Abstract
Mbd4 (methyl-CpG binding domain 4) is a novel mammalian repair enzyme that has been implicated biochemically in the repair of mismatched G-T residues at methylated CpG sites. In addition, the human protein has been shown to interact with the DNA mismatch repair protein MLH1. To clarify the role of Mbd4 in DNA repair in vivo and to examine the impact of Mbd4 inactivation on gastrointestinal (GI) tumorigenesis, we introduced a null mutation into the murine Mbd4 gene by gene targeting. Heterozygous and homozygous Mbd4 mutant mice develop normally and do not show increased cancer susceptibility or reduced survival. Although Mbd4 inactivation did not increase microsatellite instability (MSI) in the mouse genome, it did result in a 2- to 3-fold increase in C→T transition mutations at CpG sequences in splenocytes and epithelial cells of the small intestinal mucosa. The combination of Mbd4 deficiency with a germ line mutation in the adenomatous polyposis coli (Apc) gene increased the tumor number in the GI tract and accelerated tumor progression. The change in the GI cancer phenotype was associated with an increase in somatic C→T mutations at CpG sites within the coding region of the wild-type Apc allele. These studies indicate that, although inactivation of Mbd4 does not by itself cause cancer predisposition in mice, it can alter the mutation spectrum in cancer cells and modify the cancer predisposition phenotype.
The ability to maintain the integrity of the genome is essential to all organisms, and both prokaryotic and eukaryotic cells have developed a variety of repair mechanisms to cope with mutagenic and carcinogenic DNA lesions. The cellular responses that remove these potentially harmful lesions involve a number of different excision repair systems, including DNA mismatch repair (MMR), nucleotide excision (NER) and base excision repair (BER) (1–5). Recently, a candidate DNA repair gene was identified in a search of the mouse EST database by using the protein sequence of the methyl-CpG binding domain (Mbd) of methyl-CpG-binding protein 2 (MeCP2) (6). This gene, termed Mbd4, is one of four members of the Mbd gene family whose gene products bind specifically to methyl-CpG dinucleotides in the genome and are known to have a functional role in transcriptional repression. However, analysis of the Mbd4 amino acid sequence revealed that it contained a C-terminal catalytic domain that shares high homology to many known bacterial DNA glycosylases and lyases. Further biochemical analysis using oligonucleotide substrates showed that Mbd4 is a DNA glycosylase that could act in the repair of G-T or G-U mismatches at CpG sites resulting from deamination of 5-methyl cytosine or cytosine, respectively (7, 8).
Subsequently, the human homolog of Mbd4 was identified in a yeast two-hybrid screen due to its interaction with the human mismatch repair protein MLH1 (9) and was named methyl-CpG binding endonuclease 1 (MED1). For convenience, we will refer to the human MED1 gene as MBD4 throughout this study. Because of the interaction with MLH1 and the initial observation that MBD4 contained an endonuclease activity that can convert supercoiled plasmid DNA into nicked, linear molecules, it was suggested that MBD4 could function in mammalian MMR in a manner reminiscent of the MutH endonuclease in bacterial MMR (9). However, additional biochemical studies using double-stranded oligonucleotide substrates failed to detect an MBD4 endonuclease activity and therefore did not support a role of MBD4 in mammalian MMR strand discrimination (7). Rather, it was suggested that the interaction between MLH1 and MBD4 might play a role in the coordination of base excision repair and MMR to prevent potentially mutagenic repair of G-T mismatches by MBD4 that result from misincorporation of guanosine opposite a thymidine during DNA replication at CpG sites (10).
Another link between MBD4 and the MMR system was provided by the observation that between 20% and 43% of primary human colorectal carcinomas that displayed microsatellite instability (MSI) also harbored inactivating mutations in MBD4 (11–13). A substantial number of these tumors were either HNPCC tumors that carried MLH1 mutations or sporadic tumors that had lost MSH2 or MLH1 expression as assessed by immunohistochemical analysis. In addition, MBD4 mutations were also frequently observed in other microsatellite unstable cancers such as gastric, endometrial, and pancreatic carcinomas (11, 14). Sequence analysis of MBD4 in several of these tumors revealed that the vast majority of mutations occurred within a stretch of A(10) in the MBD4 coding region and were predicted to result in a truncated protein product that lacked the C-terminal catalytic domain. Interestingly, sequence analysis showed biallelic MBD4 inactivation in some tumors, suggesting that MBD4 could be a target gene of genomic instability or a tumor suppressor gene. However, the failure to detect MBD4 mutations in microsatellite stable tumors together with the lack of mutations occurring outside this mononucleotide repeat track suggested that MBD4 mutations were likely the result, rather than the cause, of MMR deficiency (12).
To examine the role of Mbd4 in DNA repair in vivo and to assess its importance for tumor suppression, we generated a mouse line with an inactivating mutation in Mbd4 by gene targeting. We found that Mbd4 inactivation did not cause MSI in the murine genome. However, we observed an increase in the mutation frequencies in the small intestine and spleen of Mbd4−/− mice that was mainly attributed to an increase in the incidence of C:G→T:A transition mutations at CpG sites. Although the mutator phenotype caused by Mbd4 deficiency was not sufficient to initiate tumorigenesis in mice, it had a profound effect on tumor progression in the gastrointestinal (GI) tract of mice carrying the Apc1638N germ line mutation. We further show that this enhanced GI tumorigenesis is specifically caused by an increase in somatic C:G→T:A transition mutations at CpG sites within the coding region of the WT Apc allele. Our results indicate that Mbd4 inactivation can modulate the mutational spectra during tumorigenesis and influence tumor progression.
Materials and Methods
Generation of Mbd4 Mutant Mice and Cell Lines.
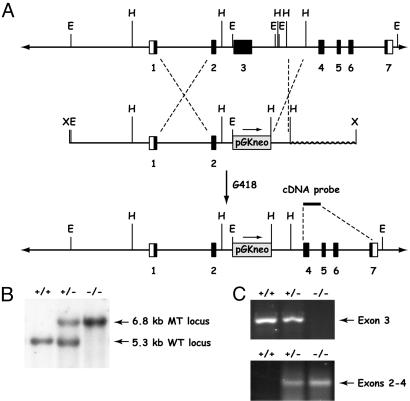
A PCR probe corresponding to nucleotides 1688 to 1873 of the Mbd4 cDNA was used to identify BAC 118O23 in the RPCI-22 mouse genomic BAC library. For inactivation of Mbd4, a targeting vector was designed to replace exon 3 and parts of intron 3 of Mbd4 with a PGKneomycin resistance cassette. A 900-bp HindIII fragment located in intron 3 was isolated and inserted into the HindIII site of pGKneomycin. Subsequently a 7.3-kb EcoRI restriction fragment spanning sequences upstream of Mbd4 to intron 2 was cloned into the EcoRI site 3′ to the PGKneomycin cassette. The targeting vector was linearized at the unique XhoI site and electroporated into WW6 embryonic stem (ES) cells (15). The cells were selected in G418 (150 μg/ml) for 12 days, and resistant colonies were isolated. Among the 240 clones screened, one positive clone was isolated. The correct targeting event was identified by Southern blot analysis of EcoRI-digested genomic DNA using a cDNA probe corresponding to exons 4 to 7. ES cells carrying the targeted Mbd4 allele were injected into C57BL/6J blastocysts to generate chimeric mice. Several male chimeras were mated to C57BL/6J females to generate Mbd4+/− F1 offspring, which were subsequently interbred to obtain F2 Mbd4−/− offspring. Genotyping of mouse tail DNA was performed by Southern blot analysis as described above for ES cell screening.
RT-PCR Analysis.
To verify the generation of an Mbd4 null mutant mouse line, total RNA was isolated from ES cells and various tissues including the spleen, kidney, and liver of Mbd4 mutant mice by using Trizol (GIBCO). RT-PCR reactions were carried out on DNase-treated RNA by using exon 3 internal primers (forward primer, 5′-TCAAGACACGAAGCAAGTGG-3′; and reverse primer, 5′-CCCTTTCTGTCTCCCTTCG-3′) to verify the loss of exon 3. Primers situated in exon 2 (forward primer, 5′-TACCACAGCGACAGAAGGC-3′) and exon 4 (reverse primer, 5′-CTTGTGTCCGTGGGATGC-3′) were used to detect the possible formation of alternative splice variants. Reverse transcription and PCR amplification were performed by using the Titan One Tube System (Roche).
Generation of Mbd4−/−, Mbd4−/−/Big Blue, and Mbd4−/−/Apc1638N/+ Mice.
F1 mice heterozygous for the Mbd4 null mutation were intercrossed to generate F2 WT, Mbd4+/− and Mbd4−/− mice. The animals used in this study were on a mixed genetic background estimated to be 62.5% C57BL/6J, 35% 129Sv, and 2.5% SJL/J. F1 heterozygous Mbd4 null mutant mice were crossed with either Big Blue (Stratagene; ref. 16) or Apc1638N mice (17), which were both on a C57BL/6J background. To generate Mbd4−/−/Big Blue or Mbd4−/−/Apc1638N/+ mice, either Mbd4+/−/Big Blue or Mbd4+/−/Apc1638N/+ offspring were subsequently intercrossed. Littermates derived from these crosses were used for the analyses of mutation frequencies and GI tumor phenotypes.
MSI Analysis.
Mutations in microsatellite sequences were assayed by PCR of single target molecules. Equal amounts of tail DNA from three mice per mouse strain (Mbd4+/+, Mbd4−/−, and Mlh1−/−) were pooled and diluted to 0.5–1.5 genome equivalents. Cycling reactions for the three markers analyzed (, D7Mit91, and D17Mit123) were performed as described (18).
Big Blue Mutational Analysis.
The in vivo mutation frequency in WT and Mbd4−/− mice was assessed by using the target lacI transgene in the Big Blue Transgenic Rodent Mutagenesis Assay System (Stratagene) according to the manufacturer's guidelines. At least three mice each from WT/Big Blue and Mbd4−/−/Big Blue mouse strains were killed at ≈10 wk of age. Single cell suspensions of splenocytes were isolated, and high-molecular weight DNA was extracted. For analysis of the mutation frequency in the small intestine, the epithelial cells of the mucosa in the jejunum were scraped off the submucosa for DNA extraction. Genomic DNA was packaged, and the phage particles were plated in the presence of 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal; Roche). Approximately 200,000 plaque-forming units were plated per DNA sample. The mutation frequency was calculated by dividing the number of blue plaques by the total number of plaques plated. To characterize lacI mutations, a segment of the lacI gene (+15 to +510, relative to the transcriptional start site) in the mutant phage particles was PCR amplified and sequenced. To determine whether Mbd4 deficiency resulted in an increase in mutations of a specific type, prevalence ratios were estimated; dependence between mutational events arising from the same mouse were adjusted by a generalized estimating equations approach (19).
Analysis of Tissues and Tumors in Mbd4 Mutant Mice.
After killing, various tissues were removed and inspected (including kidney, liver, brain, heart, skeletal muscle, skin, lung, pancreas, thymus, spleen, lymph nodes, urinary bladder, ovary, uterus, testis, prostate, esophagus, stomach, and small and large intestines). For analysis of gastrointestinal tumors in Apc1638N mice, the GI tract was opened longitudinally and examined under a dissecting microscope for the presence of tumors. The number of microadenomas was sampled from 10 serial tissue sections (4 μm) of the stomach, duodenum, jejunum, cecum, and proximal and distal colon of each mouse. Some small intestinal tumors were frozen in liquid nitrogen for subsequent molecular analysis. Tissues and tumors were processed for paraffin embedding, and sections were prepared for hematoxylin/eosin staining according to standard procedures.
Analysis of Apc Truncation Mutations.
PCR amplification of the WT Apc allele from tumor DNA and subsequent in vitro transcription and translation and sequence characterizations were performed as described (20). Statistical analyses of Apc truncation mutation incidence in GI tumors were performed by using the Fisher's exact test.
Results
Homozygous Mbd4 Null Mice Are Viable.
We generated Mbd4 mutant mice by replacing exon 3 of the murine Mbd4 gene with a PGKneomycin expression cassette in ES cells (Fig. 1A). The gene-targeting event was designed to delete the catalytic domain of Mbd4 encoded by exon 3 (21). The correct gene targeting event in ES clones and mice was verified by Southern blot hybridization (Fig. 1B). The deletion of exon 3 in the mouse was further confirmed by RT-PCR analysis of RNA isolated from multiple tissues. This analysis showed that exon 3 was lost in Mbd4−/− mice but detectable in Mbd4+/− and WT mice. Further analysis of RNA isolated from Mbd4+/− and Mbd4−/− mice revealed that exons 2 and 4 were spliced together, creating a previously uncharacterized low abundance RNA transcript (Fig. 1C). Sequence analysis of the modified RNA showed that this splice event created a frameshift mutation resulting in a polypeptide of 125 aa, of which the first 98 residues correspond to Mbd4 (data not shown). If the modified message is translated, the truncated product predicted would lack any known functional domains. This analysis indicates that we have generated a functionally null Mbd4 mutant.
Fig 1.
Strategy for the generation of Mbd4 null mutant mice. (A) Gene-targeting strategy. E, EcoRI; H, HindIII; X, XhoI. (B) Southern blot analysis of tail DNA from WT (+/+), Mbd4 heterozygous (+/−), and homozygous (−/−) mutant mice. The WT allele is indicated by a 5.3-kb fragment and the Mbd4 mutant allele (MT) by a 6.8-kb fragment. DNA was digested with EcoRI and hybridized with a 500-bp cDNA probe spanning exons 4–7 of Mbd4. (C) RT-PCR analysis of DNase-treated kidney RNA amplifying a 535-bp region within exon 3 (Upper) and a splice variant between exons 2 and 4 (Lower).
Genotyping of 118 F2 animals derived from 22 litters of 10 mating pairs showed that the Mbd4 mutation was transmitted in a normal Mendelian pattern of inheritance with 37 WT, 47 Mbd4+/−, and 34 Mbd4−/− mice. All Mbd4+/− and Mbd4−/− mice were viable and fertile. Detailed histopathological examination of a large panel of tissues (see Materials and Methods) in adult Mbd4−/− mice did not reveal any significant histopathological abnormalities.
Mbd4 inactivation did not result in increased cancer susceptibility. We carefully followed a cohort of 30 WT, 45 Mbd4+/−, and 29 Mbd4−/− mice for a period of 24 mo and noticed only one uterine leiomyosarcoma in a 13-mo-old female and one Peyer's patch lymphoma in the small intestine of a 16-mo-old male. Similarly, we did not observe a difference in survival between WT, Mbd4+/−, and Mbd4−/− mice.
Absence of MSI in Mbd4-Deficient Mice.
The mutation frequencies at microsatellite loci in the genome of Mbd4−/− mutant mice were studied by a limiting dilution PCR-based method. We examined one mononucleotide repeat marker, , and two dinucleotide repeat markers, D7Mit91 and D17Mit123. A total of 244 alleles in WT mice and 220 alleles in Mbd4−/− mice were analyzed for size alterations at the locus. There was no significant difference in the frequencies of mutant alleles in Mbd4−/− (11/220 alleles, 5%) and WT mice (7/244 alleles, 3%) at this locus (P > 0.3). We also analyzed 124 WT and 123 Mbd4−/− alleles at D7Mit91, as well as 134 WT and 111 Mbd4−/− alleles at D17Mit123. As with the mononucleotide repeat marker, we did not observe significant differences in the mutation frequencies for both dinucleotide markers: D7Mit91 (Mbd4−/−, 3/123, 2%; and WT, 3/124, 2%; P = 1.0); D17Mit123 (Mbd4−/−, 2/111, 2%; and WT, 8/134, 6%; P > 0.1). In contrast, we observed significant MSI in Mlh1−/− mice (22) at all three loci (mutation frequencies: 12/73, 16% for ; 19/123, 15% for D7Mit91; and 19/69, 28% for D17Mit123; for all three markers, P < 0.0005).
Increased Frequency of Transition Mutations in Mbd4−/− Mice.
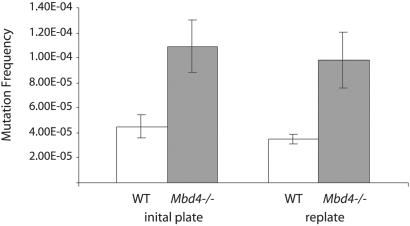
We next performed a quantitative and qualitative analysis of spontaneous mutations in mouse tissues by using the Big Blue mutation detection system (16). Three or four mice of each genotype were killed, and the mutation frequencies at the lacI locus in spleen and small intestinal epithelium were determined. Mutant plaques were verified in a second round of plating at low density. The results for spleen before and after replating are shown in Fig. 2. The splenocytes in Mbd4−/− mice displayed an average mutation frequency of 9.83 × 10−5, an almost 3-fold increase compared with 3.47 × 10−5 in WT mice. A similar trend was detected in the small intestinal epithelial cells, where we observed more than a 2-fold increase in the mutational frequency in Mbd4−/− mice (6.34 × 10−5) compared with WT mice (3.00 × 10−5).
Fig 2.
lacI mutation frequency in the spleen of WT and Mbd4−/− mice. Mutational frequencies before and after replating are shown, along with the corresponding standard error bars.
We also determined the spectra of lacI mutations in the small intestine in WT and Mbd4−/− mice. In both WT and Mbd4−/− mice, the majority of lacI mutations were caused by C:G→T:A transitions (Table 1). However, this type of mutation was almost twice as prevalent in the Mbd4−/− mice compared with WT mice (64% vs. 37%; P = 0.01). In addition, almost all of the C:G→T:A mutations in the Mbd4−/− mice were found at CpG sites (96%; 45/47). The overall frequency of C:G→T:A mutations occurring specifically at CpG sites was more than twice as high in Mbd4−/− mice than in WT mice (62% and 28%, respectively; P = 0.001).
Table 1.
Mutation spectrum of lacI mutants
| WT (%) | Mbd4−/− (%) | |
|---|---|---|
| Base substitutions | 41 (76) | 60 (82) |
| Transitions | 23 (43) | 51 (70) |
| C:G→T:A, all | 20 (37) | 47 (64) |
| At CpG sites | 15 (28) | 45 (62) |
| At non-CpG sites | 5 (9) | 2 (3) |
| T:A→C:G | 3 (6) | 4 (5) |
| Transversions | 18 (33) | 9 (12) |
| C:G→A:T | 9 (17) | 4 (5) |
| C:G→G:C | 4 (7) | 1 (1) |
| T:A→G:C | 2 (4) | 1 (1) |
| T:A→A:T | 3 (6) | 3 (4) |
| Insertions/deletions | 13 (24) | 13 (18) |
| Insertions | 13 (24) | 12 (16) |
| Deletions | 0 (0) | 1 (1) |
| Total sequences | 54 (100) | 73 (100) |
Compared with WT mice:
, P = 0.01;
, P = 0.001; χ2 test.
Mbd4 Deficiency Accelerates Apc-Driven GI Tumorigenesis.
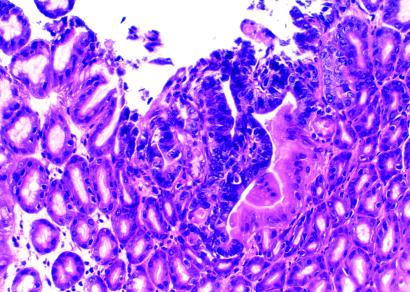
To specifically study the effect of the increased mutational frequency in Mbd4−/− mice on GI tumorigenesis, we introduced the Apc1638N allele (17) into the Mbd4-deficient background. Mbd4−/−/Apc1638N/+ and WT/Apc1638N/+ were killed at 10 mo of age, and the tumor incidence and multiplicity in the GI tract were determined. The majority of WT/Apc1638N/+ mice and all of the Mbd4−/−/Apc1638N/+ had developed GI tumors at this age (Table 2). We did not observe any tumors outside the digestive tract based on gross inspection of other organs. The overall tumor multiplicity in the Mbd4−/−/Apc1638N/+ was 2.4 times that of Apc1638N/+ single mutant mice at a borderline significance (P = 0.074). However, when specific regions of the GI tract were examined, we found statistically significant (P < 0.05) increases in tumor numbers in the jejunum and ileum (Table 3). Subsets of tumors from Mbd4−/−/Apc1638N/+double mutant and Apc1638N/+ single mutant mice were histologically classified. In both strains of mice, a spectrum of benign and malignant tumors was seen (Table 4). A comparison of the histologic types of the tumors between the two mouse strains showed on average three times as many adenocarcinoma in Mbd4−/−/Apc1638N/+ mice as compared with Apc1638N/+ mice. However, this difference did not reach statistical significance because of the relatively high standard deviation due to the limited number of mice available for analysis. In addition, closer microscopic inspection of the GI tract revealed a 10-fold increase in the number of microadenomas in the Mbd4−/−/Apc1638N/+ mice (2.70 ± 2.58 per mouse) compared with Apc1638N/+ single mutant mice (0.25 ± 0.46 per mouse; P = 0.025; Fig. 3).
Table 2.
Tumor incidence of Mbd4−/−/Apc1638N/+ mice
| Genotype (n)
|
Age, wk
|
Sex (M:F)
|
No. (%) of mice with tumors
|
GI tract | ||
|---|---|---|---|---|---|---|
| Stomach (%) | Small intestine (%) | Large intestine (%) | ||||
| WT/Apc1638N/+ (8) | 39.7 ± 3.0 | 1:1.7 | 7 (88) | 3 (38) | 7 (88) | 0 (0) |
| Mbd4−/−/Apc1638N/+ (10) | 40.4 ± 2.5 | 1:1 | 10 (100) | 8 (80) | 10 (100) | 0 (0) |
Mean ± SD. n, Number of mice studied.
Table 3.
Multiplicity of GI tumors of Mbd4−/−/Apc1638N/+ mice
| Genotype
|
Total
|
Stomach
|
Small intestine | |||
|---|---|---|---|---|---|---|
| Subtotal | Duodenum | Jejunum | Ileum | |||
| WT/Apc1638N/+ | 5.02 ± 4.47 | 0.63 ± 0.74 | 4.39 ± 4.50 | 2.63 ± 0.74 | 1.63 ± 2.33 | 0.13 ± 0.35 |
| Mbd4−/−/Apc1638N/+ | 12.20 ± 9.87 | 1.80 ± 1.75 | 10.40 ± 9.11 | 2.70 ± 1.70 | 6.30 ± 7.48 | 1.40 ± 1.65 |
Excluding microadenomas. Mean ± SD. Compared with Apc1638N/+ mice:
, P = 0.074,
, P < 0.05; Mann–Whitney test.
Table 4.
Histologic types of GI tumors in Mbd4 mutant mice
| Genotype
|
Total no. of tumors found
|
No. (%) of tumors examined
|
Histologic tumor types per mouse | |||
|---|---|---|---|---|---|---|
| Adenocarcinoma | Tubulovillous adenoma | Tubular adenoma | Microadenoma | |||
| WT/Apc1638N/+ | 40 | 27 (68) | 1.00 ± 1.20 | 0.50 ± 0.76 | 1.88 ± 1.89 | 0.25 ± 0.46 |
| Mbd4−/−/Apc1638N/+ | 122 | 85 (70) | 3.40 ± 4.62 | 0.40 ± 0.52 | 4.70 ± 3.59 | 2.70 ± 2.58 |
Mean ± SD. Compared with Apc1638N/+ mice:
, P = 0.072,
, P = 0.025; ANOVA test.
Fig 3.
Microadenoma in the GI tract of an Mbd4−/− /Apc1638N/+ mouse. Tumor located in the stomach with characteristic features of microadenoma, composed of a cluster of neoplastic glands of varying sizes and shapes. The tumor cells in the glands are crowded and show enlarged nuclei and hyperchromatic staining. Hematoxylin/eosin ×200.
Mbd4 Deficiency Increases Somatic Apc Mutations in GI Tumors.
We next examined the status of the WT Apc allele in the GI tumors from both groups of mice. A total of 22 WT/Apc1638N/+ and 29 Mbd4−/−/Apc1638N/+ tumors were analyzed for Apc truncation mutations by in vitro transcription and translation analysis. Truncated Apc products were detected in 23 of 29 (79%) Mbd4−/−/Apc1638N/+ tumors whereas only 7 of 22 (32%) were found in WT/Apc1638N/+ tumors. In eight of the Mbd4−/−/Apc1638N/+ tumor samples, more than one truncated polypeptide was identified by in vitro transcription and translation (Table 5). The 2.5-fold increase in somatic Apc mutations in the double mutant Mbd4−/−/Apc1638N/+ mice was highly significant (P = 0.001).
Table 5.
Analysis of Apc truncation mutations in intestinal tumors from Mbd4−/−/Apc1638N/+ mice
| WT/Apc1638N/+ (%) | Mbd4−/−/Apc1638N/+ (%) | |
|---|---|---|
| Tumors | 22 (100) | 29 (100) |
| With Apc truncations | 7 (32) | 23 (79) |
| With >1 mutant allele | 0 (0) | 8 (28) |
| Total Apc mutations | 7 (100) | 34 (100) |
Combined with data from ref. 20.
Compared with Apc1638N/+ mice: P = 0.001; Fisher's exact test.
Prevalence of C:G→T:A Transitions at CpG Sites in Apc.
The Apc truncation mutations in the GI tumors were further characterized by sequencing. A summary of the Apc mutations together with the surrounding sequences is shown in Table 6. The vast majority of Apc mutations in Mbd4−/−/Apc1638N/+ tumors were comprised of C:G→T:A transitions (28/34, 82%) whereas a smaller proportion of WT/Apc1638N/+ tumors were found to carry this type of Apc mutation (4/7, 57%). We also detected a small number of other mutations, including C:G→A:T and G:C→A:T transitions, as well as insertions or deletions of two or more nucleotides (Table 6). Examination of the sequences surrounding each Apc mutation site in Mbd4−/−/Apc1638N/+ tumors showed that all of the C:G→T:A mutations had occurred at CpG sites within the Apc coding region.
Table 6.
Sequence Analysis of Apc mutations in Apc1638N/+ and Mbd4−/−/Apc1638N tumors
| Codon | Mutation | Consequence | Wild-type sequence | WT/Apc1638N/+ | Mbd4−/−/Apc1638N/+ |
|---|---|---|---|---|---|
| 803 | C→T | Arg→Stop | GCC AAT CGA CAT GAT | — | 3 |
| 854 | C→T | Arg→Stop | GAG AGA GAG CGA GGT | — | 8 |
| 874 | C→T | Arg→Stop | TCA AAA CGA GGT CTG | 1 | 2 |
| 921 | C→T | Arg→Stop | GCG GCA CGA AGA AGC | 2 | 5 |
| 934 | ins TACA | frameshift | AAC ACA TAC AAC TTC | 1 | — |
| 939 | G→T | Glu→Stop | AAG TCG GAA AAT TCA | 1 | — |
| 956 | C→T | Arg→Stop | TAT AAA CGA TCT TCA | — | 7 |
| 989 | G→T | Glu→Stop | GAT GAT GAA AGT AAA | — | 1 |
| 992 | del 8bp, ins A | frameshift | AAA TTT TGC AGT TAT | 1 | — |
| 993 | C→A | Cys→Stop | AAA TTT TGC AGT TAT | — | 1 |
| 1018 | G→T | Glu→Stop | GAT GGA GAA CTG GAT | — | 1 |
| 1047 | G→A | Trp→Stop | GAA AGG TGG GCA AGA | 1 | — |
| 1112 | C→T | Arg→Stop | ACA AAT CGA ATG GGT | — | 2 |
| 1128 | del GT | frameshift | TCT CTG TGT CAG GAA | — | 1 |
| 1195 | C→A | Ser→Stop | TCA TTT TCA TTC TCA | — | 1 |
| 1287 | del 8bp | frameshift | GGA TGT GAT CAG ACA | — | 1 |
| 1334 | C→T | Arg→Stop | CCC AGC CGA CTC CAG | — | 1 |
| Total | 7 | 34 |
Site of mutation is shown in bold.
Combined with data from ref. 20. ins, insertion; del, deletion.
Discussion
We have generated a mouse line with a null mutation in Mbd4 to study the in vivo role of this gene in DNA repair and tumorigenesis. The phenotypic analysis of this mouse line showed that loss of Mbd4 function was compatible with normal development. We did not observe any reduction in the survival of Mbd4+/− or Mbd4−/− mutant mice as compared with WT mice. Similarly, there was no significant increase in the cancer susceptibility phenotype of the Mbd4 mutant mice even at an old age of up to 24 mo. These results suggest that inactivating mutations in Mbd4 alone do not have a significant impact on the initiation of tumorigenesis in mammals.
We next assessed the impact of Mbd4 inactivation on MMR in vivo by studying the mutation rates at microsatellite loci in the genomes of Mbd4−/− mice. The genomes of Mbd4-deficient mice remained stable at all loci tested, whereas the genomes of Mlh1-deficent mice displayed MSI at high frequencies. These results indicate that inactivation of Mbd4 does not impair the repair of frameshift mutations in vivo and are consistent with in vitro studies that showed that loss of MBD4 function does not affect MMR in a biochemical assay system (23). Furthermore, these findings support the notion that the observed mutations in the short mononucleotide repeat tract in the coding region of MBD4 of microsatellite unstable colorectal tumors are secondary to a mutator phenotype that is caused by MMR-deficiency or other repair deficiencies. Similar short repeat tract mutations in microsatellite unstable colorectal cancers are frequently found in the coding regions of a number of other genes that are important for tumor formation, including TGF-βRII, IGF-IIR, and BAX, and the MMR genes MSH3 and MSH6 (14, 24). If Mbd4 inactivation causes a mutator phenotype in cancer cells, it could potentially impact tumorigenesis. Consequently, the analysis of the mutation frequencies at the lacI gene in the Big Blue reporter system showed that inactivation of Mbd4 led to a small but significant increase in somatic mutations in the spleen and small intestine. Strikingly, the majority of lacI mutations in the small intestinal epithelium that had occurred were C:G→T:A transition mutations at CpG sites.
The lack of an increased cancer susceptibility phenotype in Mbd4−/− mice even at a very old age suggests that the mutator phenotype caused by Mbd4 inactivation is not sufficient to initiate tumorigenesis by itself. A comparison with Msh2-, Msh6-, and Mlh1-deficient mice, which display strong cancer phenotypes, shows that MMR-deficiency results in small intestinal mutation frequencies that are almost an order of magnitude higher than WT (26–28). However, it is also possible that loss of other functions of MMR proteins, such as the induction of apoptosis in response to DNA damage, can contribute to the cancer predisposition phenotype in MMR-deficient animals (29). The relatively moderate mutator phenotype in Mbd4−/− mice may also be explained by the more limited substrate specificity of Mbd4 as compared with MMR proteins. In addition, spontaneous deamination of 5-methylcytosine occurs at a very high frequency in the genome (30), and organisms appear to have evolved redundant repair mechanisms to respond to this type of DNA lesion. For example, biochemical analysis indicates that thymine DNA glycosylase (TDG) shares overlapping enzymatic activity with Mbd4 for the repair of T-G mismatches (31). It will be interesting to test whether the combined loss of Mbd4 and TDG will further increase the incidence of C:G→T:A mutations and initiate tumorigenesis.
The small increase in mutation frequency and the change in the mutation spectrum caused by loss of Mbd4 function had a significant impact on Apc-driven GI tumorigenesis. We observed significant increases in tumor multiplicity in the jejunum and ileum of Mbd4−/−/Apc1638N/+ mice. The increase in tumor multiplicity in the jejunum parallels the observed increase in mutation frequency in this tissue. It will be interesting to investigate whether the other parts of the GI tract show a similar phenomenon. In addition, Mbd4 deficiency had a considerable impact on the tumor spectrum. When comparing the histologic types of tumors found in Mbd4−/−/Apc1638N/+ mice to those of Apc1638N/+ mice, the number of tumors that had progressed into adenocarcinomas increased in Mbd4−/−/Apc1638N/+ mice. The most dramatic difference, however, was seen in the number of microadenomas, microscopically detected early-stage neoplasm with invasive potential (Fig. 3). The Mbd4−/−/Apc1638N/+ double mutant displayed a 10-fold increase in the number of microadenomas as compared with the single Apc1638N/+ mutant. These results indicate that loss of Mbd4 function in the context of a cancer susceptibility allele can modify the tumor distribution in the GI tract and has a significant effect on tumor development.
The differences in the GI tumor phenotype between Mbd4−/−/Apc1638N/+ mice and Apc1638N/+ mice are associated with the genome-wide accumulation of C:G→T:A mutation at CpG sites, as indicated by the increase in somatic lacI mutations. However, one of the earliest events in GI tumor formation in mice is the inactivation of the Apc tumor suppressor function (32–34). Our analysis of the impact of Mbd4 deficiency on mutations in the remaining WT Apc allele in Mbd4−/−/Apc1638N/+ GI tumors showed that a large majority of these tumors had acquired somatic truncation mutations in the WT Apc allele. In comparison, somatic Apc truncation mutations were detected in only a small proportion of Apc1638N/+ tumors. The latter finding is consistent with the previous observation that 70–80% of Apc1638N/+ tumors lose the WT Apc allele by a loss of heterozygosity event (35) and indicates that the mutator phenotype caused by Mbd4 deficiency had a major impact on the genetic mechanism leading to Apc gene inactivation.
Sequence analysis of the Apc mutations in the Mbd4−/−/Apc1638N/+ tumors showed that the increase in the incidence of somatic Apc mutations was largely caused by an increase in C:G→T:A mutations at CpG sites. A calculation taking into account the increase in the incidence of somatic Apc mutations (2.5-fold) as well as the increase in C:G→T:A mutations (2-fold) suggests that mutations at CpG sites in the Apc coding region occur with a 5-fold higher likelihood in the small intestinal mucosa of Mbd4−/− mutant mice as compared with WT mice. Interestingly, applying the same calculation for the lacI region tested resulted in a similar 5-fold increase of CpG mutations. Based on this assumption, we speculate that loss of Mbd4 function can also increase the probability of CpG mutations in the coding region of other genes that effect GI tumor formation. For example, nearly 50% of somatic TP53 mutations in colorectal cancers are comprised of C:G→T:A transitions at CpG sites (36, 37), suggesting that Mbd4 inactivation might also have a significant impact on TP53 mutations.
In summary, this study provides evidence that Mbd4 functions in vivo in the suppression of C:G→T:A mutations at CpG sites in mammalian genomes. Our results further suggest that, although the mutator phenotype caused by Mbd4 inactivation by itself is not sufficient to initiate tumorigenesis in mice, it can have a modifying effect on tumor formation in cancer-predisposing genetic backgrounds. Similarly, it is possible that loss of Mbd4 function will have significant impact on the response to environmental or carcinogenic challenges, an idea that can be tested in future studies of Mbd4 mutant mice.
Note
While this manuscript was in preparation, another study was published that reported the effect of a mutagenic insertion in intron 1 of the mouse Mbd4 gene on the mutation frequencies at the λ phage cII locus in liver and spleen (25). The analysis in these tissues showed a comparable 3.3-fold increase in mutation frequencies, as well as in the incidence of C:G→T:A transitions at CpG dinucleotides. Bisulfite sequencing of the cII locus further showed that the majority of CpG sites in liver and spleen were methylated, implying that the high incidence of C:G→T:A mutations was the result of failure to repair spontaneous 5-methylcytosine deamination at methylated CpG residues. Our observation that the lacI locus in the epithelial cells of the small intestine displays C:G→T:A transitions almost exclusively at CpG sites further supports this notion.
Acknowledgments
This work was supported by grants to W.E. (CA76329 and CA93484), and A.M.C.B. (CA47207), Center Grant CA13330 to the Albert Einstein College of Medicine from the National Institutes of Health, and an American Association for Cancer Research–Cancer Research Foundation of America Fellowship (to M.K).
Abbreviations
Mbd4, methyl-CpG binding domain 4
MMR, DNA mismatch repair
MSI, microsatellite instability
GI, gastrointestinal
ES, embryonic stem
References
- 1.Kolodner R. (1996) Genes Dev. 10, 1433-1442. [DOI] [PubMed] [Google Scholar]
- 2.Modrich P. & Lahue, R. (1996) Annu. Rev. Biochem. 65, 101-133. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg E. C. (2001) Nat. Rev. Cancer 1, 22-33. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T. (2000) Mutat. Res. 462, 129-135. [DOI] [PubMed] [Google Scholar]
- 5.Hoeijmakers J. H. (2001) Nature 411, 366-374. [DOI] [PubMed] [Google Scholar]
- 6.Hendrich B. & Bird, A. (1998) Mol. Cell. Biol. 18, 6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendrich B., Hardeland, U., Ng, H. H., Jiricny, J. & Bird, A. (1999) Nature 401, 301-304. [DOI] [PubMed] [Google Scholar]
- 8.Petronzelli F., Riccio, A., Markham, G. D., Seeholzer, S. H., Genuardi, M., Karbowski, M., Yeung, A. T., Matsumoto, Y. & Bellacosa, A. (2000) J. Cell. Physiol. 185, 473-480. [DOI] [PubMed] [Google Scholar]
- 9.Bellacosa A., Cicchillitti, L., Schepis, F., Riccio, A., Yeung, A. T., Matsumoto, Y., Golemis, E. A., Genuardi, M. & Neri, G. (1999) Proc. Natl. Acad. Sci. USA 96, 3969-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellacosa A. (2001) Cell Death Differ. 8, 1076-1092. [DOI] [PubMed] [Google Scholar]
- 11.Riccio A., Aaltonen, L. A., Godwin, A. K., Loukola, A., Percesepe, A., Salovaara, R., Masciullo, V., Genuardi, M., Paravatou-Petsotas, M., Bassi, D. E., et al. (1999) Nat. Genet. 23, 266-268. [DOI] [PubMed] [Google Scholar]
- 12.Bader S., Walker, M., Hendrich, B., Bird, A., Bird, C., Hooper, M. & Wyllie, A. (1999) Oncogene 18, 8044-8047. [DOI] [PubMed] [Google Scholar]
- 13.Miyaki M., Iijima, T., Shiba, K., Aki, T., Kita, Y., Yasuno, M., Mori, T., Kuroki, T. & Iwama, T. (2001) Oncogene 20, 5215-5218. [DOI] [PubMed] [Google Scholar]
- 14.Yamada T., Koyama, T., Ohwada, S., Tago, K. I., Sakamoto, I., Yoshimura, S., Hamada, K., Takeyoshi, I. & Morishita, Y. (2002) Cancer Lett. 181, 115-120. [DOI] [PubMed] [Google Scholar]
- 15.Ioffe E., Liu, Y., Bhaumik, M., Poirier, F., Factor, S. M. & Stanley, P. (1995) Proc. Natl. Acad. Sci. USA 92, 7357-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohler S. W., Provost, G. S., Fieck, A., Kretz, P. L., Bullock, W. O., Sorge, J. A., Putman, D. L. & Short, J. M. (1991) Proc. Natl. Acad. Sci. USA 88, 7958-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fodde R., Edelmann, W., Yang, K., van Leeuwen, C., Carlson, C., Renault, B., Breukel, C., Alt, E., Lipkin, M., Khan, P. M., et al. (1994) Proc. Natl. Acad. Sci. USA 91, 8969-8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuraguchi M., Yang, K., Wong, E., Avdievich, E., Fan, K., Kolodner, R. D., Lipkin, M., Brown, A. M., Kucherlapati, R. & Edelmann, W. (2001) Cancer Res. 61, 7934-7942. [PubMed] [Google Scholar]
- 19.Zeger S. L., Liang, K. Y. & Alberts, P. S. (1998) Biometrics 44, 1049-1060. [PubMed] [Google Scholar]
- 20.Kucherlapati M., Yang, K., Kuraguchi, M., Zhao, J., Lia, M., Heyer, J., Kane, M. F., Fan, K., Russell, R., Brown, A. M., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 9924-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendrich B., Abbott, C., McQueen, H., Chambers, D., Cross, S. & Bird, A. (1999) Mamm. Genome 10, 906-912. [DOI] [PubMed] [Google Scholar]
- 22.Edelmann W., Cohen, P. E., Kane, M., Lau, K., Morrow, B., Bennett, S., Umar, A., Kunkel, T., Cattoretti, G., Chaganti, R., et al. (1996) Cell 85, 1125-1134. [DOI] [PubMed] [Google Scholar]
- 23.Drummond J. T. & Bellacosa, A. (2001) Nucleic Acids Res. 29, 2234-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peltomaki P. (2001) Hum. Mol. Genet. 10, 735-740. [DOI] [PubMed] [Google Scholar]
- 25.Millar C. B., Guy, J., Sansom, O. J., Selfridge, J., MacDougall, E., Hendrich, B., Keightley, P. D., Bishop, S. M., Clarke, A. R. & Bird, A. (2002) Science 297, 403-405. [DOI] [PubMed] [Google Scholar]
- 26.Andrew S. E., McKinnon, M., Cheng, B. S., Francis, A., Penney, J., Reitmair, A. H., Mak, T. W. & Jirik, F. R. (1998) Proc. Natl. Acad. Sci. USA 95, 1126-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mark S. C., Sanderock, L. E., Luchman, H. A., Baross, A., Edelmann, W. & Jirik, F. R. (2002) Oncogene 21, 7126-7130. [DOI] [PubMed] [Google Scholar]
- 28.Baross-Francis A., Makhani, N., Liskay, R. M. & Jirik, F. R. (2001) Oncogene 20, 619-625. [DOI] [PubMed] [Google Scholar]
- 29.Fishel R. (2001) Cancer Res. 61, 7369-7374. [PubMed] [Google Scholar]
- 30.Laird P. W. & Jaenisch, R. (1996) Annu. Rev. Genet. 30, 441-464. [DOI] [PubMed] [Google Scholar]
- 31.Neddermann P. & Jiricny, J. (1993) J. Biol. Chem. 268, 21218-21224. [PubMed] [Google Scholar]
- 32.Levy D. B., Smith, K. J., Beazer-Barclay, Y., Hamilton, S. R., Vogelstein, B. & Kinzler, K. W. (1994) Cancer Res. 54, 5953-5958. [PubMed] [Google Scholar]
- 33.Luongo C., Moser, A. R., Gledhill, S. & Dove, W. F. (1994) Cancer Res. 54, 5947-5952. [PubMed] [Google Scholar]
- 34.Oshima M., Oshima, H., Kitagawa, K., Kobayashi, M., Itakura, C. & Taketo, M. (1995) Proc. Natl. Acad. Sci. USA 92, 4482-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smits R., Kartheuser, A., Jagmohan-Changur, S., Leblanc, V., Breukel, C., de Vries, A., van Kranen, H., van Krieken, J. H., Williamson, S., Edelmann, W., et al. (1997) Carcinogenesis 18, 321-327. [DOI] [PubMed] [Google Scholar]
- 36.Schmutte C., Yang, A. S., Beart, R. W. & Jones, P. A. (1995) Cancer Res. 55, 3742-3746. [PubMed] [Google Scholar]
- 37.Kleihues P., Schauble, B., zur Hausen, A., Esteve, J. & Ohgaki, H. (1997) Am. J. Pathol. 150, 1-13. [PMC free article] [PubMed] [Google Scholar]