methylation assays
A double-stranded oligonucleotide with 3′ terminal single A overhangs, and harbouring adjacent zinc finger (underlined) and HpaII (bold) recognition sites (core sequence 5′-CTCCGGCTTCCATGGAGACGCAGAAGCCCT-3′) was ligated into the pCR 2.1-Topo vector (Invitrogen) to generate the vector pZMol. A target site vector was made containing a larger insert but still containing only a single HpaII site. This was accomplished by PCR of pZMol using primers specific for the pCR 2.1-Topo vector at regions close to, but not including, the nearest flanking HpaII sites to the site introduced by oligonucleotide insertion described above (5′-AGCGGGCGCTAGGGCGCTGGCAAGTGT-3′ and 5′-GTAGTTGTGTGGAATTGTGAGCGGAT-3′). The PCR product was then ligated into the pCR 2.1-Topo vector, to generate ZMTopo.
Ex vivo methylation analysis involved incubation of 0.5 µg ZMTopo vector with increasing concentrations of recombinant purified targeted or non-targeted HpaII enzyme, in HpaII methylase buffer (see above) in the presence of 5 µM AdoMet, in a reaction volume of 10 µl. After either a 30 or 60 min incubation at 37°C, the reaction was stopped by heating the samples at 85°C for 10 min. The samples were then subjected to digestion by R.HpaII (0.2, 0.5 or 10 U) for 90 min at 37°C. Samples were analysed on a 1% agarose gel.
RESULTS
Plasmids coding for targeted methyltransferases are fully methylated in vivo
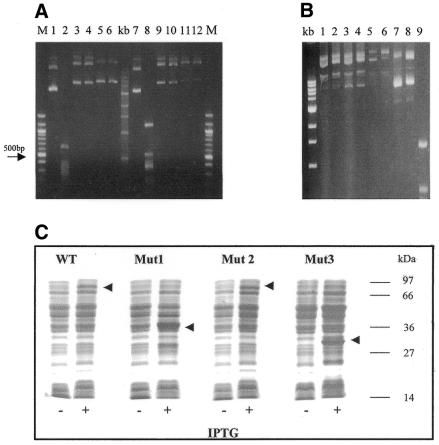
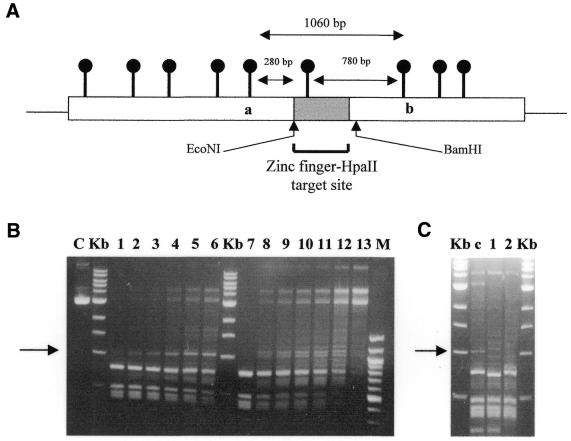
As a preliminary analysis of the targeted methylation properties of zinc finger–methyltransferase fusion enzymes, pGEX-based plasmids harbouring the genes for the GST–3Zf:HhaI or GST–3Zf:HpaII fusion proteins (henceforth referred to as Zf.M.HhaI and Zf.M.HpaII) and which contained multiple HhaI and HpaII restriction sites, were digested with HhaI or HpaII, respectively (Fig. 1A). Both these plasmids were shown to be fully resistant to cleavage, demonstrating that the Mtases had retained biological activity (lanes 6 and 12). Our expectation had been that the presence of the zinc finger component would reduce the level of wild-type methylation observed by the Mtase component. In order to further examine the nature of the methylation that occurred during bacterial growth, a number of mutant targeted HpaII constructs were made. Firstly, removal of the methionine codon at position 1 in the HpaII gene of the targeted HpaII construct generated a mutant (Mut2) that similarly and predictably produced a fully methylated plasmid (see Fig. 1B, lane 5 compared to 6). However, additional mutations, in which frameshifts were introduced into the GST–3Zf:HpaII vector, either at the start of the zinc finger gene of Mut2 (Mut3, Zf.HpaII gene out of frame with GST gene) or between the zinc finger and HpaII genes in the wild-type construct (Mut1, HpaII gene out of frame with GST–Zf gene), also gave rise to fully protected plasmids (Fig. 1B, lane 7 compared to 8 and lane 3 compared to 4). SDS–PAGE analysis of induced protein expression for each of these mutants revealed protein products of the predicted size (see Fig. 1C). Taken together, these results confirm that while the correct transcription and translational signals are being used in the cell under inducing conditions, internal start codons are also being used in E.coli, which give rise to methyltransferases lacking a fully functional zinc finger targeting component and at levels sufficient to fully methylate plasmid DNA under normal growth conditions. As such, further in vivo analysis of these enzymes in E.coli would have been uninformative under these conditions.
Figure 1.
Restriction protection analysis of in vivo methylated plasmid DNA. (A) Plasmids encoding targeted HhaI and HpaII were isolated from E.coli and subjected to restriction by restriction enzymes HhaI and HpaII respectively, as described in Materials and Methods. Lane 1, uncut control pGEX5X-3 vector; lane 2, R.HhaI restricted pGEX5X-3 vector; lane 3, uncut pGHhaI vector; lane 4, R.HhaI restricted pGHhaI vector; lane 5, uncut pGZfHhaI vector; lane 6, R.HhaI restricted pGZfHhaI vector; lane 7, as lane 1; lane 8, R.HpaII restricted pGEX5X-3 vector; lane 9, uncut pGHpaII vector; lane 10, R.HpaII restricted pGHpaII vector; lane 11, uncut pGZfHpaII vector; lane 12, R.HpaII restricted pGZfHpaII vector; lane m, 100 bp marker (NEB); lane kb, 1 kb ladder (NEB). (B) Plasmids encoding mutant targeted HpaII were isolated from E.coli and subjected to restriction by HpaII. Mutants are: Mut1, wild-type vector cut with EcoRI and filled in to generate HpaII out of frame with GST–Zf; Mut2, wild-type vector with first codon (methionine) of HpaII removed; Mut3, Mut2 vector cut with NdeI and filled in to generate Zf-HpaII out of frame with GST (for details see text). Lanes 1 and 2, uncut and R.HpaII cut wild-type targeted HpaII vector, respectively; lanes 3 and 4, uncut and R.HpaII cut Mut1 vector; lanes 5 and 6, uncut and R.HpaII cut Mut2 vector; lanes 7 and 8, uncut and R.HpaII cut Mut3 vector; lane 9, R.HpaII cut pGEX empty vector. (C) SDS–PAGE analysis of protein induction for wild-type and mutant targeted HpaII enzymes. Molecular weights are indicated on the right of the gel. Induced protein products are arrowed.
In the following series of experiments, gel retardation and methylation analyses were used to characterise the interaction between targeted C5-Mtases (i.e. with a zinc finger moiety) and control, non-targeted C5-Mtase (i.e. no zinc finger moiety) and DNA (oligonucleotides listed in Table 1). All experiments were carried out with glutathione S-transferase (GST) fused to the N-terminus of the recombinant enzymes via the zinc finger moiety (see Materials and Methods). The presence of GST has previously been demonstrated to have no effect on the activity of the C5-Mtase MspI, for example (24–26), or on the binding of zinc fingers to DNA (27,28).
Table 1. Oligodeoxynucleotides used during protein–DNA binding and methylation studies.
| Oligonucleotide | Sequence (5′→3′) (top strand) |
|---|---|
| Zf | TTCCATGGAGACGCAGAAGCCCTTCAGTTCCAG |
| ZfHhaI | GATGGCGAGGGC*GCCTTCCATGGAGACGCAGAAGCCCTTCAGCGGCCA |
| ZfHpaII (Sep16) | GATGGCGAGGCC*GGCACACATGGAGACGCAGAAGCCCTTCAGCGGCCA |
| HhaI | GGGATGGCCAGGGCGCCTTCCATGGAGAC |
| HpaII | GATGGCGAGGCCGGCTTCCATGGAGACGCAGGTGAGTTCCTCACGCCA |
| HpaII* | GTACGAGCAGCTCCCGGGTCAGTCTGCCTA |
| Non-Sp | GCTGATTGCGAGGGTACCTTCCATGTAACTATTCTCTAACTTCAGG |
| ZhpaII | GATGGCGAGGCCGGAAGCCCTTCAGCGGCCA |
| Sep 2 | GATGGCGAGGCCGGCAGAAGCCCTTCAGCGGCC |
| Sep 5 | GATGGCGAGGCCGGACGCAGAAGCCCTTCAGCGGCCA |
| Sep 6 | GATGGCGAGGCCGGGACGCAGAAGCCCTTCAGCGGCCA |
| Sep 11 | GATGGCGAGGCCGGATGGAGACGCAGAAGCCCTTCAGCGGCC |
| Sep 17 | GATGGCGAGGCCGGTGTGCTGTAGCTACGCAGAAGCCCTTCAGCGGCCA |
| Sep 22 | GATGGCGAGGCCGGTGTGCTGTAGCTGGAGGACGCAGAAGCCCTTCAGCGGCCA |
| Sep 25 | GCTCATCCACTGCGCACTCATGTCAGACTACCCGGTGTGCTGTAGCTACTAAGTGACGCAGAAGCCCCTCAGTGC |
| Sep 33 | GCTGCTCATCCATCCCGGCGCTTACTGGTGTAGCTAAGTCTAAGTGACGCAGAAGCCCCTCAGTGC |
| Sep 39 | GCTCATCCACTCCCGGTGTCAGACTAGCGCTGTCTGTAGCTACTAAGTGACGCAGAAGCCCCTCAGTGC |
| Sep 44 | GCTCATCCACTCCCGGACTCATGTCAGACTAGCGCTGTGCTGTAGCTACTAAGTGACGCAGAAGCCCCTCAGTGC |
Both the C5-Mtase and zinc finger recognition sites are underlined. ZfHhaI and ZfHpaII were also synthesised with 5-methyl-2′-deoxycytosine at the target cytosine in the C5-Mtase recognition site. The target base is asterisked in the respective sequences. Where indicated in the text, 5-methyl-2′-deoxycytosine was also present in the bottom strand at the corresponding symmetrically opposite target cytosine. [Only the top (5′→3′) strands are shown.] Base separation between subsites is measured from (and includes) the target cytosine in the HpaII site up to the first base before the zinc finger site.
Targeted C5-Mtases preferentially methylate DNA duplexes containing adjacent zinc finger and C5-Mtase recognition sites
The ability of Zf.M.HhaI to methylate DNA in a targeted manner was investigated by oligodeoxynucleotide methylation and competition–methylation assays. In order to discriminate between targeted and non-targeted methylation in the competition methyltransferase assays described, C5-Mtase site-containing oligodeoxynucleotides were synthesised as 29mers or 30mers (compared to 48mer target site oligodeoxynucleotides). This size difference enhances resolution of the two oligonucleotides during electrophoresis.
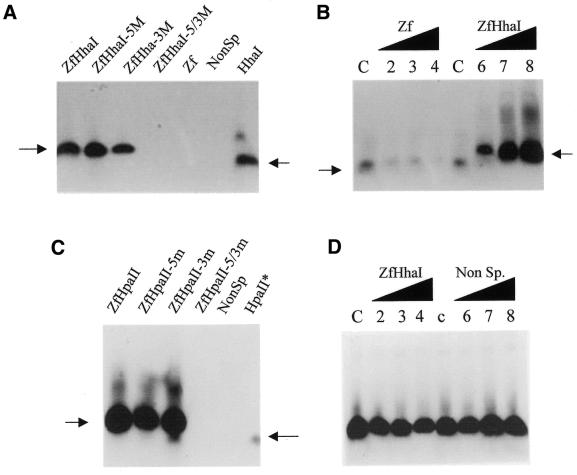
The results of methylation assays designed to confirm the ability of Zf.M.HhaI to methylate both strands of the target sequence are shown in Figure 2A. (The designation of ‘top’ strand for all experiments described subsequently is defined by the sequence orientation of oligonucleotides listed in Table 1.) The enzyme is clearly able to transfer tritiated methyl groups from [3H-methyl]AdoMet to hemi-methylated cognate DNA duplexes (Fig. 2A, lanes 2 and 3). Duplexes that contained no C5-Mtase sites (lanes 5 and 6) or duplexes that were fully methylated (lane 4) were not subsequently methylated, confirming that the methylation observed for non-methylated or hemi-methylated duplex target site DNA had occurred only at the HhaI site. A significant level of methylation was observed by Zf.M.HhaI for duplexes that contained a HhaI site only (Fig. 2A, lane 7).
Figure 2.
Methylation analysis of targeted and non-targeted HhaI and HpaII enzymes. (A) Oligodeoxynucleotide methylation assays were performed to confirm double-strand methylation by Zf.M.HhaI. All reactions contained 1 pmol Zf.M.HhaI protein and 3 pmol duplex DNA as indicated in each lane. Reactions were carried out as described in Materials and Methods. The designations 5M, 3M and 5/3M, in this and other figures, refer to the oligonucleotides being pre-methylated at the target cytosine on either the top, bottom or both DNA strands respectively. (B) Methylation competition assays. All lanes contained 1 pmol Zf.M.HhaI protein and 3 pmol HhaI oligodeoxynucleotide and competitor DNA at the following levels: lane C, no competitor DNA; lanes 2–4, Zf oligodeoxynucleotide added as competitor at 1-, 5- and 10-fold molar excess, respectively; lanes 6–8, ZfHhaI oligodeoxynucleotide added similarly at 1-, 5- and 10-fold molar excess. The arrows indicate the different mobilities of the oligonucleotides used. (C) Oligodeoxynucleotide methylation assays were performed to confirm double-strand methylation by Zf.M.HpaII. All lanes contained 1 pmol Zf.M.HpaII protein and 3 pmol duplex DNA as indicated in each lane. (D) Competition methylation analysis. All reactions contained 1 pmol Zf.M.HpaII, 3 pmol ZfHpaII oligonucleotide and competitor DNA at the following levels: lane c, no competitor DNA; lanes 2–4, as lane c except for the addition of 1-, 5- and 10-fold molar excess of ZfHhaI competitor oligodeoxynucleotide (i.e. effectively zinc finger only site); lanes 6–8, as lane c but with the addition of 1-, 5- and 10-fold molar excess of non-specific (Non-Sp.) oligodeoxynucleotide.
The results of experiments designed to assess the ability of competitor oligonucleotides to modulate the methylation by Zf.M.HhaI of DNA substrates containing a HhaI site are presented in Figure 2B. Addition of target competitor duplex ZfHhaI (lanes 6–8) or duplexes that contain the zinc finger recognition site alone (lanes 2–4) cause a readily observable reduction in non-targeted methylation of duplexes containing only a M.HhaI site (lanes C), accompanied by a steady increase in degree of methylation of the competitor duplex (arrowed on the right hand side of Fig. 1B). Effective and complete competition is observed even at equimolar levels when the competing duplex contained the zinc finger recognition and C5-Mtase sites, rather than duplexes containing a zinc finger recognition site alone.
In a repeat of the experiments described in Figure 2A for Zf.M.HhaI, the degree of non-targeted methylation of a 30mer oligonucleotide harbouring a single M.HpaII site by Zf.M.HpaII was only just detectable under conditions where the signal acquired by the target site-containing oligodeoxynucleotides was extremely intense (Fig. 2C, compare lanes 1–3 with lane 6). Indeed, in three additional repeats of this assay, non-targeted methylation of the HpaII* oligonucleotide by Zf.M.HpaII was not detectable at all (data not shown). Similarly to Zf.M.HhaI, Zf.M.HpaII is able to methylate either strand of the target duplex (Fig. 2C, lanes 2 and 3).
Due to the extremely low levels of non-targeted DNA methylation produced by Zf.M.HpaII, the competition experiments described for Zf.M.HhaI in Figure 2B could not be performed with this enzyme. However, a series of alternative competition experiments were conducted using the Zf.M.HpaII enzyme, in order to examine the ability of either zinc finger site-containing or non-specific oligodeoxynucleotide sequences to compete with methylation of a target oligonucleotide (ZfHpaII). Results are shown in Figure 2D. Specific methylation of the target oligodeoxynucleotide is not significantly affected by even a 10-fold molar excess of zinc finger site-containing oligodeoxynucleotide (Fig. 2D, lane 4). Similar results were obtained for competition by non-specific duplex (lanes 6–8). These data further confirm that the C5-Mtase component of the targeted enzymes contributes significantly to the specificity of the targeted enzyme as a whole.
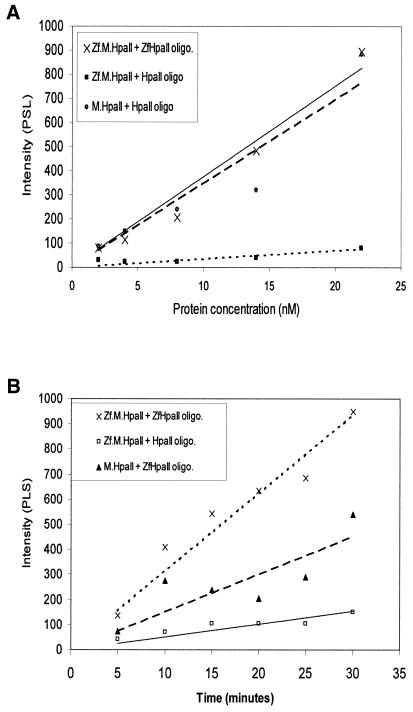
Time course and titrational methylation assays were performed on Zf.M.HpaII in order to examine the relative catalytic activity at targeted and non-targeted Mtase sites. The results of titrational assays are shown in Figure 3A. The targeted enzyme is shown to catalyse methylation of the target oligonucleotide to an ∼11-fold higher level than a duplex harbouring an M.HpaII site only. The relative catalytic activity of Zf.M.HpaII for a duplex harbouring a zinc finger and adjacent C5-Mtase site is similar to that of non-targeted HpaII for an oligonucleotide harbouring a C5-Mtase site.
Figure 3.
Titrational and time course analysis of targeted and non-targeted HpaII methylation for different oligonucleotide substrates. (A) Increasing concentrations of Zf.M.HpaII and M.HpaII enzyme were incubated with ZfHpaII or HpaII oligonucleotides (final concentration 150 nM) over the concentration range shown. The relative levels of methylated oligonucleotide, as evaluated by phosphorimager analysis, were plotted against protein concentration. The intensity of the tritiated oligonucleotides are plotted as PSL values (photo-stimulated luminescence), which are directly proportional to the radioactivity of the samples being measured. The best fit line for Zf.M.HpaII binding to ZfHpaII oligonucleotide is denoted by a solid line. (B) Time course methylation profile for the interaction of targeted and non-targeted HpaII with oligonucleotide substrates. Reactions contained 20 fmol protein and 3 pmol DNA (for details see Materials and Methods).
The results of time course methylation analyses, shown in Figure 3B, reveal a similar trend, with an 8-fold difference in activity for Zf.M.HpaII methylating target site- versus M.HpaII site-containing oligonucleotides. Only an ∼2-fold difference in catalytic activity was observed between targeted and non-targeted HpaII for ZfHpaII DNA.
Targeted HpaII C5-Mtase binds specifically and in cis to a recognition site comprising adjacent zinc finger and Mtase subsites
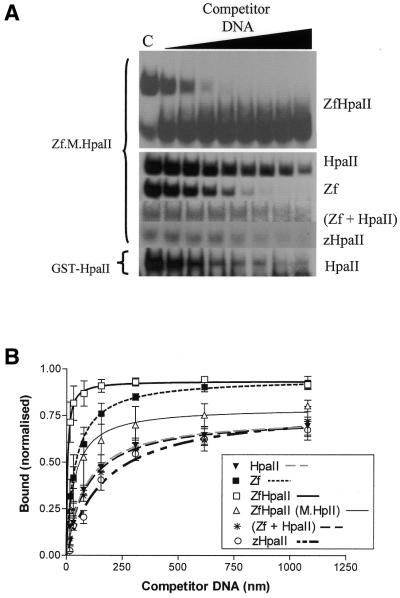
In order to obtain more quantitative binding data describing the interactions of such targeted enzymes with cognate and related sites, kinetic analyses of competition gel shift assays were performed on the Zf.M.HpaII and M.HpaII enzymes, as described in Materials and Methods. The competition approach ensures that within each set of comparative experiments identical levels of protein and probe are used, while the more readily quantifiable competitor DNA level is varied. (For all subsequent band shift assays reported the labelled duplex in each case was ZfHpaII; see Table 1.) The gel shifts are shown in Figure 4A. For the self-competition experiment, a full gel is shown. (For all subsequent experiments throughout this work, only the region of the gel showing retarded probe is shown. Each gel figure shown represents one of a triplicate of experiments from which binding data were calculated.) While absolute Kd values per se cannot realistically be determined via oligonucleotide competition approaches, highly reproducible relative Kd values can be determined that are a proportional reflection of the binding affinity of the enzyme for the various DNA substrates.
Figure 4.
(A) Gel shift competition assays. ZfHpaII probe (1.0 nM) was incubated with M.HpaII or Zf.M.HpaII enzymes (1.7 nM) (see Materials and Methods). Lane C, no competitor DNA; subsequent lanes contain competitor DNA at final concentrations of 15.5, 30.8, 77, 154, 308, 616 and 1078 nM. The competitor DNA used is indicated on the right of each gel; the protein assayed in each case is indicated on the left of each gel. For the self-competition experiment, the full retardation gel is shown. For all subsequent experiments, only that portion of the gel containing the retarded probe is shown. (B) Analysis of triplicate binding data described in (A) above.
Binding analysis of the gel shifts shown in Figure 4A are shown in Figure 4B and reveal that Zf.M.HpaII binds to its target site with an apparent Kd of 4.77 (± 0.93) × 10–9 M, but binds to zinc finger or C5-Mtase sites alone with Kd of 38.6 (± 3.9) and 86.8 (± 10.8) × 10–9 M, respectively, ∼8- and 18-fold differences in binding affinity, respectively. This result clearly demonstrates specific binding to a sequence comprising adjacent zinc finger and Mtase recognition sites. Under identical conditions, the three zinc finger protein (as a fusion to GST only) binds to its recognition site with an apparent Kd of ∼14 × 10–9 M (data not shown). M.HpaII binds to a M.HpaII site-containing oligonucleotide with a Kd of 35.08 (± 7.9) × 10–9 M, significantly weaker than for Zf.M.HpaII binding its target site, but stronger than for Zf.M.HpaII binding just a HpaII site. Thus the targeted HpaII enzyme displays both an enhanced binding affinity compared to its individual components and a reduced affinity for individual subsites associated with each component of the fusion enzyme. Double competition binding experiments for Zf.M.HpaII, using a combination of the competitor duplexes HpaII and Zf, which contain single M.HpaII and zinc finger protein recognition sites, respectively, yielded an apparent Kd of 93.65 (± 24.6) × 10–9 M. This is a much larger value than found for binding target site and zinc finger site-containing oligonucleotides alone, despite the opportunity for interaction of each component of the fusion protein with its own recognition subsite, and suggests that both components of the targeted enzyme preferentially bind to linear DNA, with subsites arranged in cis. Competition experiments using an oligonucleotide in which the HpaII site replaced the first 3 bp of the zinc finger subsite (zHpaII, bottom gel in Fig. 3A) gave an apparent Kd of 169.4 (± 29.7) × 10–9 M, again much larger than that found for binding to just a HpaII site-containing oligonucleotide. All these results taken together confirm that the binding affinity obtained for the interaction of the enzyme with the target site sequence is a result of both the C5-Mtase and zinc finger subunits of the targeted enzyme, acting in concert and in cis on the same DNA substrate.
Targeted HpaII binding is modulated by the methylation status of the target site
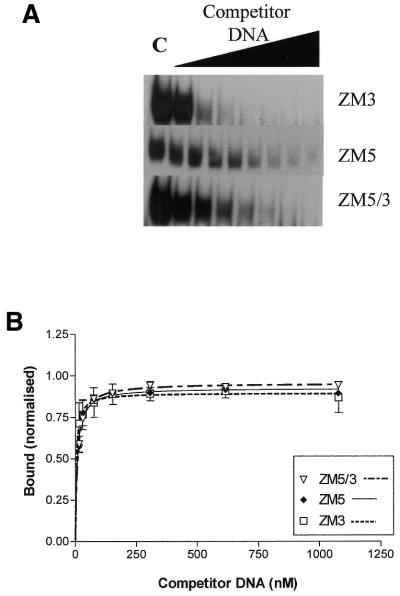
In this experiment, competitor oligonucleotides were synthesised in which either the top, bottom or both top and bottom strand target cytosine positions contained methylcytosine. Representative gel mobility titrations showing the effect of methylation status of the target site on binding of Zf.M.HpaII are shown in Figure 5A. Kinetic analysis of these data is shown in Figure 5B. The enzyme binds to the ZM3 oligonucleotide (bottom cytosine methylated) with an apparent Kd of 4.28 (± 1.2) × 10–9 M and to the ZM5 oligonucleotide (top strand methylated) with a Kd of 7.6 (± 0.87) × 10–9 M, nearly 2-fold weaker. The protein bound to the fully methylated target site ZM5/3 with an apparent Kd of 9.25 (± 0.53) × 10–9 M. Taken together, these results indicate a slight preference for the Mtase component of Zf.M.HpaII binding the top strand, presumably as a consequence of motional constraints placed upon the targeted enzyme by the zinc finger–DNA interactions or of the flexibility of the (Gly4Ser)3 linker between the zinc finger and C5-Mtase components of the targeted enzyme.
Figure 5.
Gel retardation competition analysis of the interaction of Zf.M.HpaII with pre-methylated target site DNA. (A) ZfHpaII probe was incubated with Zf.M.HpaII enzyme as in Figure 3. Lane C, no competitor DNA; subsequent lanes contain competitor DNA at final concentrations of 15.5, 30.8, 77, 154, 308, 616 and 1078 nM. The competitor DNA used is indicated on the right of each gel. (B) Analysis of triplicate binding data described in (A). Oligonucleotides ZM3, ZM5 and ZM5/3 are identical to oligonucelotide ZfHpaII but contain methylcytosine at the target site (i.e. mCCGG) on the top, bottom and both strands, respectively.
Targeted HpaII C5-Mtase binds and methylates specifically at recognition sites with variable subsite spacing
The ability of the targeted HpaII C5-Mtase enzyme to bind specifically to oligodeoxynucleotides with an increasing base pair separation between the zinc finger and M.HpaII subsites was examined by gel retardation competition and oligonucleotide methyltransferase assays. Figure 6A shows the gel shift assays resulting from competition with duplexes harbouring zinc finger and M.HpaII subsites separated over the range 2–44 bp (oligonucleotides Sep2–Sep44, see Table 1).
Figure 6.
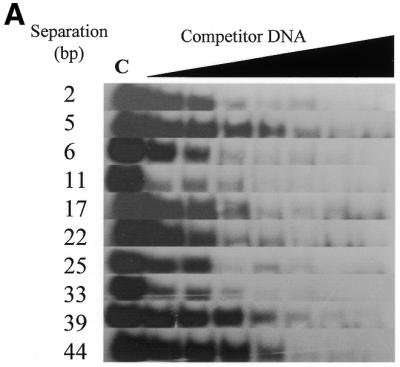
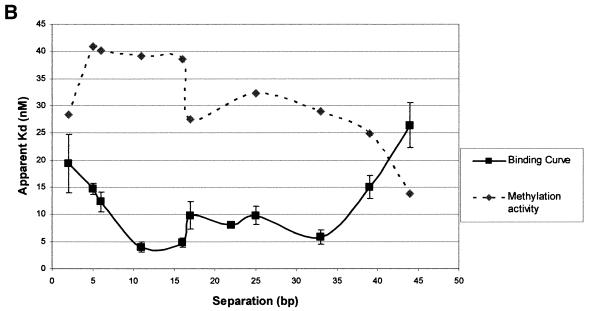
Zf.M.HpaII binds specifically to oligodeoxynucleotides with variable ‘subsite’ spacings. (A) Gel retardation analysis. Lane C in all cases contains Zf.M.HpaII enzyme and ZfHpaII labelled probe at the same levels as shown in Figure 3. Competitor DNA was added to the same final concentrations as given in Figure 3. The base pair separation between subsites for each competitor oligonucleotide is shown to the left of each gel. (B) The apparent Kd values for triplicate competition binding experiments for each subsite spacing were plotted against the subsite spacing. The methylation signal obtained from incubation of each oligonucleotide (30 pmol) with Zf.M.HpaII protein (0.1 pmol) for 30 min in the presence of [3H]AdoMet is also plotted on the same graph. The curve shown is representative of a triplicate set, which all followed the same trend. The methylation intensities have been normalised to fit on the graph.
Figure 6B shows a plot of the binding affinity of the enzyme for each subsite spacing. Also presented on the graph are the results of methylation assays for each oligonucleotide (for details see figure legend). The values have been normalised against the apparent Kd values, so that they can be presented in the same figure. It is immediately apparent that binding and methylation curves correlate extremely well, being nearly mirror images of each other, i.e. as the apparent Kd reduces (tighter binding) the methylation signal increases. A general area of tight binding and high methylation rate can be seen to occur from ∼10 bp up to ∼40 bp. Two local optima, coincident for both binding and methylation, centred at ∼13 bp and at 34 bp separation, may also be present. An extremely sharp transition point can be seen to occur between the 16 and 17 bp subsite separations, where both binding and methylation become weaker. This is a significantly larger change than is seen for the 5 to 6 bp subsite spacing transition and probably reflects a ‘stress site’, at which flexibility of the fusion protein linker and the related orientational preference of the methylase component directly conflict with the spatial presentation of the target cytosines at the Mtase subsite.
Targeted HpaII specifically methylates plasmid DNA harbouring a target sequence ex vivo, against a background of non-targeted HpaII sites
The vector ZMTopo was constructed, harbouring a single targetable site (see Materials and Methods) but containing an additional 23 M.HpaII recognition sites. In the absence of any methylation, the target plasmid should yield a characteristic restriction pattern after digestion by R.HpaII. Specific methylation at the target site would result in an alteration of this restriction pattern, giving rise to a unique 1060 bp fragment. A schematic describing this rationale is shown in Figure 7A.
Figure 7.
Ex vivo methylation analysis of Zf.M.HpaII interacting with target site-containing plasmid DNA. (A) Schematic outline of the key elements of the ZMTopo vector used as substrate in the ex vivo studies described below (see also Materials and Methods). The region of the plasmid harbouring the zinc finger and single flanking HpaII site is shown as a grey box with a lollipop on it. In the scenario where this site has been preferentially methylated (designated by a black lollipop, compared to unmethylated vector HpaII sites, shown in grey), the nearest cleavage by R.HpaII will only occur at flanking HpaII sites a and b, resulting in the generation of a 1060 bp fragment. If this site is not methylated, derivative fragments of ∼280 and 780 bp will be produced. Restriction enzyme sites for EcoNI and BamHI are also shown. These sites are unique to DNA fragments harbouring the target sequence. (B) Target site-containing vector ZMTopo was incubated with increasing amounts of Zf.M.HpaII or M.HpaII enzyme for 30 min prior to digestion with R.HpaII. Lanes 1–6 and 7–12 represent digestion of vector preincubated with 25, 50, 75, 125, 175 and 225 fmol Zf.M.HpaII and M.HpaII protein, respectively. The expected 1060 bp DNA fragment indicative of targeted methylation is arrowed. Lane U, unrestricted ZMTopo DNA; lane C, unmethylated vector DNA restricted with R.HpaII; lane M, 100 bp ladder (NEB). Key size bands are indicated. (C) All lanes were identical to those described for lane 4 in (B), but with the inclusion of 10 U of the restriction enzyme EcoNI (lane 1) or BamHI (lane 2) or just water (lane c). The 1060 bp DNA fragment present in lane c is arrowed.
A titrational ex vivo methylation assay was performed, incubating target site plasmid ZMTopo with increasing amounts of recombinant Zf.M.HpaII, in order to simulate the in vivo scenario of methylating more complex substrates. The DNA was subsequently digested with R.HpaII and the digestion products analysed by agarose gel electrophoresis. The results are shown in Figure 7B. A band corresponding to the expected size of 1060 bp can be clearly seen, increasing in intensity in those lanes that had been incubated with Zf.M.HpaII (lanes 1–6), but not in lanes that had been incubated just with M.HpaII (lanes 7–12). The identification of this band as being the target site-containing fragment was confirmed by further digestion of the DNA with restriction enzymes whose sites only occurred within the target fragment (see Fig. 7A). As can be seen in Figure 7C, upon restriction with EcoNI and BamHI, the 1060 bp fragment disappears.
The appearance of a unique band from the target site vector in this assay only when incubated with targeted enzyme and corresponding in size to that expected from the result of targeted methylation, and the confirmation of predicted restriction enzyme sites within this fragment, confirms our conclusion that targeted methylation is occurring under these conditions.
DISCUSSION
We have presented data describing the evaluation of fusion proteins comprising a three zinc finger protein covalently linked to either HpaII or HhaI Mtase via a flexible peptide linker. The principle behind this approach was to bias the action of the Mtase component to specific recognition sites than were proximal to the 9 bp DNA sequence recognised by the zinc finger protein. The expectation of this initial approach would be that the intrinsic specificity of each component of the fusion protein should remain unaltered, although one would expect changes in general binding affinities for each component for its subsite, as part of a fusion protein. We have evaluated whether this was indeed the case in detail for the targeted HpaII enzyme, as well as examining orientational and subsite spacing preferences.
The initial discovery that plasmids encoding targeted methyltransferases were fully methylated suggested initially that fusion of a Mtase to the zinc finger protein did not modulate either Mtase activity or specificity in E.coli. However, the observation of similar methylation by constructs specifically mutated so as not to express a functional targeted methyltransferase suggested that internal transcription and/or translation occurred from vectors coding for the targeted enzymes in E.coli. Given the repetitive nature of the DNA linker sequence joining the zinc finger and methyltransferase genes, the possibility of both translational and transcriptional stalling present themselves, allowing alternative ‘re-start’ points for both events to re-occur along the template. Due to the nature of methyltransferases, only a small number of non-targeted molecules would need to be produced to fully protect the coding plasmid under typical overnight growth conditions. This property suggests methyltransferases as excellent reporters for constitutive and low level expression/translation studies. However, any evaluation of targeted methylation efficacy in bacteria using this current expression system was not possible, due to the considerations mentioned above.
Comparative in vitro analysis of the catalytic activity of purified Zf.M.HhaI and Zf.M.HpaII enzymes revealed that Zf.M.HhaI in fact catalyses a significant level of methylation of duplexes harbouring a M.HhaI site alone (i.e. it is able to function as a discrete methyltransferase at some level, irrespective of its zinc finger component). Binding studies, however, revealed no interaction between targeted HhaI and this duplex (data not shown). Such non-targeted DNA methylation has also been described for the only other report of a targeted C5-Mtase, a fusion between a three zinc finger protein and the 5′-CpG-3′-specific prokaryotic SssI C5-Mtase (5). Consequently, this enzyme has limited use in in vivo applications. Non-specific methylation associated with the targeted M.SssI enzyme may be related to the observed processive action of this enzyme, proceeding along CpG-containing substrate molecules and methylating one strand of DNA at a time. Such activity would exacerbate the level of non-targeted methylation expected. The HhaI and HpaII C5-Mtases, however, do not display the same processive characteristics as described for M.SssI (6).
The results of methylation competition experiments described for targeted HhaI and HpaII broadly correlate with data from gel shift assays and provide confirmation that both the DNA binding and DNA methylation events are targeted phenomena. The degrees of targeted and non-targeted methylation observed between these two enzymes might reflect differences in their 3-dimensional structures, which would modulate the interaction between the C5-Mtase and zinc finger moieties. It may be significant that the N-terminal regions of each C5-Mtase differ in length before the start of homology block I (1,2). M.HhaI has 13 amino acids and M.HpaII has 34 amino acids in this region, which are fused directly to the linker sequence between the zinc finger and C5-Mtase components. Contributions made by each region to the overall nature and flexibility of the existing linker region would be expected to modulate the characteristics of each enzyme and represent a potential avenue of exploration for additional studies aimed at further enhancing the specificity of these enzymes. An earlier targeted HhaI prototype enzyme, containing a (Gly)6 linker, was in fact non-functional (data not shown), and confirms the importance of the linker sequence between the components of a fusion enzyme, as demonstrated in other studies (5,28–31).
Titrational and time course methylation assays on the targeted HpaII enzyme interacting with target and C5-Mtase site substrate oligonucleotides confirmed methylation specificity and that the enzymatic properties of the C5-Mtase component of the targeted enzyme had not been impaired as a consequence of its fusion protein status. Interestingly, methylation of the shorter (30mer) duplexes by targeted M.HpaII was significantly lower than for the longer DNA substrates (48mers) used in time course and titration assays, and may reflect a requirement for an initial non-specific interaction between Zf.M.HpaII and DNA before methylation proceeds at the specific site.
Binding studies on the active targeted HpaII enzyme are necessarily complicated by the fact that the Mtase component of the fusion enzyme has an intrinsic catalytic activity. During the course of a normal binding assay, for example, one would expect large protein conformational changes to occur as well as forced dissociation of the protein from the DNA subsequent to target site methylation. An additional aspect would be the generation of fully methylated and hemi-methylated DNA products that would re-enter the general pool of DNA to be re-bound. A further complication lies in the proposed base flipping mechanism associated with C5-Mtases, which may have an associated DNA ‘tracking’ component and which might also be expected to distort the DNA. To counter the problems associated with catalysis interfering with accurate measurement of binding affinities, we have performed all binding assays in the presence of the commonly used cofactor analogue AdoHcy, which lacks a methyl donor group. Thus catalytic acceleration of dissociation during the binding assay has been addressed to some degree.
Kinetic analysis of Zf.M.HpaII binding to a series of different oligonucleotides representing target sequence permutations revealed that the highest affinity interaction was found for Zf.M.HpaII binding to its full target site, which was 8-fold tighter than to a zinc finger site oligonucleotide and 18-fold tighter than to a Mtase site oligonucleotide. These results confirm that the targeted enzyme displays a binding affinity greater than either of its component parts, although the greater contribution is most likely from the zinc finger moiety and suggests a degree of cooperativity between each component in the binding event. Evidence of cooperativity in binding also exists, from the results of double competition experiments using separate Zf- and Mtase site-containing competitor oligonucleotides. The resultant apparent Kd values are significantly larger than for the protein binding to target site or zinc finger site oligonucleotides alone and are in fact more comparable to that of binding to a methylase site-containing oligonucleotide. This is an unusual result, since one would expect the zinc finger oligonucleotide component of the double titration assay to exert a similar competitive effect as in previous experiments, when it was sole competitor. It can only be inferred that the fusion enzyme cannot interact with subsites located on separate DNA molecules with the same efficiency as each protein would in its wild-type form, or that such interactions are now short lived due to steric problems. For example, crystal structure analysis of the HhaI Mtase and its complex with DNA shows that significant conformational changes occur upon DNA binding (3), and it is conceivable that such movement in the methyltransferase component of the targeted enzyme would lead to a reduction in zinc finger–DNA interactions in a trans binding scenario. Similarly, competition by the mutated zinc finger site oligonucleotide zHpaII, in which the HpaII site directly encroaches into the zinc finger site to the extent that only 6 of the original 9 bp of the zinc finger site are present, is significantly less effective than by a simple HpaII site-containing oligonucleotide, and again argues for a cooperative (albeit negative in this instance) effect.
Similar ‘bi-modal’ binding systems, such as covalently linked zinc finger multi-arrays or zinc finger–transcription factor fusion enzymes, have also been shown to demonstrate cooperativity in binding between component proteins when similarly analysed (32,33). Such cooperativity as seen in these and our own study, where the binding affinity for the hybrid target site is significantly greater than the affinity of each protein component for its respective site, is thought to be largely due to an enhanced local concentration of each protein component on the DNA. This is due to ‘tethering’ of each component near the DNA, as a consequence of binding of the other component to its recognition subsite. Cooperative interactions might also arise due to direct interaction between component proteins of the fusion enzyme, resulting in an enhancement of their DNA binding affinity. Linker-associated interactions may also contribute to the binding event.
Binding studies using pre-methylated competitor duplexes showed that the C5-Mtase component of Zf.M.HpaII preferentially interacts with the top strand of the target duplex. This is most likely attributable to the flexibility of the linker peptide joining the zinc finger and C5-Mtase components. The lower binding affinity observed for the fully methylated duplex reflects the fact that a fully methylated Mtase site is no longer a substrate for methylation and thus not an optimal binding site. Significantly, DNA methylation analysis of both the Zf.M.HhaI and Zf.M.HpaII enzymes revealed that these enzymes are capable of transferring methyl groups to both strands of the target sequence at the respective C5-Mtase subsites. In contrast to gel shift competition assays, however, under the conditions of these methylation experiments no obvious strand preference can be inferred.
The ability of Zf.M.HpaII to specifically bind duplexes with variable spacing between the zinc finger and C5-Mtase subsites was shown to correlate with the methylation activity of the enzyme for each oligonucleotide. In both instances, maximum activity/affinity occurred over the range 10–40 bp separation between subsites. Within this region, evidence for the existence of two local optima is observed at 13 and 32 bp. These fluctuations fall at an interval corresponding to whole helical turns of DNA and are not inconsistent with the target cytosines being optimally positioned for interaction with the methylase component of the fusion enzyme at these particular subsite spacings. The ability of the C5-Mtase component to specifically recognise and bind its subsite up to 44 bp away from the anchored zinc finger subunit suggests great potential for this enzyme construct in delivering methylation over a significant length of DNA. The binding affinity at this separation (26 nM) is still stronger than that of the protein binding to just a zinc finger site (38 nM). Interestingly, binding of the enzyme to DNA with a subsite separation of only 2 bp was still significantly tighter than to oligonucleotides with ‘infinite’ spacing (i.e. the Zf and HpaII oligonucleotides). This suggests, rather surprisingly, that even at such a close subsite separation, where in fact the HpaII and zinc finger sites overlap but are maintained (see Table 1), both components of the fusion enzyme are still contributing to the binding event in a cooperative manner. This conclusion is also in accord with results obtained for binding of the protein to the zHpaII oligonucleotide previously discussed. Given that when the linker can be considered either relaxed (at 2 bp subsite separation) or extended (at 44 bp separation) cooperative binding is still observed, it seems unlikely that the linker contributes significantly to the cooperative binding event.
Preliminary titrational methylation assays had suggested that the DNA:protein ratio may represent a critical factor for the successful function of these targeted enzymes. This conclusion was confirmed when methylation analysis of a target site plasmid revealed successful targeted methylation at the target site and where the majority of vector DNA was unmethylated. Such targeted methylation occurred at specific ratios of protein:DNA concentration, which we have calculated to be in the range 1:10–1:50. This observation is also significant, given that in vivo studies on the zinc finger protein activation of target promoters has shown that at high cellular protein concentrations, non-specific binding and transcriptional activation from non-target site-containing promoters was observed (34).
Here we have constructed targeted C5-Mtases that can methylate DNA in a targeted manner. In the absence of a target site, these enzymes are not absolutely mono-functional, although Zf.M.HpaII is close to being so. When the enzymes are presented with both target site- and non-target site-containing DNA, methylation occurs at the target site in preference to any other. The targeted HpaII enzyme displays the most specific binding and methylation attributes and has been shown to be able to deliver targeted methylation over at least a 35 bp region. Methylation analyses using complex substrates, such as target site vectors, more closely approach in vivo conditions and have shown that targeted methylation is achievable at specific ratios of protein to DNA.
However, a key problem still exists with these enzymes, which is that the level of normal, i.e. non-targeted, methylation is quite high. In the absence of target sites, the affinity of the targeted enzyme for Mtase sites alone is high enough for significant methylation to occur. Second generation targeted enzymes are therefore currently being evaluated, in which the C5-Mtase moieties harbour mutations that result in reduced DNA binding affinity at the Mtase recognition site and reduced catalytic activity (27). Such lower activity would be compensated for by the tethering effect of the zinc finger component, which should occur only at the target site. These approaches should significantly reduce, if not eliminate, non-targeted methylation. Additionally, the zinc finger protein used in this study has only moderate binding affinity for its recognition site and targeting could certainly be further enhanced by use of tighter binding zinc finger proteins or larger zinc finger arrays.
While there is still room for significant advances to be made in this area, these current and future enzymes represent valuable tools for the study of DNA methylation spread and control in in vivo studies, as well as potential effectors for targeted promoter gene silencing induced through a DNA methylation-mediated cellular response.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the Leukaemia Research Fund of the UK, the BBSRC and the Royal Society.
REFERENCES
- 1.Posfai J., Bhagwat,A.S., Posfai,G. and Roberts,R.J. (1989) Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res., 17, 2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson G. (1992) Amino acid sequence arrangements of DNA-methyltransferases. Methods Enzymol., 216, 259–278. [DOI] [PubMed] [Google Scholar]
- 3.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 4.Reinisch K.M., Chen,L., Verdine,G.L. and Lipscomb,W.N. (1995) The crystal structure of HaeIII methyltransferase covalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell, 82, 143–153. [DOI] [PubMed] [Google Scholar]
- 5.Xu G.-L. and Bestor,T.H. (1997) Cytosine methylation targetted to predetermined sequences. Nature Genet., 17, 376–378. [DOI] [PubMed] [Google Scholar]
- 6.Renbaum P. and Razin,A. (1992) Mode of action of the Spiroplasma CpG methylase M.SssI. FEBS Lett., 313, 243–247. [DOI] [PubMed] [Google Scholar]
- 7.Matsuo K., Gramatikoff,K. and Schaffner,W. (1994) The CpG-specific methylase SssI has topoisomerase activity in the presence of Mg2+. Nucleic Acids Res., 22, 5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toth M., Lichtenberg,U. and Doerfler,W. (1989) Genomic sequencing reveals a 5-methylcytosine-free domain in active promoters and the spreading of preimposed methylation patterns. Proc. Natl Acad. Sci. USA, 86, 3728–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindsay H. and Adams,R.L.P. (1996) Spreading of methylation along DNA. Biochem. J., 320, 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tollefsbol T.O. and Hutchison,C.A.,III (1997) Control of methylation spreading in synthetic DNA sequences by the murine DNA methyltransferase. J. Mol. Biol., 269, 494–504. [DOI] [PubMed] [Google Scholar]
- 11.Okano M., Xie,S. and Li,E. (1998) Cloning and characterisation of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genet., 19, 219–220. [DOI] [PubMed] [Google Scholar]
- 12.Komura J.-I., Okada,T. and Ono,T. (1995) Repression of transient expression by DNA methylation in transcribed regions of reporter genes introduced into cultured human cells. Biochim. Biophys. Acta, 1260, 73–78. [DOI] [PubMed] [Google Scholar]
- 13.Remus R., Kammer,C., Heller,H., Schmidtz,B., Schell,G. and Doerfler,W. (1999) Insertion of foreign DNA into an established mammalian genome can alter the methylation of cellular DNA sequences. J. Virol., 73, 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyszynski M.W., Gabbara,S. and Bhagwat,A.S. (1992) Substitutions of a cysteine conserved among DNA cytosine methylases result in a variety of phenotypes. Nucleic Acids Res., 20, 319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mi S. and Roberts,R.J. (1993) The DNA-binding affinity of HhaI methylase is increased by a single amino acid substitution in the catalytic center. Nucleic Acids Res., 21, 2459–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Razin A. (1998) CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBO J., 17, 4905–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng H.-H. and Bird,A. (1999) DNA methylation and chromatin modification. Curr. Opin. Genet. Dev., 9, 158–163. [DOI] [PubMed] [Google Scholar]
- 18.Jones P.L., Veenstra,G.-J.C., Wade,P.A., Vermaak,D., Kass,S.U., Landsberger,N., Strouboulis,J. and Wolffe,A.P. (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet., 19, 187–191. [DOI] [PubMed] [Google Scholar]
- 19.Nan X., Ng,H.-H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenman,R.E. and Bird,A. (1998) Transcriptional repression by the methy-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y., Ng,H.-H., Erdjument-Bromage,H., Tempst,P., Bird,A. and Reinberg,D. (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev., 13, 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Choo Y., Sanchez-Garcia,I. and Klug,A. (1994) In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence. Nature, 372, 642–645. [DOI] [PubMed] [Google Scholar]
- 23.Smith D.B. (1993) Purification of glutathione S-transferase fusion proteins. Methods Mol. Cell. Biol., 4, 220–239. [Google Scholar]
- 24.Taylor C., Ford,K.G., Connolly,B.A. and Hornby,D.P. (1993) Determination of the order of addition of substrates to MspI DNA methyltransferase using a novel mechanism-based inhibitor. Biochem. J., 291, 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ford K., Taylor,C., Connolly,B. and Hornby,D. (1993) Effects of co-factor and deoxycytidine substituted oligodeoxynucleotides upon sequence-specific interactions between MspI DNA methyltransferase and DNA. J. Mol. Biol., 230, 779–786. [DOI] [PubMed] [Google Scholar]
- 26.Hurd P.J., Whitmarsh,A.J., Baldwin,G.S., Kelly,S.M., Waltho,J.P., Price,N.C., Connolly,B.A. and Hornby,D.P. (1999) Mechanism-based inhibition of C5-cytosine DNA methyltransferases by 2-H pyrimidinone. J. Mol. Biol., 286, 389–401. [DOI] [PubMed] [Google Scholar]
- 27.McNamara A.R. and Ford,K.G. (2000) A novel four zinc-finger protein targeted against p190BcrAbl fusion oncogene cDNA: utilisation of zinc-finger recognition codes. Nucleic Acids Res., 28, 4865–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pomerantz J.L., Sharp,P.A. and Pabo,C.O. (1995) Structure based design of transcription factors. Science, 267, 93–96. [DOI] [PubMed] [Google Scholar]
- 29.Li L. and Chandrasegaran,S. (1993) Alteration of the cleavage distance of FokI restriction endonuclease by insertion mutagenesis. Proc. Natl Acad. Sci. USA, 90, 2764–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J.-S., Kim,J., Cepek,K.L., Sharp,P.A. and Pabo,C.O. (1997) Design of TATA box-binding protein/zinc-finger fusions for targeted regulation of gene expression. Proc. Natl Acad. Sci. USA, 94, 3616–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Y.-G., Kim,P.S., Herbert,A. and Rich,A. (1997) Construction of a Z-DNA specific restriction endonuclease. Proc. Natl Acad. Sci. USA, 94, 12875–12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J.-S. and Pabo,C.O. (1998) Getting a handhold on DNA: design of poly-zinc finger proteins with femtomolar dissociation constants. Proc. Natl Acad. Sci. USA, 95, 2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pomerantz J.L., Sharp,A.P. and Pabo,C.O. (1995) Structure-based design of transcription factors. Science, 267, 93–96. [DOI] [PubMed] [Google Scholar]
- 34.Choo Y., Castellanos,A., Garcia-Hernandez,B., Sanchez-Garcia,I. and Klug,A. (1997) Promoter-specific activation of gene expression directed by bacteriophage-selected zinc-fingers. J. Mol. Biol., 273, 525–532. [DOI] [PubMed] [Google Scholar]