Abstract
The adult mouse gastric epithelium undergoes continuous renewal in discrete anatomic units. Lineage tracing studies have previously disclosed the morphologic features of gastric epithelial lineage progenitors (GEPs), including those of the presumptive multipotent stem cell. However, their molecular features have not been defined. Here, we present the results of an analysis of genes and pathways expressed in these cells. One hundred forty-seven transcripts enriched in GEPs were identified using an approach that did not require physical disruption of the stem cell niche. Real-time quantitative RT-PCR studies of laser capture microdissected cells retrieved from this niche confirmed enriched expression of a selected set of genes from the GEP list. An algorithm that allows quantitative comparisons of the functional relatedness of automatically annotated expression profiles showed that the GEP profile is similar to a dataset of genes that defines mouse hematopoietic stem cells, and distinct from the profiles of two differentiated GEP descendant lineages (parietal and zymogenic cell). Overall, our analysis revealed that growth factor response pathways are prominent in GEPs, with insulin-like growth factor appearing to play a key role. A substantial fraction of GEP transcripts encode products required for mRNA processing and cytoplasmic localization, including numerous homologs of Drosophila genes (e.g., Y14, staufen, mago nashi) needed for axis formation during oogenesis. mRNA targeting proteins may help these epithelial progenitors establish differential communications with neighboring cells in their niche.
Keywords: stem cells, growth factor signaling, protein turnover, mRNA localization , bioinformatics
The mammalian gastrointestinal tract is lined by an epithelium that is constantly renewed. Although multipotent stem cells are known to fuel this renewal, the molecular properties of these cells are poorly understood.
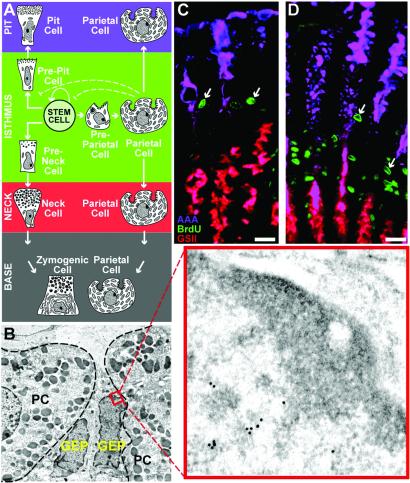
Tritiated thymidine/EM autoradiographic lineage tracing studies have delineated the morphological features of the stem cell niche in the adult mouse stomach (1). The glandular epithelium is composed of tubular invaginations termed gastric units. In the corpus (central region) of the stomach, each unit contains an average of ≈200 cells, representing three predominant lineages: pit, parietal, and zymogenic. The multipotent stem cell (undifferentiated granule-free progenitor) resides in the unit's isthmus (ref. 1; Fig. 1A). One of its committed daughters, the granule-free prepit cell precursor, produces mucus-secreting pit cells, which differentiate as they climb from the isthmus to the orifice of the unit (2). Another daughter, the granule-free preneck cell precursor, gives rise to pepsinogen-producing neck cells, which differentiate to zymogenic cells as they descend to the base of the unit (3). Unlike the pit and zymogenic lineages, acid-producing parietal cells (PCs) differentiate within the isthmus from granule-free preparietal cell progenitors and then migrate either up or down the unit (Fig. 1 A and B; ref. 4).
Fig 1.
Parietal cell ablation produces GEP amplification in tox176 mice. (A) Schematic representation of a gastric unit in the middle third of a normal adult mouse stomach. The unit contains four compartments: pit, isthmus, neck, and base. The multipotent stem cell in the isthmus gives rise to three principal epithelial lineages (pit, parietal, and zymogenic). Only PCs differentiate within the isthmal stem cell niche. They then undergo a bidirectional migration to the pit and base regions. (B) EM immunohistochemical study showing juxtaposition of two mitochondria-rich PCs and two granule-free GEPs in the isthmus of a normal adult mouse gastric unit. PCs are outlined by dashes. The boxed region in one GEP (and the higher power Inset) shows a portion of the nucleus labeled with goat anti-PCNA and 18-nm-diameter gold particle-conjugated donkey anti-goat Ig. (C and D) Multilabel study of a 16-week-old normal germ-free mouse (C) and an age-matched conventionally raised tox176 animal (D). Each mouse received an i.p. injection of BrdUrd 90 min before sacrifice. Purple, pit cells labeled with Alexafluor 647-AAA; red, neck cells tagged with biotinylated GSII and Alexafluor 594- streptavidin; green, isthmal S-phase progenitors detected with goat anti-BrdUrd and Alexafluor 488-donkey anti-goat Ig. Note marked expansion of S-phase cells (e.g., arrows) in PC-ablated tox176 gastric units. (Bars, 25 μm.)
We have shown that there is increased proliferation of isthmal gastric epithelial progenitors (GEPs) in adult transgenic mice with genetically engineered, mutant diphtheria toxin A fragment (tox176)-mediated ablation of PCs (Fig. 1 C and D; refs. 5 and 6). Analysis of tox176 pedigrees with a mosaic pattern of transgene expression showed that GEP amplification occurs only in units lacking PCs (6), suggesting that PCs produce locally acting factors that regulate GEP census.
We reasoned that the increase in GEPs in tox176 mice offered an opportunity to identify molecular regulators of GEP biology, including those that mediate interactions with PCs, without having to disrupt the isthmal niche where they reside. Here, we present the results of a “dissection-free” approach that used a three-way comparison of gene expression profiles in the intact stomachs of (i) normal adult mice where GEPs represent <3% of the total epithelial population; (ii) tox176 adult mice where GEPs constitute ≈20% of the total; and (iii) embryonic day 18 (E18) mice where >90% of the developing gastric epithelium is composed of GEPs (7). Genes whose expression was increased in both ii and iii relative to i formed the GEP dataset. Several generally applicable experimental and computational methods, ranging from a new application of laser capture microdissection to an algorithmic approach for comparing the functional features of entire gene expression profiles, were used to validate and extend these results. The 147-member GEP dataset has prominent representation of genes involved in insulin-like growth factor (IGF) signaling, in regulating protein turnover (ubiquitin/proteosomal, sumoylation, and neddylation), and in controlling RNA processing and localization.
Materials and Methods
Mice.
Conventionally raised FVB/N tox176 transgenic mice (6) were maintained in microisolator cages in a specified pathogen-free state. Nontransgenic, germ-free FVB/N mice were raised in plastic gnotobiotic isolators (8).
GeneChip Comparisons.
Whole stomachs were excised and RNA extracted (midi-RNeasy kit; Qiagen, Valencia, CA). Duplicate cRNA targets were prepared from pooled RNAs (see below). Each cRNA was hybridized to a set of Mu11K GeneChips (Affymetrix, Santa Clara, CA). Overall fluorescence across each GeneChip set was scaled to a target intensity of 150 and pairwise comparisons performed using Affymetrix microarray suite software (V.4.0).
SYBR-Green-Based Real-Time Quantitative RT-PCR (qRT-PCR).
Assays were performed in triplicate as described (9), using the gene-specific primers listed in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. Normalization was to 18S rRNA (ΔΔCT method; ref. 9).
Navigated Laser Capture Microdissection (n-LCM).
Stomachs were removed from tox176 mice (n = 4), flushed with PBS, and divided in half along the cephalocaudal axis, and each half-stomach was rinsed with OCT compound. Half-stomachs were placed in a cryomold, overlaid with OCT compound, and frozen (Cytocool II, Richard-Allan Scientific, Kalamazoo, MI). Cryosections (7 μm thick) were cut and processed using protocols described in ref. 9. Well oriented gastric units, containing a continuous column of epithelial cells from the base region to the tip of the pit region, were targeted for LCM by using the PixCell II system (Arcturus, Mountain View, CA; 7.5-μm-diameter laser spot) and CapSure HS LCM Caps (Arcturus). Epithelial cells were recovered using n-LCM (see below for details; total of ≈25,000 cells per compartment per experiment; material pooled from two mice per experiment; n = 3 independent experiments). “Navigation slides” were stained with horseradish peroxidase (HRP)-tagged Griffonia simplicifolia II (GSII) lectin and Vector VIP, then with HRP-Anguilla anguilla agglutinin (AAA) and Vector SG (Vector Laboratories). RNA was isolated from captured epithelial populations [PicoPure RNA Isolation Kit (Arcturus) with on-column DNase digestion].
Results and Discussion
Identifying Genes Expressed Preferentially in Gastric Epithelial Progenitors.
Quantitative light and EM microscopic studies of tox176 mice with complete PC ablation showed that by 6–8 weeks of age, GEPs account for 10% of the total gastric unit epithelial cell census, and by 20 weeks 25% (≈20-fold higher than in age-matched normal littermates; Fig. 1 C and D; ref. 5). Guided by the strategy outlined in the Introduction, we isolated RNA from the intact stomachs of conventionally raised 16-week-old tox176 mice (equal amounts of RNA pooled from five stomachs), age-matched normal germ-free mice (n = 4), and normal embryonic day 18 animals (n = 31). Because tox176 mice lose the acid barrier to microbial colonization, they inevitably develop a mild diffuse chronic gastritis associated with bacterial overgrowth. Because a comparison of gene expression in tox176 versus normal adult germ-free stomachs would likely yield a dataset substantially enriched for GEP-associated mRNAs but “contaminated” by immune response genes, we referenced the “tox176 versus normal” dataset to the “E18 versus normal” dataset (where E18 represents a GEP-enriched, gastritis-free state).
Duplicate cRNA targets, independently generated from each RNA, were used to probe GeneChips representing ≈11,000 mouse genes and EST clusters. Genes with enhanced expression in duplicate comparisons between tox176 and normal stomachs and in duplicate comparisons between E18 and normal stomachs were culled, yielding a list of 147 genes and 6 uncharacterized ESTs with quadruplicate “Increased” calls by GeneChip software (see Table 2, which is published as supporting information on the PNAS web site).
Initial Validation of the GEP Dataset.
The three RNAs used for the GeneChip comparisons were used as templates for qRT-PCR analysis of the expression of seven genes, selected from the GEP dataset based on their known roles in regulating proliferation, differentiation, and polarity in other systems. They encoded proliferating cell nuclear antigen (PCNA; ref. 10), retinoblastoma binding protein 7 (together with RbAP48 increases histone acetyltransferase activity; ref. 11), colony stimulating factor 1 receptor (tyrosine kinase receptor important for hematopoietic and mammary epithelial development; ref. 12), ephrin receptor B4 (receptor tyrosine kinase expressed in undifferentiated hematopoietic and mammary epithelial cells; refs. 13 and 14), mago-m (homolog of Drosophila mago nashi, involved in mRNA localization during fly oogenesis; refs. 15 and 16), lactoferrin (multiple functions including regulation of IGF bioavailability; ref. 17), and annexin-A1 (participates in endosomal trafficking; ref. 18). The qRT-PCR analysis confirmed that the level of each transcript was 2- to 9-fold higher in intact stomachs with enriched GEP populations (tox176 and/or E18) compared with normal adult germ-free stomachs (see Fig. 5, which is published as supporting information on the PNAS web site).
We next used antibodies specific for PCNA and annexin, as well as three other members of the GEP dataset, to show that all five proteins were present at higher levels in intact tox176 compared with age-matched normal stomachs (see Fig. 5). Light and electron microscopic immunohistochemical studies of normal and tox176 mice confirmed PCNA expression in GEPs (Figs. 1B and 2A).
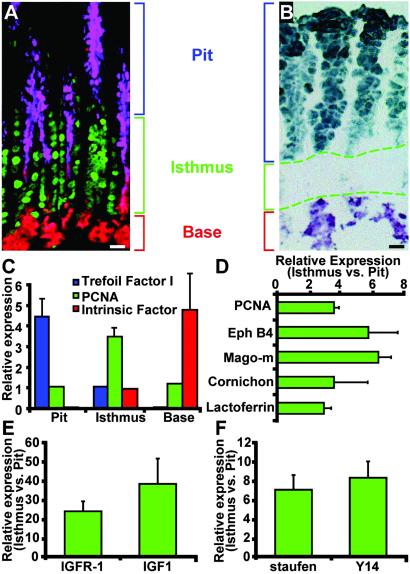
Fig 2.
qRT-PCR studies of gene expression in GEPs obtained by n-LCM. (A) Multilabel study of a section prepared from paraffin-embedded tissue, showing distribution of pit, GEP, and neck cells in gastric units from a 16-week-old tox176 mouse. Purple, pit cells visualized with AAA; green, PCNA-positive GEPs; red, GSII-positive neck cells. (B) Representative image template used for n-LCM. The cryosection was stained with AAA and GSII. Dashed lines outline isthmal regions of several adjacent gastric units. (C) qRT-PCR assays of the purity of LCM populations. For trefoil factor 1 and intrinsic factor mRNAs, mean values are expressed relative to the concentration in captured isthmal cells (set at 1). PCNA mRNA levels are expressed relative to pit cells. Results are representative of five independent dissections. (D) LCM/qRT-PCR assays of GEP database mRNAs in isthmal versus pit fractions. Mean values ± 1 SD of two to five separate experiments (n = 4 mice) are plotted. In each case, the difference in levels is statistically significant (P < 0.05, Student's t test). (E and F) LCM/qRT-PCR study of genes not in the original GEP dataset but known to be involved in IGF responses (E) and mRNA localization (F). (Bars, 25 μm.)
qRT-PCR Studies of Navigated Laser Capture Microdissected (n-LCM) GEPs.
To further verify enhanced expression of members of the dataset within the expanded tox176 GEP population, we combined qRT-PCR with a form of LCM we have termed navigated LCM (19). Immunostaining protocols requiring more than a few minutes to complete jeopardize recovery of intact mRNA. Therefore, serial cryosections were prepared from the central third of 16-week-old tox176 stomachs. Odd-numbered sections were stained with AAA to identify pit cell-specific glycans (20), GSII to visualize neck cell-specific glycans, and antibodies to PCNA to mark GEPs (Fig. 2 A and B). The multilabeled sections were used as image templates to guide (navigate) dissection of cells from the pit, isthmus, and base regions of well oriented gastric units present in adjacent, even-numbered cryosections that had been stained very briefly with methyl green and eosin Y to visualize cells yet preserve mRNA integrity. An average of 10 isthmal GEPs were recovered per sectioned gastric unit (n = 250–500 units dissected per cryosection). Dissection quality was assessed by qRT-PCR assays for known markers of the pit and neck/zymogenic cell lineages, plus GEP-associated PCNA. The results confirmed enhanced levels of trefoil factor 1 mRNA (in the pit fraction), intrinsic factor (base), and PCNA (isthmus) (Fig. 2C).
We next used n-LCM/qRT-PCR to show that expression of four genes from the GEP dataset [ephrin receptor B4, mago-m, lactoferrin, and cornichon (important for TGF/EGF vesicular trafficking and axis formation in Drosophila; ref. 21)] was 3- to 7-fold higher in isthmal tox176 cells compared with their differentiated pit cell neighbors (Fig. 2D).
Functional Comparison of the GEP Expression Profile with Profiles Obtained from Two Descendant Lineages and Mouse Hematopoietic Stem Cells.
Because there had been no previous molecular characterization of GEPs, we could not validate the GEP dataset by comparing it to a preexisting list of mRNAs. Therefore, we compared the functional features of the entire GEP dataset with those of gene expression profiles obtained from mouse hematopoietic stem cells (HSCs) and from two differentiated GEP descendants, parietal and zymogenic cells. To do so, we needed a tool that could automatically classify an entire dataset, irrespective of species of origin or method of data generation, so that each dataset could be viewed as the sum of its component parts, and its similarity to other lists determined independent of their length.
Terms defined by the Gene Ontology (GO) Consortium (www.geneontology.org/) provided a language for performing this automated annotation and functional comparison. Using a series of algorithms, we assigned GO terms to each gene in each dataset, analyzed the distribution of GO terms across the entire gene list, and determined the fractional representation (FR) of each term. We defined FR as the number of genes with a given GO term in a given list relative to the total number of genes in that list with assigned GO terms.
We had postulated that comparison of tox176 stomachs with normal germ-free mouse stomachs would lead to a progenitor cell expression profile “contaminated” with immune cell-derived transcripts. Thus, we tested our GO-based classification system by comparing the FR of immune/host defense functions in three datasets: the tox176 to normal germ-free stomach comparison (500 genes), the E18 to normal germ-free stomach comparison (580 genes), and the final, triangulated 147-gene GEP dataset. Table 3 (which is published as supporting information on the PNAS web site) shows that our triangulation strategy led to near elimination of immune/defense-related genes.
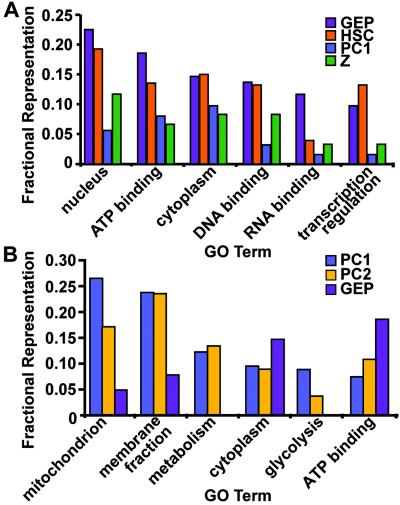
Fig. 3A compares the FR of the six most frequent GO terms in the GEP database with their FR in a previously published dataset of 767 genes expressed in fetal mouse liver-based HSCs (22), and in two GeneChip-derived datasets [one a list of 231 transcripts enriched in differentiated PCs relative to all other gastric mucosal cell types (23), the other a list of 114 zymogenic cell-enriched mRNAs (N.A., J.C.M., and J.I.G., unpublished work)]. In all six cases, the FR of the GO term in the GEP dataset was closest to that in the HSC dataset; e.g., the term “transcription regulation” was the 5th and 6th most frequently represented term in the GEP and HSC databases (10% and 13%, respectively), but only the 29th most frequent term (3%) in the zymogenic dataset, and 97th in the PC dataset (1%).
Fig 3.
Fractional representations of GO terms in GEP, PC, zymogenic (Z), and hematopoietic stem cell (HSC) datasets. (A) Six most frequent GO terms in the GEP dataset with corresponding FR in another progenitor population (HSCs) and in two terminally differentiated GEP descendant lineages (PC1 and Z). (B) Six most frequent GO terms in a previously published PC dataset (PC1; ref. 23), compared with their representation in a dataset generated from PCs that were isolated using a different method (PC2), and in the GEP dataset. Further information about the hierarchical GO-term classification scheme can be obtained from the Gene Ontology web site (www.geneontology.org/).
Fig. 3B compares the FR of the six most common GO terms in the PC dataset (PC1) with their corresponding representation in the GEP dataset and in another, 259-member GeneChip-derived dataset (PC2) generated from PCs that had been purified by elutriation as opposed to lectin panning (N.A., J.C.M., and J.I.G., unpublished work). The results reveal the functional similarity between the two differentiated PC datasets, and their distinctness from the GEP dataset; e.g., three of the most common PC GO terms (mitochondrion, metabolism, and glycolysis) were not among the top 15 terms in the GEP dataset, or among the top 12 terms in the HSC list.
Together, these results demonstrate that the automated classification scheme can be used to quantify functional similarities and differences between entire gene expression profiles (“profile surfing”). They verify that the “triangulation” strategy identified a gene expression profile in GEPs that is functionally distinct from their differentiated descendants but similar to HSCs.
Further Characterization of the Functional Features of GEPs.
Overview.
Our GO-based functional analysis prompted a more detailed inspection of the GEP dataset to identify pathways that would give us further insights about the regulation of progenitor proliferation and differentiation in the isthmal stem cell niche. Thirty-five members of the 147 member GEP list regulate and/or respond to growth factor signaling in other cellular systems. A portion of these encode components of protein modification and degradation systems. The dataset also contains a number of mouse homologs of Drosophila genes involved in RNA processing, nuclear-to-cytoplasmic trafficking, and subcellular localization.
Components of growth factor response pathways.
Several growth factors signal through their receptors to activate PI-3 kinase, which in turn activates Akt. Akt regulates β-catenin via glycogen synthase kinase-3 (GSK3). Akt, β-catenin and two β-catenin regulated genes [the transcription factor N-myc down-regulated 1 (NDRG1, involved in growth regulation and embryonic axis formation; ref. 24) and the bHLH antagonist Id2 (25)] are all in the GEP dataset. Akt regulates cell division by phosphorylating p21 (26), thereby inhibiting its association with both PCNA and cdk2, while increasing its promotion of cyclinD1-cdk4 assembly (27). PCNA, three DNA-repair-associated components (RPA3, RPA1, and Rad51), and cdk4 are in the GEP dataset.
Akt sits at the hub of several growth factor signaling pathways. However, several lines of evidence point to a prominent role for IGF signaling in regulating GEP biology. First, two GEP members, c-Myb (activated by Akt; ref. 28) and C/ebpδ, increase IGF, IGF1 receptor, and IGFBP5 expression (29–31), raising the possibility of autocrine IGF signaling in GEPs. To test this notion, we used n-LCM and qRT-PCR to assay for IGF1 and the IGF1-R expression in tox176 GEPs. The levels of these mRNAs were 34- and 22-fold higher, respectively, than in adjacent pit cells (Fig. 3E). Second, ablation of PCs in tox176 mice leads to increased proliferation of GEPs. PCs elaborate several molecules that may decrease GEP proliferation by limiting bioavailable IGF: IGF binding protein-2, and Golgi-associated, gamma adaptin ear-containing ARF binding protein 2, which controls plasma membrane delivery of the mannose- 6-phosphate receptor (32) (also known as IGF2 receptor; ref. 33). Third, GEPs express lactoferrin (Fig. 2D), which binds IGFBP3 to increase IGF bioavailability (17). Fourth, IGFs signal through Akt and mTOR to affect S6 kinase-mediated phosphorylation of the S6 ribosomal protein, increasing translation of mRNAs containing 5′ TOP (terminal oligopyrimidine tract) sequences (34). The GEP dataset contains six 5′ TOP transcripts, including eEF1α (35).
We propose that PCs and GEPs function in concert to control isthmal concentrations of bioavailable IGFs, and that IGFs are key effectors of epithelial progenitor cell biology and census in the isthmus. This conclusion is consistent with reports that forced expression of IGF2 in transgenic mice results in increased gastric size, whereas a soluble IGF2 receptor has the opposite effect (36).
Regulators of protein turnover.
Regulation of protein turnover is critical for growth factor signaling (37). Components of the ubiquitin/proteosomal protein degradation pathway are well represented in the GEP database. These include N-terminal asparagine amidase (prepares protein substrates for ubiquitination; ref. 38), the X-chromosome ubiquitin-activating (E1) enzyme, and two ubiquitin conjugating (E2) enzymes (Ubc6 and Ubc10). In addition, mRNAs encoding the epsilon, theta, and zeta subunits of the TCP-ring complex, plus elongin C, are all enriched in GEPs. The TCP-ring complex promotes folding of von Hippel Lindau (VHL) protein which, in turn, complexes with elongins B and C to form a multicomponent E3 ubiquitin ligase intimately involved in cell cycle regulation and growth suppression (39).
Covalent addition of small ubiquitin-related modifier proteins (SUMO1, -2, and -3) regulates the activities of several growth regulators (40). GEPs are enriched in both SUMO1 and SUMO2 mRNAs, and their c-Myb and β-catenin targets. Neddylation involves the covalent linkage of Nedd8 (81 residue ubiquitin-like polypeptide) to target proteins, including ROC/SCF-cullin (E3 ubiquitin complex critical for cell cycle control), as well as the GEP-associated E3 VHL-elongin complex described above (41). Amyloid beta precursor protein-binding protein 1, whose mRNA is enriched in GEPs, is part of the multisubunit complex that catalyzes neddylation (42).
The GEP list contains three proteosomal subunits (Pad1, regulatory subunit S10, and Psmb3), as well as subunit 3 of the COP9 signalosome. The signalosome is a multisubunit complex that may serve as an alternate 19S proteosomal lid to regulate turnover of multiple cell division- and growth factor signaling-related proteins; e.g., it inhibits degradation of the Akt target p27, activates Jun, and de-neddylates ROC/SCF-cullin (43).
mRNA processing and cytoplasmic localization.
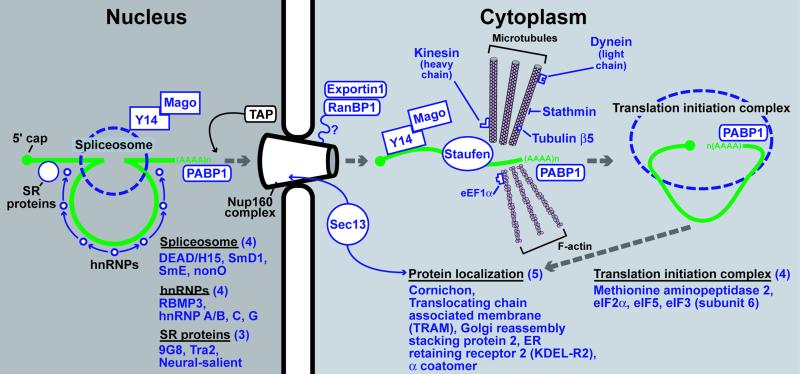
Mago-m, whose enriched expression in GEPs was validated by LCM/qRT-PCR (Fig. 2D), is a mouse homolog of Drosophila mago nashi. Studies in flies and mammals indicate that Mago associates with certain nascent mRNAs through its interaction with Y14 RNA binding protein and remains affiliated with these transcripts throughout spliceosome-mediated processing, nuclear export, and cytoplasmic localization (Fig. 4). Mago nashi means “without grandchildren” in Japanese and refers to the fact that the maternal effect mago nashi null allele results in daughters that are sterile because their oocytes cannot localize mRNAs critical for axis formation (15, 16).
Fig 4.
GEP-enriched transcripts encoding proteins involved in nuclear mRNA processing and export, cytoplasmic localization, and translation. Schematic representation of the processing of a single intronic sequence from an idealized RNA polymerase II transcript and movement of the product through the nuclear pore to a cytoplasmic site of translation and protein localization. Green, mRNA transcript; blue, present in the GEP dataset and/or confirmed by qRT-PCR analysis of laser capture microdissected GEPs; black, components of the pathway.
The GEP dataset is enriched in several other transcripts encoding proteins that help direct and integrate nuclear mRNA processing, nuclear export, and cytoplasmic localization (Fig. 4). These mRNAs encode polyA binding protein 1, four proteins associated with the spliceosome [DEAD-box helicase 15, SmD1, SmE, and the non-POU domain containing octamer binding protein (nonO)] and four hnRNPs (RBP3, hnRNPA/B, hnRNPC, and hnRNPG), plus three serine/arginine-rich (SR) proteins (9G8, Tra2, and neuro-salient). GEP-enriched transcripts also specify three nuclear pore proteins that participate in RNA export from the nucleus (exportin 1, Ran binding protein 1, and the mouse ortholog of yeast Sec13p). Microtubule-associated proteins are involved in mRNA segregation (44). The GEP dataset contains tubulin β5, the tubulin-bound motor proteins kinesin heavy chain 5 (KIF5B) and Tctex-1 (one of three dynein light chains), plus stathmin (a cell cycle-regulated, tubulin-binding protein). Actin microfilaments have also been implicated in RNA targeting; the GEP member eEF1α binds both mRNA and F-actin to localize translation in specific subcellular domains (45). GEPs also express several components of the translation initiation apparatus [eIF2γ, eIF3 subunit 6, eIF5, and methionylaminopeptidase 2 (removes initiator Met residues and blocks phosphorylation of eIF2α, promoting translation; ref. 46)].
Interestingly, null mutations of Drosophila genes involved in each of the mRNA processing/localization steps described above impede establishment of polarity in developing oocytes. These mutations involve half pint (encodes a spliceosome component; ref. 47), squid (hnRNPA/B-like; ref. 48), tsunagi (Y14 ortholog; ref. 49), and mago nashi. Similar defects are produced by mutations in genes encoding components of the translation apparatus [vasa (eIF4A homolog; ref. 50), aubergine (eIF2C; ref. 51)], and that regulate microtubule-mRNA interactions [staufen, dKhc1 (kinesin heavy chain; ref. 44), and dDlc (dynein light chain; ref. 52)]. In addition, mutations of genes such as cornichon, which targets proteins to specific cellular surfaces, lead to aberrant axis formation.
To further establish a link between mRNA processing/targeting and GEP biology, we used n-LCM to harvest GEPs and pit cells from tox176 gastric units and qRT-PCR to assay for transcripts specifying two recently characterized homologs of fly genes not present in our GEP dataset. The transcript encoding Y14, which, as noted above, binds nascent mRNAs with mago-m, is increased 8-fold in GEPs. Staufen 1, a conserved double-stranded RNA binding protein that coordinates interaction of certain mRNAs with microtubules for proper transcript localization in neurons and oocytes (53), is enriched 6-fold (Fig. 2F).
In the Drosophila oocyte, some mRNA transcripts are targeted anteriorly, and others posteriorly (54). Presumably, the direction of specific mRNA placement reflects the nature of the expressed cellular complement of mRNA-associated targeting proteins. Stem cells in different tissues likely have differing needs for orienting mRNAs relative to other cellular components of their niche. To test the hypothesis that the pattern of expression of RNA localizing genes might be different in progenitor populations distributed at different points along the length of the normal mouse gut, we combined LCM with qRT-PCR to assay for mago-m, Y14, and staufen expression in small intestinal and colonic epithelial progenitor niches located at or near the base of crypts of Lieberkühn (19, 55, 56). Epithelial cells were microdissected from each progenitor niche, and from adjacent compartments containing their differentiated descendants (the villus epithelium in the case of the small intestine, the hexagonal surface epithelial cuff demarcating the orifice of each crypt in the case of the colon; 10,000 cells harvested per compartment per mouse; n = 3 mice). Mago-m mRNA levels were 5-fold higher in the small intestinal crypt base compared with villus epithelium, and 6-fold higher in the colonic crypt base versus surface cuff. Y14 mRNA levels were 3- and 2-fold higher, whereas staufen 1 mRNA, which is present in GEPs, was undetectable in both small intestine and colon.
BLASTn searches against the entire HSC EST database (http://stemcell.princeton.edu/) identified Y14, but not mago-m nor staufen, suggesting that HSCs may exhibit a pattern of mRNA localization distinct from gut progenitors.
These results underscore the need to understand the role of mRNA targeting and storage in epithelial progenitor cell biology. Other non-germ cell lineages use mRNA targeting to store nascent proteins and allow rapid, subcellular-specific responses to various stimuli, e.g., depolarization of neurons (57). mRNA targeting proteins may help mammalian gut epithelial progenitors divide asymmetrically and establish differential communications with neighboring cells in their niche. An intriguing possibility is that differences in expression of certain mRNA targeting proteins may help distinguish normal from preneoplastic or fully transformed gastric epithelial progenitors.
Supplementary Material
Acknowledgments
We thank Sabrina Wagoner and Jaime Dant for technical assistance, David O'Donnell and Maria Karlsson for maintaining mice, Jung Oh for advice with qRT-PCR assays, and Diane Redmond for computer graphics. This work was supported by National Institutes of Health Grant DK58529. J.C.M. is a Howard Hughes Medical Institute physician-scientist postdoctoral fellow.
Abbreviations
AAA, Anguilla anguilla agglutinin
FR, fractional representation
GEP, gastric epithelial lineage progenitor
GO, Gene Ontology
GSII, Griffonia simplicifolia II
HSC, hematopoietic stem cell
IGF, insulin-like growth factor
PC, parietal cell
PCNA, proliferating cell nuclear antigen
qRT-PCR, quantitative RT-PCR
References
- 1.Karam S. M. & Leblond, C. P. (1993) Anat. Rec. 236, 259-279. [DOI] [PubMed] [Google Scholar]
- 2.Karam S. M. & Leblond, C. P. (1993) Anat. Rec. 236, 280-296. [DOI] [PubMed] [Google Scholar]
- 3.Karam S. M. & Leblond, C. P. (1993) Anat. Rec. 236, 297-313. [DOI] [PubMed] [Google Scholar]
- 4.Karam S. M. (1993) Anat. Rec. 236, 314-332. [DOI] [PubMed] [Google Scholar]
- 5.Li Q., Karam, S. M. & Gordon, J. I. (1996) J. Biol. Chem. 271, 3671-3676. [PubMed] [Google Scholar]
- 6.Syder A. J., Guruge, J. L., Li, Q., Hu, Y., Oleksiewicz, C. M., Lorenz, R. G., Karam, S. M., Falk, P. G. & Gordon, J. I. (1999) Mol. Cell 3, 263-274. [DOI] [PubMed] [Google Scholar]
- 7.Karam S. M., Li, Q. & Gordon, J. I. (1997) Am. J. Physiol. 272, G1209-G1220. [DOI] [PubMed] [Google Scholar]
- 8.Hooper L. V., Mills, J. C., Roth, K. A., Stappenbeck, T. S., Wong, M. H. & Gordon, J. I. (2002) Methods Microbiol. 31, 559-589. [Google Scholar]
- 9.Stappenbeck T. S., Hooper, L. V., Manchester, J. K., Wong, M. H. & Gordon, J. I. (2002) Methods Enzymol. 356, 168-196. [DOI] [PubMed] [Google Scholar]
- 10.Warbrick E. (2000) BioEssays 22, 997-1006. [DOI] [PubMed] [Google Scholar]
- 11.Yarden R. I. & Brody, L. C. (1999) Proc. Natl. Acad. Sci. USA 96, 4983-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sapi E. & Kacinski, B. M. (1999) Proc. Soc. Exp. Biol. Med. 220, 1-8. [DOI] [PubMed] [Google Scholar]
- 13.Suenobu S., Takakura, N., Inada, T., Yamada, Y., Yuasa, H., Zhang, X. Q., Sakano, S., Oike, Y. & Suda, T. (2002) Biochem. Biophys. Res. Commun. 293, 1124-1131. [DOI] [PubMed] [Google Scholar]
- 14.Munarini N., Jager, R., Abderhalden, S., Zuercher, G., Rohrbach, V., Loercher, S., Pfanner-Meyer, B., Andres, A. C. & Ziemiecki, A. (2002) J. Cell Sci. 115, 25-37. [DOI] [PubMed] [Google Scholar]
- 15.Newmark P. A., Mohr, S. E., Gong, L. & Boswell, R. E. (1997) Development (Cambridge, U.K.) 124, 3197-3207. [DOI] [PubMed] [Google Scholar]
- 16.Kataoka N., Diem, M. D., Kim, V. N., Yong, J. & Dreyfuss, G. (2001) EMBO J. 20, 6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumrucker C. R. & Erondu, N. E. (2000) J. Mammary Gland Biol. Neoplasia 5, 53-64. [DOI] [PubMed] [Google Scholar]
- 18.Rescher U., Zobiack, N. & Gerke, V. (2000) J. Cell Sci. 113, 3931-3938. [DOI] [PubMed] [Google Scholar]
- 19.Wong M. H., Saam, J. R., Stappenbeck, T. S., Rexer, C. H. & Gordon, J. I. (2000) Proc. Natl. Acad. Sci. USA 97, 12601-12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falk P., Roth, K. A. & Gordon, J. I. (1994) Am. J. Physiol. 266, G987-G1003. [DOI] [PubMed] [Google Scholar]
- 21.Powers J. & Barlowe, C. (1998) J. Cell Biol. 142, 1209-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips R. L., Ernst, R. E., Brunk, B., Ivanova, N., Mahan, M. A., Deanehan, J. K., Moore, K. A., Overton, G. C. & Lemischka, I. R. (2000) Science 288, 1635-1640. [DOI] [PubMed] [Google Scholar]
- 23.Mills J. C., Syder, A. J., Hong, C. V., Guruge, J. L., Raaii, F. & Gordon, J. I. (2001) Proc. Natl. Acad. Sci. USA 98, 13687-13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly C., Chin, A. J., Leatherman, J. L., Kozlowski, D. J. & Weinberg, E. S. (2000) Development (Cambridge, U.K.) 127, 3899-3911. [DOI] [PubMed] [Google Scholar]
- 25.Rockman S. P., Currie, S. A., Ciavarella, M., Vincan, E., Dow, C., Thomas, R. J. & Phillips, W. A. (2001) J. Biol. Chem. 276, 45113-45119. [DOI] [PubMed] [Google Scholar]
- 26.Zhou B. P., Liao, Y., Xia, W., Spohn, B., Lee, M. H. & Hung, M. C. (2001) Nat. Cell Biol. 3, 245-252. [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Dowbenko, D. & Lasky, L. A. (2002) J. Biol. Chem. 277, 11352-11361. [DOI] [PubMed] [Google Scholar]
- 28.Lauder A., Castellanos, A. & Weston, K. (2001) Mol. Cell. Biol. 21, 5797-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Umayahara Y., Billiard, J., Ji, C., Centrella, M., McCarthy, T. L. & Rotwein, P. (1999) J. Biol. Chem. 274, 10609-10617. [DOI] [PubMed] [Google Scholar]
- 30.Werner H., Shalita-Chesner, M., Abramovitch, S., Idelman, G., Shaharabani-Gargir, L. & Glaser, T. (2000) Mol. Genet. Metab. 71, 315-320. [DOI] [PubMed] [Google Scholar]
- 31.Tanno B., Negroni, A., Vitali, R., Pirozzoli, M. C., Cesi, V., Mancini, C., Calabretta, B. & Raschella, G. (2002) J. Biol. Chem. 277, 23172-23180. [DOI] [PubMed] [Google Scholar]
- 32.Zhu Y., Doray, B., Poussu, A., Lehto, V. P. & Kornfeld, S. (2001) Science 292, 1716-1718. [DOI] [PubMed] [Google Scholar]
- 33.Nakae J., Kido, Y. & Accili, D. (2001) Endocr. Rev. 22, 818-835. [DOI] [PubMed] [Google Scholar]
- 34.Schmelzle T. & Hall, M. N. (2000) Cell 103, 253-262. [DOI] [PubMed] [Google Scholar]
- 35.Kimball S. R., Shantz, L. M., Horetsky, R. L. & Jefferson, L. S. (1999) J. Biol. Chem. 274, 11647-11652. [DOI] [PubMed] [Google Scholar]
- 36.Zaina S. & Squire, S. (1998) J. Biol. Chem. 273, 28610-28616. [DOI] [PubMed] [Google Scholar]
- 37.Weissman A. M. (2001) Nat. Rev. Mol. Cell. Biol. 2, 169-178. [DOI] [PubMed] [Google Scholar]
- 38.Kwon Y. T., Balogh, S. A., Davydov, I. V., Kashina, A. S., Yoon, J. K., Xie, Y., Gaur, A., Hyde, L., Denenberg, V. H. & Varshavsky, A. (2000) Mol. Cell. Biol. 20, 4135-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baba M., Hirai, S., Kawakami, S., Kishida, T., Sakai, N., Kaneko, S., Yao, M., Shuin, T., Kubota, Y., Hosaka, M. & Ohno, S. (2001) Oncogene 20, 2727-2236. [DOI] [PubMed] [Google Scholar]
- 40.Muller S., Hoege, C., Pyrowolakis, G. & Jentsch, S. (2001) Nat. Rev. Mol. Cell. Biol. 2, 202-210. [DOI] [PubMed] [Google Scholar]
- 41.Ohh M., Kim, W. Y., Moslehi, J. J., Chen, Y., Chau, V., Read, M. A. & Kaelin, W. G., Jr. (2002) EMBO Rep. 3, 177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osaka F., Kawasaki, H., Aida, N., Saeki, M., Chiba, T., Kawashima, S., Tanaka, K. & Kato, S. (1998) Genes Dev. 12, 2263-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X., Menon, S., Lykke-Andersen, K., Tsuge, T., Di, X., Wang, X., Rodriguez-Suarez, R. J., Zhang, H. & Wei, N. (2002) Curr. Biol. 12, 667-672. [DOI] [PubMed] [Google Scholar]
- 44.Cha B. J., Serbus, L. R., Koppetsch, B. S. & Theurkauf, W. E. (2002) Nat. Cell Biol. 4, 592-598. [DOI] [PubMed] [Google Scholar]
- 45.Liu G., Grant, W. M., Persky, D., Latham, V. M., Jr., Singer, R. H. & Condeelis, J. (2002) Mol. Biol. Cell 13, 579-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cutforth T. & Gaul, U. (1999) Mech. Dev. 82, 23-28. [DOI] [PubMed] [Google Scholar]
- 47.Van Buskirk C. & Schupbach, T. (2002) Dev. Cell 2, 343-353. [DOI] [PubMed] [Google Scholar]
- 48.Lall S., Francis-Lang, H., Flament, A., Norvell, A., Schupbach, T. & Ish-Horowicz, D. (1999) Cell 98, 171-180. [DOI] [PubMed] [Google Scholar]
- 49.Mohr S. E., Dillon, S. T. & Boswell, R. E. (2001) Genes Dev. 15, 2886-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghabrial A. & Schupbach, T. (1999) Nat. Cell Biol. 1, 354-357. [DOI] [PubMed] [Google Scholar]
- 51.Harris A. N. & Macdonald, P. M. (2001) Development (Cambridge, U.K.) 128, 2823-2832. [DOI] [PubMed] [Google Scholar]
- 52.Schnorrer F., Bohmann, K. & Nusslein-Volhard, C. (2000) Nat. Cell Biol. 2, 185-190. [DOI] [PubMed] [Google Scholar]
- 53.Tang S. J., Meulemans, D., Vazquez, L., Colaco, N. & Schuman, E. (2001) Neuron 8, 463-475. [DOI] [PubMed] [Google Scholar]
- 54.Johnstone O. & Lasko, P. (2001) Annu. Rev. Genet. 35, 365-406. [DOI] [PubMed] [Google Scholar]
- 55.Bjerknes M. & Cheng, H. (1981) Am. J. Anat. 160, 51-63. [DOI] [PubMed] [Google Scholar]
- 56.Bjerknes M. & Cheng, H. (1981) Am. J. Anat. 160, 77-91. [DOI] [PubMed] [Google Scholar]
- 57.Krichevsky A. M. & Kosik, K. S. (2001) Neuron 32, 683-696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.