Abstract
Peptides corresponding to the C-terminal heptad repeat of HIV-1 gp41 (C-peptides) are potent inhibitors of HIV-1 entry into cells. Their mechanism of inhibition involves binding in a helical conformation to the central coiled coil of HIV-1 gp41 in a dominant–negative manner. Short C-peptides, however, have low binding affinity for gp41 and poor inhibitory activity, which creates an obstacle to the development of small drug-like C-peptides. To improve the inhibitory potency of short C-peptides that target the hydrophobic pocket region of gp41, we use two strategies to stabilize the C-peptide helix: chemical crosslinking and substitution with unnatural helix-favoring amino acids. In this study, the short linear peptide shows no significant inhibitory activity, but a constrained peptide (C14linkmid) inhibits cell–cell fusion at micromolar potency. Structural studies confirm that the constrained peptides bind to the gp41 hydrophobic pocket. Calorimetry reveals that, of the peptides analyzed, the most potent are those that best balance the changes in binding enthalpy and entropy, and surprisingly not those with the highest helical propensity as measured by circular dichroism spectroscopy. Our study reveals the thermodynamic basis of inhibition of an HIV C-peptide, demonstrates the utility of constraining methods for a short antiviral peptide inhibitor, and has implications for the future design of constrained peptides.
Peptides corresponding to the C-terminal heptad repeat of HIV-1 gp41 (C-peptides) are potent inhibitors of HIV-1 infection in vitro (1, 2) and in vivo (3). Previous work suggests C-peptides, when added exogenously, inhibit HIV-1 entry into cells by binding to the trimeric coiled coil formed in a fusion intermediate by the N-terminal heptad repeat (N-region) of HIV-1 gp41 (4–8). This binding event prevents the endogenous C-terminal heptad repeat (C-region) of gp41 from binding to the N-region, thereby blocking the formation of the six-helix bundle necessary to juxtapose and fuse the viral and cellular membranes.
When bound to the gp41 N-region, the C-peptides are α helical. In isolation, however, the C-peptides are in a random coil configuration (9). The C-peptide-binding event exacts a large energetic penalty due to the loss of conformational entropy, going from an unstructured ensemble of many configurations to a fixed helical conformation. Even for the relatively weak helix–coil transition (10), this penalty can be substantial (11). The entropic penalty is especially significant for short C-peptides, which bind with small interfaces and therefore gain less enthalpy. For example, in two separate studies, a 19-residue C-peptide shows poor inhibitory activity (12), and a 14-residue C-peptide shows no measurable activity (13). The low inhibitory potency of short C-peptides remains an impediment to developing a small orally bioavailable HIV-1 entry inhibitor.
One method for reducing the loss of binding energy is to stabilize the helical conformation of the C-peptide in the unbound state. Stabilizing the C-peptide helix decreases the number of nonhelical conformations available to the peptide, thereby reducing the loss of conformational entropy on binding to gp41. Many helix-stabilizing methods have been described (10, 14). Two strategies explored in this report are the use of unnatural helix-favoring amino acids and chemical crosslinkers.
For helix-favoring amino acids, we focus on α-aminoisobutyric acid (Aib), a well studied unnatural amino acid that contains two methyl groups attached to the α carbon (15). Its helical propensity matches that of Ala (16), and its α,α disubstituted structure restricts its conformation to α and 310 helices. Substitution of Aib into a peptide sequence can dramatically increase its helical propensity. An alternative helix-stabilizing strategy is to covalently link the side chains of two residues separated in sequence but spatially close in a helix. Of the many crosslinkers studied (10), we use a crosslinker that connects two glutamic acid residues at (i, i + 7) spacing via an α,ω diaminoalkane group (17). The diaminoalkane crosslinker was shown to be effective in helix stabilization compared with other crosslinkers. Although both strategies are chemically accessible and effective in helix stabilization, previous studies have focused on model peptides and relied mostly on circular dichroism (CD) and NMR spectroscopy to assess helical content (10). There are surprisingly few demonstrations of such chemistry being applied to peptide inhibitors in a biological setting (13, 18).
We report here the use of constraining methods to improve the inhibitory potency of a 14-residue C-peptide (called C14) that targets the hydrophobic pocket region of HIV-1 gp41. The hydrophobic pocket is an attractive drug target, because it may be less prone to drug-resistant mutations (2, 4, 19, 20). Furthermore, peptides selected by phage display to bind to the hydrophobic pocket effectively inhibit HIV-1 entry into cells (19). As such, short C-peptides that target the hydrophobic target would be attractive starting points for the development of a new class of small anti-HIV peptides. However, in previous studies, short C-peptides corresponding to the pocket-binding region failed to inhibit HIV-1 entry (12, 13), most likely due to weak binding to the target. Thus, proper helix stabilization of C14 may be important for obtaining inhibitory activity. The challenge is made greater here than in model systems, because the C14 sequence does not have a high intrinsic helical propensity.
We explore the effectiveness of the Aib and crosslinker strategies in promoting the inhibitory activity of a C14 peptide that targets the HIV-1 gp41 hydrophobic pocket, and investigate their mechanism of action by structural studies and energetic measurements.
Materials and Methods
Peptide Synthesis and Purification.
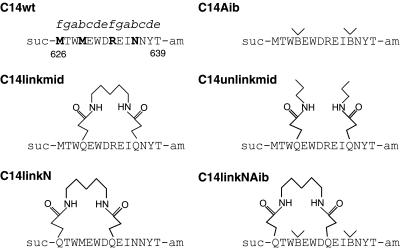
Peptides C14wt and C14Aib were synthesized by standard Fmoc chemistry with succinylated N termini and amidated C termini (sequences shown in Fig. 1). The purity and mass of the two peptides were verified by liquid chromatography–mass spectroscopy (Finnigan-Mat, San Jose, CA; LCQ). Peptides C14linkmid, C14linkN, and C14linkNAib were synthesized essentially as described (21). However, before forming the “lock” on resin, the peptide N termini were blocked by coupling to succinic acid mono t-butyl ester. Deprotection and purification were as described (21). All peptides eluted as single peaks from C18 HPLC columns (>95% purity); identities were confirmed by electrospray mass spectrometry. To synthesize C14unlinkmid, Fmoc-l-Glu α-Ο-t-butyl ester (Nova Biochem) was activated with one equivalent of carbonyldiimidazole in acetonitrile, then coupled to n-propyl amine. After removal of solvent and chromatography on silica gel (1:1 ethyl acetate/hexanes), the amide was treated with trifluoroacetic acid to remove the t-butyl protecting group. The resulting Fmoc amino acid was lyophilized from water then used directly for synthesis of C14unlinkmid.
Fig 1.
Peptides used in this study: C14wt (wild-type sequence); C14Aib (Met-629 and Asn-636 mutated to Aib); C14linkmid (crosslinked in the middle of the peptide at positions 629 and 636); C14unlinkmid (N-ɛ-propylglutamine at positions 629 and 636 to mimic the hydrophobicity of the crosslinker); C14linkN (crosslinked at the N terminus at positions 626 and 633); and C14linkNAib (crosslinked at the N terminus and Aib substitutions in the middle of the peptide). “B” denotes Aib residue, “suc” denotes succinimide, and “am” denotes amide. The amino acids mutated in C14wt are shown in bold. The heptad position according to the HIV-1 gp41 sequence and the residue number are shown, respectively, above and below the C14wt peptide sequence.
Cell–Cell Fusion Assay.
Inhibition of cell–cell fusion (syncytia formation) was assayed by coculturing 3 × 104 Chinese hamster ovary (CHO) cells expressing HXB2 envelope, tat, and rev with 5 × 104 HeLa cells expressing CD4 and long terminal repeat-driven β-galactosidase (19). The cells were mixed in the presence of varying concentrations of C14 peptides. Peptide stocks were dissolved in dimethyl sulfoxide and their concentrations determined by absorbance at 280 nm in 6 M GuHCl (22). The final concentration of dimethyl sulfoxide in tissue media was 1% in all experiments. The CHO and HeLa cells were incubated in the presence of the C14 peptides for 20 h and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactoside to detect syncytia, which were visualized by microscopy and counted manually. The IC50, the concentration needed to inhibit 50% of cell–cell fusion, was calculated from fitting the data to the equation, y = k/(1 + [peptide]/IC50), where y = number of syncytia, and k is a scaling constant.
NMR Spectroscopy.
1H NMR experiments were performed by using a Bruker (Billerica, MA) AMX spectrometer operating at 500 MHz and 25°C. Data were processed with Felix 98.0 (Molecular Simulations, Waltham, MA) on Silicon Graphics (Mountain View, CA) computers, and all spectra were referenced to 2,2-dimethyl-2-silapentane-5-sufonic acid. Complexes of C14 peptides with IQN17 were of 1:1 stoichiometry, with final solute concentrations of each component ranging from 0.5 to 1.0 mM, depending on solubility. All samples were dissolved in 100 mM NaCl/50 mM sodium phosphate (pH 7.5); the buffers used were >99.7% D2O to remove overlapping resonances from solvent-exchangeable backbone and side-chain protons. Some samples that contained a complex of C14 peptide with IQN17 displayed slight precipitation, which was not observed for the separate IQN17 and C-peptide components.
X-Ray Crystallography.
Equimolar amounts of C14linkmid and IQN17 were mixed to reach a concentration of 1.2 mM. The complex was crystallized by mixing 1 μl of the protein solution with 1 μl of 16% isopropanol/0.1 M Tris, pH 8.6/1 M (NH4)2SO4 by using the hanging drop method. Before data collection, the crystal was flash-frozen with 20% ethylene glycol added as cryoprotectant. A native data set was collected at the National Synchrotron Light Source at Brookhaven National Laboratory (beam X4A) at 1.9-Å resolution. The data were processed by using the programs denzo and scalepack (23). The crystals belong to the space group P63 with unit cell dimensions of a = b = 38.4 Å, c = 169.7 Å. The structure was solved by using the molecular replacement method with the program amore (24) and a resolution range of 8–2.5 Å. The structure of unbound IQN17 (V.N.M. and P.S.K., unpublished data) and a model of the last 12 residues of C14linkmid bound to the hydrophobic pocket [in the same conformation as C34 in the gp41 core (4)] was used as a search model. There were two monomers in the asymmetric unit, with the 3-fold axis of the IQN17 trimer coinciding with the crystallographic c-axis. Interpretation of the electron density map and model building was done with the program o (25). The structure was subsequently refined with cns (26) by using a twinning fraction of 0.50 (27) (details of the twinning are published as supporting information on the PNAS web site, www.pnas.org).
In addition to electron density maps calculated with coefficients 2Fo − Fc and Fo − Fc, composite omit maps were used to minimize model bias. Refinement of temperature factors, cycles of conjugate gradient minimization, and cycles of rebuilding were performed. The final model consists of all residues of both peptides except for the two N-terminal residues of C14linkmid. Structural analysis with procheck (28) shows that 97.1% of residues occupy the most favored regions of the Ramachandran plot, with the remaining residues occupying allowed regions. Data collection and refinement statistics are shown in Table 2.
Table 2.
Data collection and refinement statistics
| Data collection and processing | |
| Resolution range, Å | 20.0–1.86 |
| Total reflections | 59,821 |
| Unique reflections | 11,190 |
| Completeness, % | 94.3 (80.2) |
| Rsym, % | 4.4 (15.0) |
| Average I/σ | 13.6 (7.2) |
| Refinement statistics | |
| Resolution range, Å | 20.0–1.86 |
| Reflections (working/test) | 10,090/687 |
| Number of nonhydrogen protein atoms | 1,016 |
| Number of water molecules | 77 |
| Rcryst, % | 20.8 (29.7) |
| Rfree, % | 24.3 (27.1) |
| Average protein B, Å2 | 36.0 |
| rms deviation bonds, Å | 0.009 |
| rms deviation angles, ° | 1.0 |
Values in parentheses correspond to the highest-resolution shell (1.94–1.86 Å).
Rsym = ΣΣj|Ij − 〈I〉|/ΣΣ|〈I〉|, where Ij is the recorded intensity of the reflection j, and 〈I〉 is the mean recorded intensity over multiple recordings.
I is the integrated intensity of a measured reflection, and σ is the estimated error in the measurement.
413 reflections are rejected because their twin-related reflections are not observed (26).
Rcryst, Rfree = Σ∥Fo − Fc∥/|F|, where the Rcryst and Rfree are calculated using the working and free reflection sets, respectively.
CD Spectroscopy.
CD spectra were measured on an Aviv (Lakewood, NJ) 62DS spectrapolarimeter at 25°C in PBS (10 mM sodium phosphate/150 mM NaCl, pH 7.0) and 50 μM total peptide concentration. CD spectra were also recorded in the presence of 0–50% (vol/vol) trifluoroethanol (TFE). In these samples, the final buffer and salt concentrations were the same, and the CD spectrum of the appropriate solvent reference was subtracted from the peptide spectrum.
Isothermal Titration Calorimetry.
Isothermal titration calorimetry was performed on a VP-ITC microcalorimeter (MicroCal, Amherst, MA). All solutions were degassed before the experiment. Experiments were carried out in PBS at 37°C. C14 peptides ranging from 0.6 to 0.8 mM were injected in 6 μl of increments into the reaction cell (1.49-ml volume) containing IQN17 ranging from 60 to 80 μM. The titration of IQN17 with C14 peptides was carried out until a C14:IQN17 concentration ratio of about 2.5:1 was reached. The heat of dilution of the C14 peptides was measured by making additional injections after complete saturation. This value was subtracted from the heat of reaction to obtain the heat of binding. The data were fit to a single binding site model by using the Microcal (Amherst, MA) origin analysis software to obtain the thermodynamic parameters.
Results
C14 Peptides and Inhibitory Activities.
We used two strategies, substitution with Aib and a diaminoalkane crosslinker, intended to stabilize the helical conformation of a 14-residue C-peptide whose sequence corresponds to HIV-1 gp41 beginning with Met-626 and ending with Thr-639 (using the gp160 numbering from the HXB2 strain) (Fig. 1). On the basis of the crystal structure of the HIV-1 gp41 core, this peptide is expected to target the gp41 hydrophobic pocket (2, 4, 6, 7). We produced six peptides of different modifications at different amino acid positions for our analysis (Fig. 1).
We used a cell–cell fusion experiment to assay the biological activity of the peptides (19). As shown in Table 1, of the six peptides we produced, C14linkmid is the most potent inhibitor of syncytia formation (IC50 = 35 μM), followed by C14Aib (IC50 = 144 μM). C14linkmid is also the only peptide that shows measurable inhibitory activity in a luciferase-based viral infectivity assay (2), although at a higher IC50 (approximately 500 μM; data not shown) than in the cell–cell fusion assay. C14wt and C14unlinkmid show very weak inhibition of cell–cell fusion with IC50 values exceeding 500 μM, and C14linkN and C14linkNAib show no inhibitory activity.
Table 1.
Inhibition of cell–cell fusion by C14 peptides
| Peptide | IC50 for cell–cell fusion, μM |
|---|---|
| C14linkmid | 35 |
| C14Aib | 144 |
| C14wt | >500 |
| C14unlinkmid | >500 |
| C14linkN | No activity |
| C14linkNAib | No activity |
C14wt and C14unlinkmid display very weak inhibition; the lower boundaries of their IC50 values are estimated.
Structural Studies.
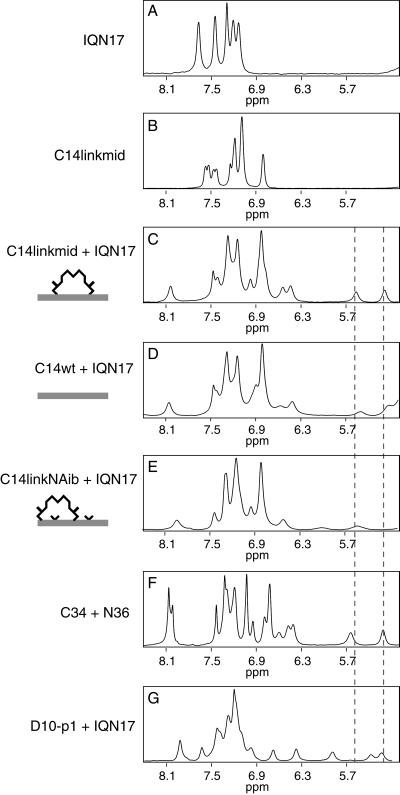
To confirm that peptides bind to the hydrophobic pocket, NMR spectroscopy was performed on C14linkmid, C14wt, and C14linkNAib in the presence of IQN17 (Fig. 2). IQN17 is a soluble peptide that contains the HIV-1 gp41 hydrophobic pocket fused to a trimeric coiled coil derived from the GCN4 leucine zipper (19). The NMR spectra of IQN17 and all three C14 peptides in isolation reveal resonances in the aromatic region but not in the 5.0- to 6.5-ppm region, termed the “fingerprint” region (see Fig. 2 A and B for representative spectra). When mixed, however, C14linkmid/IQN17 produces distinct resonances in the fingerprint region (Fig. 2C), corresponding to Trp residues in the hydrophobic pocket that experience a significant change in the magnetic environment on formation of the complex (19). This region of the NMR spectrum is sensitive even to small rotations of the Trp side chains because of the strong ring-current effect. The resonances are virtually identical with those from the gp41 core (Fig. 2F) but different from those of IQN17 with a D-peptide (Fig. 2G). This comparison suggests that the detailed structure of the hydrophobic pocket for C14linkmid/IQN17 resembles that of the gp41 core (4, 6) and not of the D-peptide–IQN17 complex (19).
Fig 2.
One-dimensional 1H-NMR spectra of various peptides bound to the gp41 hydrophobic pocket presented by IQN17. The dashed lines in the fingerprint region highlight the distinct resonances of Trp residues in the hydrophobic pocket. A–G are described in Results. For F and G, the peptides C34 and N36 (4) and D10-p1 and IQN17 (19) have been previously described.
Even though C14wt is a weak biological inhibitor, the NMR spectrum of C14wt/IQN17, under high peptide concentrations, is also similar in the fingerprint region to that of the gp41 core (Fig. 2D). In contrast, C14linkNAib, which exhibits no biological inhibitory activity, produces weak resonances with different chemical shifts in the fingerprint region (Fig. 2E), which may be due to chemical shift averaging or a different conformation of the Trp residues in the hydrophobic pocket.
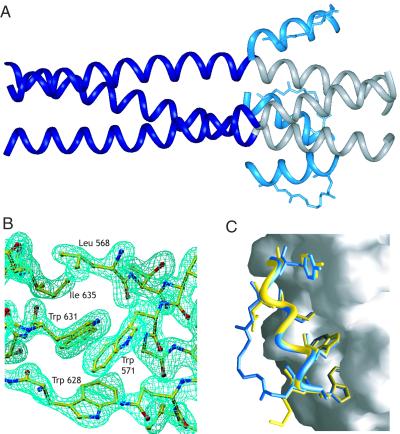
To more precisely analyze the structure of the constrained peptides, we obtained a 1.9-Å resolution x-ray crystal structure of the most biologically potent peptide, the crosslinked peptide C14linkmid, bound to IQN17 (Table 2, Fig. 3). Although the crystal showed merohedral twinning, the structure was refined with excellent statistics by using detwinning methods (see Materials and Methods and supporting information on the PNAS web site). As expected, IQN17 forms a continuous and regular three-stranded coiled coil, and the crosslinked peptide binds only to the gp41 region (Fig. 3A). The structure reveals that C14linkmid binds to the HIV-1 gp41 hydrophobic pocket in essentially the same conformation as the pocket-binding region of a linear C-peptide, C34 (Fig. 3 B and C). In particular, the orientations of the side chains in the hydrophobic pocket (Trp-628, Trp-631, Ile-635, Leu-568, Trp-571) are virtually identical in the two structures. The main chain of C14linkmid is also similar to C34 (rms deviation of 0.4 Å), demonstrating that the crosslink imparts no detectable distortion on the backbone of the C14 peptide in the bound conformation. Moreover, the crosslinker exhibits a high B-factor (60–67 Å2), suggesting it is either unstructured or adopts multiple discrete conformations.
Fig 3.
Crystal structure of the crosslinked peptide, C14linkmid, bound to the HIV-1 gp41 hydrophobic pocket. (A) Ribbon representation of C14linkmid bound to IQN17. For IQN17, the GCN4-derived portion is shaded dark blue, and the gp41 portion is shaded gray. C14linkmid is shaded light blue. The crosslinker of C14linkmid is also shown. In this crystal form, the three C14linkmid/IQN17 chains are related by crystallographic symmetry. (B) A 2Fo − Fc electron density map of the hydrophobic pocket. The map is contoured at 2.0 σ. (C) An overlay of C14linkmid (light blue) and C34 (yellow) bound to the hydrophobic pocket. The structure of C34, taken from the gp41 core (PDB ID code ) (4), is superimposed onto the C14linkmid/IQN17 structure. For the two C-peptides, the side chains of residues in the hydrophobic pocket (Trp-628, Trp-631, Ile-635, and Tyr-638) and at the crosslinking positions (629 and 636) are shown. IQN17 is shown as a surface representation. The figure is prepared by using the program GRASP (39).
Mechanism of Helix Stabilization.
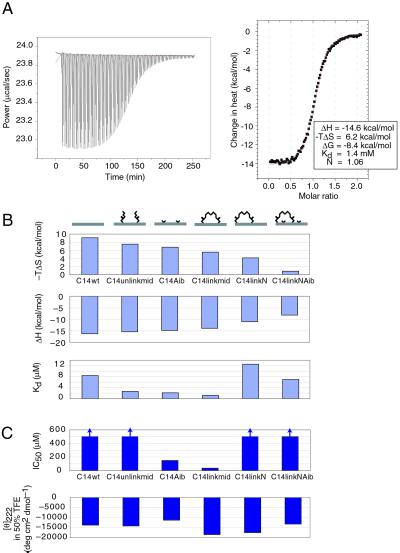
CD spectroscopy indicated that none of the peptides in isolation were significantly helical, exhibiting [θ]222 values of –5,000 to –10,000 deg⋅cm2⋅dmol−1 [theoretical value of a 100% helix is approximately –30,700 deg⋅cm2⋅dmol−1 for a 14-residue peptide (29)]. We also examined the helical propensity of the C14 peptides by titration with TFE (30), a helix-stabilizing solvent, and followed their helicity by CD spectroscopy (see supporting information on the PNAS web site for CD spectra of C14linkmid in different TFE concentrations). Even at 50% (vol/vol) TFE, the C14 peptides are poorly helical (Fig. 4C). Although C14linkmid, the most potent inhibitor, has the highest helical content in the presence of TFE, the helical propensities of the other peptides do not correlate with their inhibitory potencies or structural modifications.
Fig 4.
Isothermal titration calorimetry of IQN17 with C14 peptides. Experiments were carried out in PBS at 37°C. (A) Titration of IQN17 with C14linkmid. The thermodynamic parameters after fitting to a two-state transition are given (N is the molar ratio of titration). (B) Thermodynamic parameters of binding for C14 peptides: binding entropy, binding enthalpy, and dissociation constant. Binding entropies are given as (–TΔS), which are expressed in free energy units (kcal/mol) and can therefore be directly compared with ΔH. (C) Inhibitory activity for cell–cell fusion (IC50) and helical propensity [[θ]222 by CD spectroscopy at 50% (vol/vol) TFE]. For the C14 peptides showing poor inhibition, the IC50 values of 500 μM in this graph are estimated lower boundaries as indicated with the arrows (see Table 1).
To investigate the thermodynamics of binding to the hydrophobic pocket, isothermal titration calorimetry was performed on the binding of C14 peptides to IQN17 at 37°C. All titrations are consistent with two-state binding (Fig. 4A), and all C14 peptides bind to IQN17 in the expected ratio of 1:1 (three peptides per IQN17 trimer; the molar ratio of titration ranges from 0.88 to 1.09). C14linkmid, the most potent inhibitor, exhibits the strongest binding to IQN17 with a Kd of 1.2 μM. Next in binding affinity are C14Aib and C14unlinkmid with Kd values of 2.1 and 2.6 μM, respectively. The weakest binders are C14linkNAib, C14wt, and C14linkN, which have Kd values of 7.0, 8.4, and 12.6 μM, respectively. As a general trend, the peptides that exhibit a large gain of binding enthalpy also exhibit a large loss of binding entropy, and vice versa (Fig. 4B). Interestingly, peptides at the two extremes show relatively weak binding, whereas peptides with the strongest binding affinities are those in the middle of the enthalpy–entropy spectrum.
Discussion
Efficacy of the Modifications.
Both the crosslinking and Aib modifications confer significant biological activity to a HIV-1 C-peptide that would otherwise exhibit weak inhibition. Compared with C14wt, C14linkmid binds ≈7 times more tightly to the hydrophobic pocket and inhibits cell–cell fusion ≈15 times more potently, whereas the effects of C14Aib are weaker (gains of ≈4-fold activity in both categories) (Table 1 and Fig. 4). [Note that in another study, a crosslinked peptide similar to C14linkmid showed no inhibitory activity (13)].
In a previous study, a group of D-peptides was selected by phage display to bind to the gp41 hydrophobic pocket and inhibited cell–cell fusion with IC50 values in the range of 3.6–130 μM (19). An advantage of the current study is that it circumvents the need for selection experiments, provided it can constrain natural peptide sequences into effective biological inhibitors. In the current study, the Kd of the most potent constrained C14 peptide matches that of the most potent D-peptide (31) (1.2 μM for both C14linkmid and D10-p5–2K), and the IC50 values of the two constrained C14 peptides are in the range of activities of the D-peptides. The thermodynamics of binding are also similar for both groups of peptides. The binding of both the C14 peptides and D-peptides to the hydrophobic pocket is driven by a favorable change in enthalpy, which is approximately –15 kcal/mol in both cases (31). Overall, the efficacy of the constrained peptides approaches that of the D-peptides.
For the crosslinked C14 peptides, the efficacy of the crosslinker depends on its position in the peptide sequence. In particular, C14 peptides crosslinked at the N terminus (C14linkN and C14linkNAib) do not inhibit cell–cell fusion, whereas the C14 peptide crosslinked in the middle of the peptide (C14linkmid) inhibits cell–cell fusion at micromolar potency. Calorimetry shows that the N-terminal crosslinker results in entropically more favorable binding but incurs a greater enthalpic penalty and results in a lower binding affinity. The low binding affinity and small gain of enthalpy may stem from the N-terminal linker constraining the key pocket-binding residues Trp-628 and Trp-631 into an incorrect conformation (2) (Trp-628 is outside the linker of C14linkmid). The increased entropic stabilization may stem from the ability of a crosslinker at the N terminus to cap the helix; the stabilization of a helix at the N terminus, which is generally more mobile than the middle of a helix, may reduce more degrees of freedom than it would in the middle of the helix, a phenomenon observed previously in a crosslinker (32).
Binding Affinity to the gp41 Hydrophobic Pocket vs. Inhibitory Potency.
In general, we observe a good correlation between binding affinity to the gp41 hydrophobic pocket and cell–cell fusion inhibitory activity (Fig. 4 B and C). Nonetheless, the IC50 in the cell–cell fusion assay is consistently >10-fold less potent than the Kd measurement in solution [a difference that holds true also for D-peptides (19, 31)]. Additional factors, other than binding affinity to the target, may be necessary for blocking viral entry.
C14unlinkmid Binds Strongly to the Hydrophobic Pocket.
An exception to the general trend is C14unlinkmid, which binds to the hydrophobic pocket relatively strongly but does not inhibit cell–cell fusion. This discrepancy suggests factors exist that prevent a good binder from being a good biological inhibitor. For example, a modification may increase nonspecific aggregation to serum proteins or proteins on the virion surface, or it may alter the proteolytic sensitivity of the peptide.
C14unlinkmid binds strongly to the hydrophobic pocket despite the severing of the covalent crosslinker. The high binding affinity stems from amino acid substitutions with N-ɛ-propylglutamine, whose chemical structure resembles amino acids containing a straight-chain aliphatic group such as Leu, Met, Lys, and Arg. Interestingly, all these amino acids highly favor the helical conformation (10, 16). Thus, the mechanism of stabilization of C14unlinkmid may be similar to that of C14Aib, which involves substitutions with helix-favoring amino acids. The effect is expected to be weaker than C14Aib, as confirmed by measurements of binding entropies and binding constants (Fig. 4B), because Aib is a strong helix-favoring amino acid. Overall, the properties of C14unlinkmid suggest the existence of two components in the mechanism of helix stabilization by the crosslinker: first, via an increased helical propensity of the two substituted amino acids; and second, further stabilization from formation of the covalent bond.
Changes in Entropy and Enthalpy.
Across the C14 peptides in our study, we observe a compensation of changes in the entropy of binding (ΔΔS) with changes in the enthalpy of binding (ΔΔH) (Fig. 4B). (ΔΔS and ΔΔH refer to the differences in ΔS and ΔH of a C14 peptide compared with the ΔS and ΔH of the C14wt peptide.) The compensation of changes in enthalpy and entropy is not perfect, resulting in a variation of binding free energies. The peptides that bind to the hydrophobic pocket with the highest affinity (C14linkmid, C14Aib, and C14unlinkmid) exhibit only modest changes in enthalpy and entropy.
In biomolecular interactions, changes in enthalpy and entropy are often similar in magnitude but opposite in their effects on free energy (31, 33, 34). One explanation for the observed enthalpy–entropy compensation of the C14 peptides is that the modifications change the binding enthalpy in a way that is not easily rationalized structurally (35), with the changes in entropy being a direct consequence via the typical compensation mechanism (34). However, we note that, significantly, the trend of entropic stabilization (ΔΔS) is not random but follows strikingly well the stabilization expected from the severity of the constraint (Fig. 4B). This observation suggests that a major component of the variation in ΔΔS is due to structural modification of the C14 peptides that result in differences in conformational entropy.
There also exists a trend between the changes in binding enthalpy (ΔΔH) and the severity of the constraint. Enthalpy can be lost in the bound state if the modifications directly or indirectly change the van der Waals and hydrogen bond contacts in the binding interface (35). For the moderately constrained peptides (C14unlinkmid, C14Aib, and C14linkmid), only small losses of binding enthalpy are observed, and the x-ray crystal structure confirms there are no gross structural differences in the bound conformation of a moderately constrained peptide compared with a linear peptide. For the highly constrained peptides (C14linkN and C14linkNAib), a larger loss of binding enthalpy is observed, which may reflect a larger difference in the structure of the bound state (35) due to conformational strain imparted by the modifications (as shown by NMR spectroscopy for C14linkNAib in Fig. 2E). There exists no trend between the conformations of the unbound states and binding enthalpy (Fig. 4 B and C).
Helical Content vs. Inhibitory Potency.
Helix-stabilization strategies are judged to be effective if they confer a large degree of helicity to model peptides as measured by CD or NMR spectroscopy (10). Their effect on biologically relevant peptides of moderate helical propensities has been less tested, and a direct correlation between induced helicity and biological activity is often lacking (one demonstration is ref 13). In the present study, CD measurements show that none of the C14 peptides, constrained or linear, are helical at room temperature (Fig. 4C). Therefore, neither the crosslinker nor Aib confer significant helical content to C14, a short peptide of moderate intrinsic helical propensity. Furthermore, a general correlation between helical content or helical propensity (as measured by TFE-induced helicity) and biological activity does not exist, suggesting that for the C14 peptides, measurements by CD spectroscopy are not a reliable indicator of the effectiveness of the constraining method.
Future Design of Helical Inhibitors.
This study has several implications for the future design of constrained, biologically active peptides (18, 21, 36–38). In particular, a focus in the development of helix-stabilization methods has been to maximize the helical content of short peptides. In this study, the peptides with the greatest inhibitory potency and binding affinity to the hydrophobic pocket (C14linkmid and C14Aib) did not exhibit significant helical content or helical propensity by CD spectroscopy. In fact, the highly constrained peptides had poor inhibitory potencies, possibly because they were constrained into an inactive conformation. For C14linkmid, the slackness of the crosslinker was also apparent in the crystal structure, because it failed to adopt a well-defined conformation. Despite the modest structural effects, however, the modifications for C14linkmid and C14Aib translated into a large reduction in conformational entropy and a significant increase in biological activity. Therefore, rather than constraining a peptide into a rigid helix, for future designs one should also consider the use of looser linkers, which can reduce the number of degrees of freedom in the unbound state without perturbing the helical conformation of the bound state.
Finally, the strategies used here are immediately applicable to the design of helical inhibitors against other viruses that share the trimer-of-hairpins motif for membrane fusion, especially those for which analogous helical C-peptides have been shown to inhibit viral entry (8).
Supplementary Material
Acknowledgments
We thank Ben Sanford and Michael Burgess for peptide synthesis; Leslie Gaffney for help with the manuscript; Dr. Debra Eckert for initiating the project; Dr. David Akey for help with x-ray crystallography; and Drs. Amy Keating and Ben Chen for helpful discussions. S.K.S. acknowledges financial support from a Howard Hughes Medical Institute Predoctoral Fellowship and National Science and Engineering Council of Canada Postgraduate Scholarships. P.A.C. acknowledges support from National Institutes of Health National Research Service Award Grant 5 F32 GM19917-0. This research was supported in part by Grant GM44162 from the National Institutes of Health.
Abbreviations
CD, circular dichroism
TFE, trifluoroethanol
N-region, N-terminal heptad repeat
C-region, C-terminal heptad repeat
Aib, α-aminoisobutyric acid
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code ).
References
- 1.Wild C. T., Shugars, D. C., Greenwell, T. K., McDanal, C. B. & Matthews, T. J. (1994) Proc. Natl. Acad. Sci. USA 91, 9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan D. C., Chutkowski, C. T. & Kim, P. S. (1998) Proc. Natl. Acad. Sci. USA 95, 15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilby J. M., Hopkins, S., Venetta, T. M., DiMassimo, B., Cloud, G. A., Lee, J. Y., Alldredge, L., Hunter, E., Lambert, D., Bolognesi, D., et al. (1998) Nat. Med. 4, 1302-1307. [DOI] [PubMed] [Google Scholar]
- 4.Chan D. C., Fass, D., Berger, J. M. & Kim, P. S. (1997) Cell 89, 263-273. [DOI] [PubMed] [Google Scholar]
- 5.Furuta R. A., Wild, C. T., Weng, Y. & Weiss, C. D. (1998) Nat. Struct. Biol. 5, 276-279. [DOI] [PubMed] [Google Scholar]
- 6.Weissenhorn W., Dessen, A., Harrison, S. C., Skehel, J. J. & Wiley, D. C. (1997) Nature 387, 426-430. [DOI] [PubMed] [Google Scholar]
- 7.Tan K., Liu, J., Wang, J., Shen, S. & Lu, M. (1997) Proc. Natl. Acad. Sci. USA 94, 12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckert D. M. & Kim, P. S. (2001) Annu. Rev. Biochem. 70, 777-810. [DOI] [PubMed] [Google Scholar]
- 9.Lu M., Blacklow, S. C. & Kim, P. S. (1995) Nat. Struct. Biol. 2, 1075-1082. [DOI] [PubMed] [Google Scholar]
- 10.Andrews M. J. I. & Tabor, A. B. (1999) Tetrahedron 55, 11711-11743. [Google Scholar]
- 11.D'Aquino J. A., Gomez, J., Hilser, V. J., Lee, K. H., Amzel, L. M. & Freire, E. (1996) Proteins 25, 143-156. [DOI] [PubMed] [Google Scholar]
- 12.Jin B. S., Ryu, J. R., Ahn, K. & Yu, Y. G. (2000) AIDS Res. Hum. Retroviruses 16, 1797-1804. [DOI] [PubMed] [Google Scholar]
- 13.Judice J. K., Tom, J. Y. K., Huang, W., Wrin, T., Vennari, J., Petropoulos, C. J. & McDowell, R. S. (1997) Proc. Natl. Acad. Sci. USA 94, 13426-13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeGrado W. F., Summa, C. M., Pavone, V., Nastri, F. & Lombardi, A. (1999) Annu. Rev. Biochem. 68, 779-819. [DOI] [PubMed] [Google Scholar]
- 15.Karle I. L. & Balaram, P. (1990) Biochemistry 29, 6747-6756. [DOI] [PubMed] [Google Scholar]
- 16.O'Neill K. T. & DeGrado, W. F. (1990) Science 250, 646-651. [DOI] [PubMed] [Google Scholar]
- 17.Phelan J. C., Skelton, N. J., Briasted, A. C. & McDowell, R. S. (1997) J. Am. Chem. Soc. 119, 455-460. [Google Scholar]
- 18.Cochran A. G. (2001) Curr. Opin. Chem. Biol. 5, 654-659. [DOI] [PubMed] [Google Scholar]
- 19.Eckert D. M., Malashkevich, V. N., Hong, L. H., Carr, P. A. & Kim, P. S. (1999) Cell 99, 103-115. [DOI] [PubMed] [Google Scholar]
- 20.Ferrer M., Kapoor, T. M., Strassmaier, T., Weissenhorn, W., Skehel, J. J., Oprian, D., Schreiber, S. L., Wiley, D. C. & Harrison, S. C. (1999) Nat. Struct. Biol. 6, 953-960. [DOI] [PubMed] [Google Scholar]
- 21.Skelton N. J., Chen, Y. M., Dubree, N., Quan, C., Jackson, D. Y., Cochran, A., Zobel, K., Deshayes, K., Baca, M., Pisabarro, M. T. & Lowman, H. B. (2001) Biochemistry 40, 8487-8498. [DOI] [PubMed] [Google Scholar]
- 22.Edelhoch H. (1967) Biochemistry 6, 1948-1954. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z., (1993) Oscillation Data Reduction Program (Science and Engineering Research Council, Daresbury Laboratory, Warrington, U.K.).
- 24.Navaza J. (2001) Acta Crystallogr. D 57, 1367-1372. [DOI] [PubMed] [Google Scholar]
- 25.Jones T. A., Zou, J. W., Cowan, S. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 26.Brunger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 27.Yeates T. O. (1997) Methods Enzymol. 276, 344-358. [PubMed] [Google Scholar]
- 28.Laskowski R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 29.Yang J. T., Wu, C. C. & Martinez, H. M. (1986) Methods Enzymol. 130, 208-269. [DOI] [PubMed] [Google Scholar]
- 30.Jasanoff A. & Fersht, A. R. (1994) Biochemistry 33, 2129-2135. [DOI] [PubMed] [Google Scholar]
- 31.Cole J. L. & Garsky, V. M. (2001) Biochemistry 40, 5633-5641. [DOI] [PubMed] [Google Scholar]
- 32.Kapurniotu A. & Taylor, J. W. (1995) J. Med. Chem. 38, 836-847. [DOI] [PubMed] [Google Scholar]
- 33.Cavanagh J. & Akke, M. (2000) Nat. Struct. Biol. 7, 11-13. [DOI] [PubMed] [Google Scholar]
- 34.Dunitz J. D. (1995) Chem. Biol. 2, 709-712. [DOI] [PubMed] [Google Scholar]
- 35.Vaughan C. K., Buckle, A. M. & Fersht, A. R. (1999) J. Mol. Biol. 286, 1487-1506. [DOI] [PubMed] [Google Scholar]
- 36.Zutshi R., Brickner, M. & Chmielewski, J. (1998) Curr. Opin. Chem. Biol. 2, 62-66. [DOI] [PubMed] [Google Scholar]
- 37.Peczuh M. W. & Hamilton, A. D. (2000) Chem. Rev. 100, 2479-2494. [DOI] [PubMed] [Google Scholar]
- 38.Cochran A. G. (2000) Chem. Biol. 7, R85-R94. [DOI] [PubMed] [Google Scholar]
- 39.Nicholls A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281-296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.