Abstract
The primary cause of Duchenne muscular dystrophy (DMD) is a mutation in the dystrophin gene leading to the absence of the corresponding RNA transcript and protein. Absence of dystrophin leads to disruption of the dystrophin-associated protein complex and substantial changes in skeletal muscle pathology. Although the histological pathology of dystrophic tissue has been well documented, the underlying molecular pathways remain poorly understood. To examine the pathogenic pathways and identify new or modifying factors involved in muscular dystrophy, expression microarrays were used to compare individual gene expression profiles of skeletal muscle biopsies from 12 DMD patients and 12 unaffected control patients. Two separate statistical analysis methods were used to interpret the resulting data: t test analysis to determine the statistical significance of differential expression and geometric fold change analysis to determine the extent of differential expression. These analyses identified 105 genes that differ significantly in expression level between unaffected and DMD muscle. Many of the differentially expressed genes reflect changes in histological pathology. For instance, immune response signals and extracellular matrix genes are overexpressed in DMD muscle, an indication of the infiltration of inflammatory cells and connective tissue. Significantly more genes are overexpressed than are underexpressed in dystrophic muscle, with dystrophin underexpressed, whereas other genes encoding muscle structure and regeneration processes are overexpressed, reflecting the regenerative nature of the disease.
The muscular dystrophies are a group of clinically and genetically heterogeneous myopathic disorders characterized by progressive degenerative changes in skeletal muscle fibers. This group of genetically distinct disorders shares clinical and pathological characteristics but varies in severity, inheritance pattern, and molecular defect. Duchenne muscular dystrophy (DMD) is the most common of these disorders, affecting 1 in 3,500 male births. DMD is caused by mutations or deletions in the dystrophin gene leading to its reduction at the mRNA level and absence at the protein level. The cloning of the dystrophin gene led to the characterization of dystrophin as a large cytoskeletal protein associated with a protein complex [the dystrophin-associated protein complex (DAPC)] that links the cytoskeleton to the extracellular matrix. The DAPC is a large multicomponent complex that includes the dystroglycans, the sarcoglycans, the syntrophins, and sarcospan. The DAPC is disrupted in many other forms of muscular dystrophy and mutations in genes encoding DAPC components other than dystrophin are associated with the limb-girdle muscular dystrophies (see refs. 1–3 for review).
In skeletal muscle fibers, the DAPC spans the sarcolemma forming a structural link between the extracellular matrix and the cytoskeleton via the laminin-binding protein, α-dystroglycan, and the actin-binding protein, dystrophin. It has been proposed that this link allows the DAPC to stabilize the membrane against contraction-induced damage. The generally accepted model for muscular dystrophy is that disruption of the DAPC breaks a mechanical linkage crucial for sarcolemmal integrity. Physical sarcolemmal breaks or calcium leak channel openings then elevate intracellular free calcium, triggering calcium-activated proteases and fiber necrosis (4). However, the DAPC has been implicated in other roles. There is evidence that the DAPC is involved in signaling via its interactions with Grb2, calmodulin, and neuronal nitric oxide synthase (NOS) (5). Other evidence suggests that the DAPC may act as a scaffold to spatially organize cell signaling network components necessary for myofiber homeostasis (6). Roles for DAPC signaling in microvascular function (7, 8) and muscle fiber type determination have also been suggested (9). The role of the DAPC in normal muscle and how its function is disrupted in dystrophic muscle, although emerging, are still unclear. Additionally, mutations in non-DAPC protein encoding genes, such as calpain 3 (10) and caveolin 3 (11), have been shown to cause muscular dystrophy, substantiating the theory that more than one molecular pathway contributes to the disease phenotype.
The histological picture of dystrophic muscle differs substantially from that of unaffected muscle. In dystrophic muscle, there is muscle fiber necrosis and incomplete regeneration, variation in fiber size, centralization of nuclei, proliferation of connective and adipose tissue, infiltration of immune cells, altered metabolic capacity, reduced blood supply, activation of apoptotic pathways, and presumably other, unknown, processes (12). Despite knowledge of the primary genetic defects and a well-documented histological pathology, the molecular pathology leading to muscle cell degeneration in the muscular dystrophies is poorly understood. None of the existing models for the role of the DAPC, or how disruption of the DAPC leads to the disease phenotype, can account for all of these features, suggesting additional processes are at work.
Large-scale parallel gene expression analysis allows the examination of the molecular pathophysiological pathways of dystrophic muscle in a physiological context and the comparison of how these pathways differ from those in normal muscle. This information allows observations from histological pathology to be contrasted with molecular pathological findings (13). Several important caveats notwithstanding (14–17), a profile of the transcriptome across the diseased and healthy states might highlight the involvement of a previously unsuspected regulatory and/or pathological pathway. Additionally, quantitatively minor components with important control functions might be identified.
This powerful tool has been the basis of a number of recent publications in which dystrophic muscle is compared with unaffected muscle. The majority of these studies have used a mouse model of muscular dystrophy (18–22), but a comparison of gene expression differences in DMD patients has also been published (23). We present a more extensive study of DMD patients. Here, the biopsies are not pooled, as has been previously published (23); instead, each muscle biopsy (12 DMD and 12 unaffected) is analyzed independently, and two separate statistical analyses are used in combination to derive a common list of differentially expressed genes. Thus the power of a large sample size is used to offset individual genetic variability and experimental noise. We confirm the differential expression of 42 genes identified in previous studies and add 63 genes previously unidentified as differentially expressed in DMD muscle.
Materials and Methods
Patient Samples.
Twelve quadriceps biopsies from DMD patients were compared with 12 quadriceps biopsies from unaffected controls. The 12 DMD biopsies were from young (5- to 7-year-old) males. The unaffected biopsies were primarily from young males (seven biopsies) but included three from adult males and two from females (one child, one adult). The unaffected controls expressed normal levels of dystrophin. The DMD patients showed clinical symptoms consistent with a DMD diagnosis, and the biopsies were shown to be dystrophin deficient by immunofluorescence and/or Western blotting. All biopsies were obtained under institutionally approved protocols.
Target Preparation and Array Hybridization.
Total RNA was extracted from muscle biopsies (70–120 mg) by using Trizol (Invitrogen). After resuspension in DEPC-treated water, RNA was prepared for hybridization to Affymetrix (Santa Clara, CA) HG-U95Av2 arrays according to the manufacturer. Double-stranded cDNA was synthesized (Superscript Double-Stranded cDNA Synthesis kit, Invitrogen) and used in an in vitro transcription (IVT) reaction with biotin-labeled nucleotides (Enzo BioArray High Yield RNA Transcript Labeling kit, Affymetrix). Purified (RNeasy kit, Qiagen, Chatsworth, CA), fragmented (35–200 nucleotides) biotinylated cRNA, together with IVT controls (according to Affymetrix recommendations) was hybridized to HG-U95Av2 GeneChips for 16–18 h at 45°C. Standard posthybridization wash and double-stain protocols used an Affymetrix Fluidics Station 400. The GeneChips were scanned in an Affymetrix/Hewlett–Packard G2500A Gene Array Scanner and the resulting signals quantified and stored.
Data Processing.
Affymetrix GeneChip Ver. 5.0 software (MAS5.0) was used for raw data processing and custom software for additional noise analysis and quality control (http://db.chip.org). MAS5.0 reports hybridization intensity by an aggregate non-negative numerical quantity, signal intensity. Duplicate experiments in which a single hybridization mixture is split and hybridized to two GeneChips were performed periodically for reproducibility assessment.
Statistical Analysis.
Two separate statistical methods were applied to assess differential gene expression between the two disease classes. Both techniques determine whether a probe set exhibits a significant change of its reported signal intensities across the two classes as compared with changes in difference or fold within the classes or amongst replicates. The term data or signal intensity henceforth denotes the normalized signal intensities.
Method 1: To identify probe sets with significant intensity differences between disease classes.
A standard two-tail unequal variance t test was applied to the unaffected control data set vs. the DMD data set (24). The threshold was set at P < 0.0001 and the results sorted by ascending P value.
Method 2: To determine the differentiability of a probe set by its signal intensity fold change.
To identify genes with significantly different fold change between disease classes, the approach described by Zhao et al. was applied (14). For each probe set, the reported signal intensities (aj in the unaffected control class and bj in the DMD class) are ordered by increasing size. The geometric fold mean (μ) and variance (σ2) of the probe set between these two classes are defined as follows:
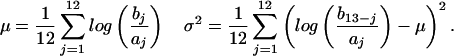 |
The natural logarithm is applied to maintain numerical symmetry. Additionally, for each probe set μcontrol and μDMD are calculated as follows
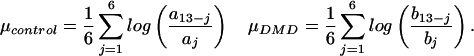 |
μcontrol and μDMD are the geometric average log fold arising from measurement variation within each disease class. μnoise is the greater of these two quantities. Because aj and bj are ordered, μnoise is non-negative. The gene is said to be overexpressed in DMD muscle with a significant fold change when:
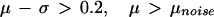 |
and underexpressed when:
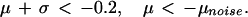 |
The choice of 0.2 is arbitrary. It was selected in association with a permutation test to determine the false discovery rate (25). For each probe set, the class labels were shuffled, the preceding analysis repeated, and the number of significantly overexpressed probe sets counted. This procedure was iterated 12,000 times, and the false discovery rate found to be 0.0023.
Quantitative TaqMan RT-PCR.
Two-step RT-PCR was performed on serial dilutions of quadriceps muscle RNA from seven of the 12 DMD biopsies used for microarray analysis and four of the 12 unaffected control biopsies. RT-PCR was performed on the ABI PRISM 7700 Sequence Detection System by using random hexamers from the TaqMan Reverse Transcription Reagents and RT Reaction Mix (Applied Biosystems) to reverse transcribe the RNA, and TaqMan Universal PCR Master Mix and Assays-on-Demand Gene Expression probes (Applied Biosystems) for the PCR step. A standard curve for serial dilutions of 18S rRNA was similarly generated. The relative standard curve method (Applied Biosystems) was used to calculate the amplification difference between DMD and unaffected controls for each primer set.
Results
Data Analysis.
The sensitivity of microarray-generated data to noise from experimental variables is well documented (26) and is compounded when the samples in the analysis are genetically heterogeneous. These variables can be offset by a sufficiently large sample size (24 in this case), appropriate experimental approach, and statistical analysis.
Background and Normalization.
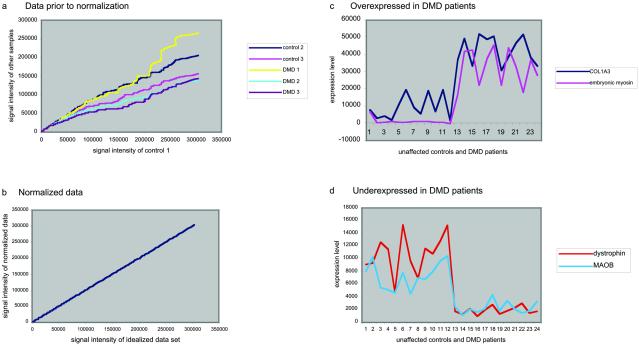
Although the MAS5.0 standardizes the overall intensity of each GeneChip to a user-defined target intensity, the resulting data sets are not normalized with respect to each other (Fig. 1 a and b). The signal intensities were therefore normalized, before any analysis, via a linear regression technique (14, 17, 27), wherein the signal intensities in each experiment are transformed so that their scatter plots have a slope one through the origin with respect to a reference data set. The reference data set is comprised of the average probe-by-probe signal intensities from an unaffected control experiment and a DMD experiment that have maximal average correlation coefficients against all other experiments within their disease class. Normalization corrects any uniform linear aberrations of the reported signal intensities between any two replicate measurements.
Fig 1.
(a) The raw signal intensity data from two control samples and three DMD samples are plotted against a third control sample demonstrating that the raw MAS5.0 standardized data sets are not normalized with respect to each other. (b) After linear regression normalization, the signal intensity plots have a linear regression with a slope of one through the origin against a reference data set. (c and d) Normalized expression values for 12 control (cases 1–12 on graph) and 12 DMD (13–24) patients are plotted, as independent data points, for two genes that are overexpressed (c) and two genes that are underexpressed (d) in DMD patients.
A difficulty in interpreting microarray data is that the signal intensity measurements reflecting mRNA expression levels can vary substantially between experiments. This is partly due to genetic heterogeneity, a factor of particular significance with human tissue, and partly due to idiosyncrasies in the probe or target preparation, hybridization and scanning. However, differences in expression between the two data sets can be identified (Fig. 1 c and d). To distinguish significant and substantial differential expression from this variation, two independent statistical methods were applied to assess differential gene expression.
Method 1: Differences.
A standard two-tail unequal variance t test was applied to the two data sets (24). The threshold was set at P < 0.0001 and the results sorted by ascending P value (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org). Two hundred forty-three probe sets, representing 235 different genes, were identified. Of these, 176 probe sets (173 genes) were overexpressed in the DMD patients vs. the unaffected class, and 67 probe sets (62 genes) were underexpressed. This method identifies genes with significant differences between the two data sets, but it does not consider the extent of the differential fold expression.
Method 2: Geometric Fold Change Analysis.
One hundred eighteen probe sets (representing 112 genes) were found to be significantly overexpressed and 21 genes underexpressed in the DMD disease class with respect to the unaffected controls (Table 4, which is published as supporting information on the PNAS web site). The false discovery rate was 0.0023. Thus, by chance alone, less than one of the 139 probe sets identified as differentially expressed may represent a false positive.
Final List of Genes.
To assure a minimal number of false positives, only the 110 probe sets (representing 105 genes) commonly yielded by the two methodologies were admitted into the list of genes differentially expressed in DMD muscle compared with unaffected muscle. These 105 genes were clustered into functional groups and ranked by probability (Table 5, which is published as supporting information on the PNAS web site).
Some genes are represented on the GeneChip by more than one probe set; e.g., collagen type I α 2 is represented by three probe sets, all of which are identified as differentially expressed by both analysis methods. However, this was not the case for all transcripts represented by multiple probe sets. A small number of genes showing consistency across their multiple probe sets were detected by only one or the other analytical method (Table 1).
Table 1.
Genes multiply identified by one statistical analysis method but not by the other
| Gene | Probe set | t test analysis: Probability | Geometric analysis: Fold change |
|---|---|---|---|
| RXRG | Probe 1 | 5.30E-06 | |
| Probe 2 | 6.99E-05 | ||
| Protein kinase C, θ | Probe 1 | 3.32E-05 | |
| Probe 2 | 5.66E-05 | ||
| EEF1A1L14 | Probe 1 | 4.87E-05 | |
| Probe 2 | 8.5E-06 | ||
| Osteopontin | Probe 1 | 45.5 | |
| Probe 2 | 11.7 | ||
| Phospholipase A2 | Probe 1 | 8.7 | |
| Probe 2 | 5.7 |
Probabilities of differential expression, from t test analysis, or fold change values, from geometric analysis, for genes multiply identified by one statistical analysis method but not identified by the other.
RT-PCR Validation.
The differential expression of 12 genes was confirmed by quantitative RT-PCR analysis of seven DMD biopsies and four unaffected biopsies. Fig. 2 shows the amplification traces for two of these genes, biglycan and embryonic myosin, in two of the DMD and two of the unaffected biopsies. RT-PCR fold change values for each gene were calculated and tabulated (Table 2), along with the fold change values obtained from geometric fold change analysis of the microarray data. The fold change values derived from each method differ slightly, partly because the sample set used for the RT-PCR analysis is different, but primarily because microarray analysis is not an absolute measure of gene expression. Interestingly, one of the genes confirmed, by RT-PCR, as overexpressed in DMD muscle was osteopontin. The differential expression of osteopontin was in some doubt, as both probe sets representing its transcript are identified as differentially expressed only by geometric fold change analysis (Table 1).
Fig 2.
TaqMan RT-PCR results for biglycan and embryonic myosin (MYH3). For each gene, profiles of product appearance from two DMD (red traces) and two unaffected biopsies (black traces) are shown. The x axis indicates the number of cycles to product appearance. In both cases, product amplification from the DMD biopsy preceded amplification from the unaffected biopsy, verifying that both biglycan and embryonic myosin are overexpressed in DMD muscle versus unaffected muscle.
Table 2.
Genes whose differential expression was verified by TaqMan RT-PCR
| Gene | Microarray fold change | RT-PCR fold change |
|---|---|---|
| Embryonic myosin | +55.0 | +100.4 |
| Perinatal myosin | +18.0 | +52.4 |
| Proenkephalin | +8.0 | +11.9 |
| Runt-related transcription factor 1 | +6.4 | +2.2 |
| Lumican | +6.1 | +2.3 |
| Biglycan | +4.2 | +5.1 |
| Transforming growth factor β | +3.0 | +1.5 |
| Guanidioacetate N-methyltransferase | −2.1 | −5.2 |
| Leiomodin 1 | −3.2 | −5.2 |
| Monoamine oxidase B | −3.3 | −3.6 |
| Phosphodiesterase 4D | −4.8 | −6.0 |
| Osteopontin, probe sets 1 and 2 | +47.5 | +15.1 |
| +11.7 |
Fold change values from geometric fold change analysis of microarray data and standard curve analysis of RT-PCR data are given for each gene.
Discussion
Overview.
Although the histological pathology of dystrophic skeletal muscle is well described, the causative molecular pathways are poorly understood. To examine the pathogenic pathways, Affymetrix HG-U95Av2 GeneChips were used to compare individual gene expression profiles of skeletal muscle biopsies from 12 DMD patients to 12 unaffected controls. Two stringent statistical analysis applications were used to evaluate the data. Geometric fold change analysis identified genes whose expression levels substantially changed between DMD and unaffected skeletal muscle (14). A two-sample t test of unequal variance confirmed that the degree of differential expression is statistically significant and assigned a probability of differential expression to each gene (24). One hundred five genes with statistically significant differential expressions were identified by both methods, and the differential expression of a subset of these genes was confirmed by quantitative RT-PCR. Over 50% of these genes have not previously been identified as differentially expressed in dystrophic versus normal skeletal muscle (Table 5). These genes increase our knowledge of DMD expression patterns and augment our understanding of the involvement of specific pathways
The histological appearance of dystrophic muscle differs substantially from normal muscle. Dystrophic muscle shows necrosis and regeneration, fiber size variation, proliferation of connective and adipose tissue, infiltration of immune cells, altered metabolic capacity, and reduced blood supply amongst other processes (12). A striking feature of the data presented is the extent to which the molecular differences correlate with the histopathological differences.
As expected, dystrophin is substantially underexpressed in DMD muscle (28, 29). Significantly more genes are overexpressed in dystrophic muscle than underexpressed, compared with unaffected muscle. This now consistent finding (20, 22) is attributed to an increase in protein turnover due to the degenerative and regenerative nature of the disease. Many muscle structure genes, and genes normally expressed only in developing muscle, are overexpressed in DMD muscle, again reflecting regeneration. Immune response signals and extracellular matrix genes are also overexpressed in dystrophic muscle reflecting infiltration of inflammatory cells and connective tissue.
In contrast to work in mdx mice, where there was an overwhelming inflammatory response (22), there was little evidence of such a response in our studies, probably due to phenotypic differences. The dystrophin-deficient phenotype of mdx hindlimb muscle is milder than that of DMD patients. mdx hindlimb muscle undergoes a period of intense necrosis and regeneration at ≈3–4 weeks of age (30, 31), after which muscle damage appears to plateau, and, despite the lack of dystrophin, displays only mild weakness.
The results presented here differ substantially from previous DMD microarray studies (23), likely due to differences in data analysis and experimental design. In this study, data are reported from a large number of individual patients, rather than from pooled samples from different patients. This approach increases the statistical power of the study and reduces the influence of single samples. As variation between patients is a significant variable (Fig. 1 a and b), pooling of samples from different patients may prevent the detection of important biological variation.
Muscle Regeneration.
Although adult skeletal muscle is considered a terminally differentiated tissue, it contains populations of mononuclear cells that can undergo several cycles of differentiation and are able to terminally differentiate into mature myofibers. Muscle damage activates these cells to proliferate and fuse forming replacement muscle fibers. Muscle regeneration can be seen at the molecular level in DMD patients by overexpression of genes encoding components of the cytoskeletal microtubules (myosins), intermediate filaments (desmin in muscle and vimentin in mesenchymal tissues), and microfilaments (actins), as well as genes encoding actin-interacting proteins (villin 2, debrin 1, and actin-regulated protein 2/3 complex). Many of the isoforms overexpressed are the same ones that are highly expressed in embryonic or developing muscle. For example, K-α-1 tubulin is overexpressed in DMD muscle and constitutively expressed throughout development (32). Embryonic and perinatal myosin heavy chains (MYH3 and MYH8) are developmental isoforms of skeletal muscle myosin heavy chains; their expression is a hallmark of muscle regeneration after birth and a hallmark of muscular dystrophies. Like MYH3 and MYH8, α-cardiac actin is overexpressed in DMD muscle and is also a specific isoform that is transiently expressed during muscle development and regeneration (23, 33).
A number of other muscle structure and regeneration genes are also overexpressed, including tubulin, myosin, myoferlin [previously shown to be up-regulated at the sarcolemma in mdx mice (34)], and calsequestrin 2. It is unclear why more muscle structure genes are not overexpressed in this or other studies. Here, it may be a result of the stringent statistical analyses.
Dystrophin and leiomodin 1 (a member of the tropomodulin family) are the only muscle genes underexpressed in DMD skeletal muscle. However, there is precedence where a change in expression at the RNA transcript level is not a prerequisite for change at the protein level. For instance, the sarcoglycans, which are decreased at the protein level in DMD patients (35–38) are not observed to be decreased at the mRNA level.
Extracellular Matrix.
Connective tissue infiltration is considered a secondary response that may further compromise muscle function in DMD. Here, 14 known extracellular matrix genes are identified as overexpressed in DMD skeletal muscle versus normal, whereas none are identified as underexpressed, consistent with the histopathology of DMD muscle and with previous reports (20, 22, 23). Up-regulation of the primary fibril-forming collagens (types I and III) is observed, as is up-regulation of additional fibrillar (type V) and nonfibrillar (type VI) collagens and genes that regulate collagen processing or encode various extracellular matrix proteoglycans. It has been reported that COL1A2, -3A1, -6A1, and -6A3 are targets of the transforming growth factor β/Smad3 pathway (39). Transforming growth factor (TGF)β is overexpressed in these 12 DMD patients. Interestingly, cathepsin K, a member of the papain family of cystein proteases that is responsible for degradation of collagen type I, is also overexpressed, perhaps indicating an attempt to counteract collagen deposition.
Chondroitin sulfate proteoglycan 2 (versican), heparin sulfate proteoglycan 2 (perlecan), biglycan, and lumican are extracellular matrix (ECM) proteoglycans that are overexpressed in DMD skeletal muscle. Versican, a member of the lectican family of ECM proteoglycans, is expressed early in muscle myogenesis as well as in regenerating muscle (40). Transcription of perlecan, a member of the testican family of ECM proteoglycans, is up-regulated by TGFβ (41). Perlecan has a function in growth promotion via its interaction with basic fibroblast growth factor, bFGF2 (42).
Biglycan and lumican are members of the small leucine-rich proteoglycan (SLRP) family of ECM proteoglycans. Lumican interacts with collagen in interstitial collagenous matrices. Biglycan also interacts with collagen (43) and is known to form a connection between the extracellular collagenous matrix and the DAPC (44). Thus the increase in biglycan expression might compensate for the absence of dystrophin by stabilizing the interaction between the DAPC and the extracellular matrix. Injured tissue has been showed to express more biglycan, collagen types I and III (and metalloproteinase I), than normal undamaged tissue (45). Biglycan, along with other SLRPs, has been suggested to have an additional signaling role via interaction with TGFβ (46).
SPARC (secreted protein, acidic, cysteine-rich; osteonectin), an extracellular calcium-binding glycoprotein that binds to several members of the ECM and is thought to regulate cell interaction with the extracellular milieu in response in injury (47), is also overexpressed in DMD skeletal muscle of these 12 patients. SPARC has been implicated in the regulation of a number of growth factors (including TGFβ and basic fibroblast growth factor, bFGF) (48, 49) and is involved in the myogenesis of skeletal muscle (50).
Secreted phosphoprotein 1 (Spp1; osteopontin), a macrophage product that enhances synthesis and turnover of extracellular matrix, is substantially elevated in dystrophin-deficient mdx muscle (22). In our study, Spp1 is identified as overexpressed by only one of the two statistical methods (Table 1), but its overexpression in DMD muscle was confirmed by RT-PCR (Table 2). Because both probe sets representing Spp1 are identified by geometric fold change analysis, we believe it is overexpressed in DMD muscle. Spp1-deficient mice show altered collagen fibrillogenesis in wound healing (51), supporting a role for Spp1 in regulating collagen synthesis and accumulation after myocardial infarction (52). The elevation of free phosphate has been shown to induce Spp1 production, which may explain its up-regulation in damaged tissue (53). A chemotactic role has also been suggested for Spp1 (54), suggesting that it may influence both fibrotic and inflammatory responses in dystrophic muscle.
Signaling Pathways Involved in DMD.
Genes encoding growth factors and growth factor related proteins are differentially expressed in DMD vs. control skeletal muscle. Overexpression of these factors may be the cause of connective tissue and extracellular matrix proliferation. One such overexpressed growth factor is TGFβ. The interaction of TGFβ with various members of the extracellular matrix has been discussed in detail. Insulin-like growth factor 2 (IGF2) has previously been reported as overexpressed in dystrophic muscle (18, 22, 23). Although we do not see overexpression of IGFs, we see an increase in expression of two IGF regulators: IGF-binding protein 4 (IGFBP4), which may inhibit IGF-mediated proliferation; and serine protease 11, which cleaves IGFBPs, thus inactivating them.
Dystrophin-deficient muscles show reduction in NOS protein (NOS1) expression (55, 56). Because NO functions as an anti-inflammatory and cytoprotective molecule, it has been proposed that loss of NOS from dystrophic muscle exacerbates muscle inflammation and fiber damage by inflammatory cells. Analysis of transgenic mdx mice expressing normal levels of NOS in muscle showed that normalization of NO production reduces macrophage concentrations in mdx muscle, substantially prevents muscle membrane injury, and decreases in serum creatine kinase concentrations (57).
A decrease of NOS mRNA expression in dystrophin-deficient muscle has not been reported here or previously (20, 22, 23), indicating that the reduction of NOS in dystrophic muscle is a posttranscriptional or -translational effect. An increase in dimethylarginine dimethylaminohydrolase 2 (DDAH2) expression is observed in DMD muscle. DDAH2 metabolizes asymmetric methylarginines, which, in turn, act to inhibit NOS, suggesting that DDAH2 may be an indirect regulator of NOS (58).
Conclusion
The development of microarray technology allows large-scale gene expression analysis projects to be undertaken enabling the examination molecular pathways in a physiological context. As in any developing field, a consensus of how best to use its power, through experimental design and analytical approach, has yet to emerge. Our approach of profiling each patient biopsy individually and using a combination of independent statistical methods to analyze the resulting data identifies differentially expressed genes while minimizing false positives. The results that have been obtained are encouraging. Not only are they verifiable by quantitative RT-PCR, but they are also consistent with the histopathology of the disease. Dystrophin is significantly underexpressed in DMD muscle; known components of the proliferating connective tissue are overexpressed, as are genes encoding components of the immune response; developmentally regulated muscle structure genes are shown to be overexpressed in DMD muscle, as predicted by the presence of regenerating muscle fibers. In short, we have elucidated molecular pathological findings characteristic of the disease and consistent with the observations from histological pathology. We have identified many genes whose function in muscle is unknown that are differentially expressed in DMD skeletal muscle. These include ESTs and genes of unknown function but also various transcription factors and genes known to encode proteins involved in signaling pathways. Our hope is to place these genes in pathophysiological pathways. Further analysis is required to achieve this and to obtain the ultimate goal of making functionally relevant conclusions about the molecular pathology of dystrophic muscle.
Note Added in Proof.
Preliminary comparative analysis of these data sets in relation to biopsies from patients with inflammatory myopathies has recently been reported (59). A similar study has recently been published by Bakay et al. (60).
Supplementary Material
Acknowledgments
We thank Drs. Hart G. W. Lidov and Emanuela Gussoni (Division of Genetics, Children's Hospital, Boston) for providing patient biopsies. This work was supported by grants from the National Institutes of Health (5 PO1NS40828-02) (I.S.K., A.H.B., and L.M.K.); the Muscular Dystrophy Association (A.H.B. and L.M.K.); the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01 AR44349) (A.H.B.); the National Heart, Lung, and Blood Institute (U01 HL066582-01) (I.S.K); and the Bernard F. and Alva B. Gimbel Foundation (L.M.K). L.M.K. is an investigator with the Howard Hughes Medical Institute.
Abbreviations
DMD, Duchenne muscular dystrophy
DAPC, dystrophin-associated protein complex
TGF, transforming growth factor
ECM, extracellular matrix
IGF, insulin-like growth factor
NOS, nitric oxide synthase
References
- 1.Monaco A. P. (1989) Trends Biochem. Sci. 14, 412-415. [DOI] [PubMed] [Google Scholar]
- 2.Matsumura K. & Campbell, K. P. (1994) Muscle Nerve 17, 2-15. [DOI] [PubMed] [Google Scholar]
- 3.Ehmsen J., Poon, E. & Davies, K. (2002) J. Cell Sci. 115, 2801-2803. [DOI] [PubMed] [Google Scholar]
- 4.Straub V. & Campbell, K. P. (1997) Curr. Opin. Neurol. 10, 168-175. [DOI] [PubMed] [Google Scholar]
- 5.Rando T. A. (2001) Muscle Nerve 24, 1575-1594. [DOI] [PubMed] [Google Scholar]
- 6.Hack A. A., Cordier, L., Shoturma, D. I., Lam, M. Y., Sweeney, H. L. & McNally, E. M. (1999) Proc. Natl. Acad. Sci. USA 96, 10723-10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coral-Vazquez R., Cohn, R. D., Moore, S. A., Hill, J. A., Weiss, R. M., Davisson, R. L., Straub, V., Barresi, R., Bansal, D., Hrstka, R. F., et al. (1999) Cell 98, 465-474. [DOI] [PubMed] [Google Scholar]
- 8.Sander M., Chavoshan, B., Harris, S. A., Iannaccone, S. T., Stull, J. T., Thomas, G. D. & Victor, R. G. (2000) Proc. Natl. Acad. Sci. USA 97, 13818-13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rafael J. A., Townsend, E. R., Squire, S. E., Potter, A. C., Chamberlain, J. S. & Davies, K. E. (2000) Hum. Mol. Genet. 9, 1357-1367. [DOI] [PubMed] [Google Scholar]
- 10.Richard I., Broux, O., Allamand, V., Fougerousse, F., Chiannilkulchai, N., Bourg, N., Brenguier, L., Devaud, C., Pasturaud, P., Roudaut, C., et al. (1995) Cell 81, 27-40. [DOI] [PubMed] [Google Scholar]
- 11.Galbiati F., Engelman, J. A., Volonte, D., Zhang, X. L., Minetti, C., Li, M., Hou, H., Jr., Kneitz, B., Edelmann, W. & Lisanti, M. P. (2001) J. Biol. Chem. 276, 21425-21433. [DOI] [PubMed] [Google Scholar]
- 12.Engel A. G. & Banker, B. Q., (1986) Myology (McGraw–Hill, New York).
- 13.Haslett J. & Kunkel, L. (2002) Int. J. Dev. Neurosci. 20, 359-365. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Q., Kho, A., Kenney, A. M., Yuk Di, D. I., Kohane, I. & Rowitch, D. H. (2002) Proc. Natl. Acad. Sci. USA 99, 5704-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo W. P., Jenssen, T. K., Butte, A. J., Ohno-Machado, L. & Kohane, I. S. (2002) Bioinformatics 18, 405-412. [DOI] [PubMed] [Google Scholar]
- 16.Tsien, C. L., Libermann, T. A., Gu, X. & Kohane, I. S. (2001) Pac. Symp. Biocomput., 496–507. [DOI] [PubMed]
- 17.Butte, A. J., Ye, J., Haring, H. U., Stumvoll, M., White, M. F. & Kohane, I. S. (2001) Pac. Symp. Biocomput., 6–17. [DOI] [PubMed]
- 18.Tkatchenko A. V., Le Cam, G., Leger, J. J. & Dechesne, C. A. (2000) Biochim. Biophys. Acta 1500, 17-30. [DOI] [PubMed] [Google Scholar]
- 19.Tkatchenko A. V., Pietu, G., Cros, N., Gannoun-Zaki, L., Auffray, C., Leger, J. J. & Dechesne, C. A. (2001) Neuromuscul. Disord. 11, 269-277. [DOI] [PubMed] [Google Scholar]
- 20.Tseng B. S., Zhao, P., Pattison, J. S., Gordon, S. E., Granchelli, J. A., Madsen, R. W., Folk, L. C., Hoffman, E. P. & Booth, F. W. (2002) J. Appl. Physiol. 93, 537-545. [DOI] [PubMed] [Google Scholar]
- 21.Rouger K., Le Cunff, M., Steenman, M., Potier, M. C., Gibelin, N., Dechesne, C. A. & Leger, J. J. (2002) Am. J. Physiol. 283, C773-C784. [DOI] [PubMed] [Google Scholar]
- 22.Porter J. D., Khanna, S., Kaminski, H. J., Rao, J. S., Merriam, A. P., Richmonds, C. R., Leahy, P., Li, J., Guo, W. & Andrade, F. H. (2002) Hum. Mol. Genet. 11, 263-272. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y. W., Zhao, P., Borup, R. & Hoffman, E. P. (2000) J. Cell Biol. 151, 1321-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Press W. H., Teukolsky, S. A., Vetterling, W. T. & Flannery, B. P., (1992) Numerical Recipes: The Art of Scientific Computing (Cambridge Univ. Press, Cambridge, U.K.).
- 25.Tusher V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pritchard C. C., Hsu, L., Delrow, J. & Nelson, P. S. (2001) Proc. Natl. Acad. Sci. USA 98, 13266-13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamayo P., Slonim, D., Mesirov, J., Zhu, Q., Kitareewan, S., Dmitrovsky, E., Lander, E. S. & Golub, T. R. (1999) Proc. Natl. Acad. Sci. USA 96, 2907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chamberlain J. S., Pearlman, J. A., Muzny, D. M., Gibbs, R. A., Ranier, J. E., Caskey, C. T. & Reeves, A. A. (1988) Science 239, 1416-1418. [DOI] [PubMed] [Google Scholar]
- 29.Chelly J., Gilgenkrantz, H., Lambert, M., Hamard, G., Chafey, P., Recan, D., Katz, P., de la Chapelle, A., Koenig, M., Ginjaar, I. B., et al. (1990) Cell 63, 1239-1248. [DOI] [PubMed] [Google Scholar]
- 30.Dangain J. & Vrbova, G. (1984) Muscle Nerve 7, 700-704. [DOI] [PubMed] [Google Scholar]
- 31.Carnwath J. W. & Shotton, D. M. (1987) J. Neurol. Sci. 80, 39-54. [DOI] [PubMed] [Google Scholar]
- 32.Miller T. A., Bulman, A. L., Thompson, C. D., Garst, M. E. & Macdonald, T. L. (1997) J. Med. Chem. 40, 3836-3841. [DOI] [PubMed] [Google Scholar]
- 33.Toyofuku T., Hoffman, J. R., Zak, R. & Carlson, B. M. (1992) Dev. Dyn. 193, 332-339. [DOI] [PubMed] [Google Scholar]
- 34.Davis D. B., Delmonte, A. J., Ly, C. T. & McNally, E. M. (2000) Hum. Mol. Genet. 9, 217-226. [DOI] [PubMed] [Google Scholar]
- 35.Campbell K. P. & Kahl, S. D. (1989) Nature 338, 259-262. [DOI] [PubMed] [Google Scholar]
- 36.Ervasti J. M., Ohlendieck, K., Kahl, S. D., Gaver, M. G. & Campbell, K. P. (1990) Nature 345, 315-319. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida M. & Ozawa, E. (1990) J. Biochem. (Tokyo) 108, 748-752. [DOI] [PubMed] [Google Scholar]
- 38.Ohlendieck K., Ervasti, J. M., Matsumura, K., Kahl, S. D., Leveille, C. J. & Campbell, K. P. (1991) Neuron 7, 499-508. [DOI] [PubMed] [Google Scholar]
- 39.Verrecchia F., Chu, M. L. & Mauviel, A. (2001) J. Biol. Chem. 276, 17058-17062. [DOI] [PubMed] [Google Scholar]
- 40.Carrino D. A., Sorrell, J. M. & Caplan, A. I. (1999) Poultry Sci. 78, 769-777. [DOI] [PubMed] [Google Scholar]
- 41.Iozzo R. V., Pillarisetti, J., Sharma, B., Murdoch, A. D., Danielson, K. G., Uitto, J. & Mauviel, A. (1997) J. Biol. Chem. 272, 5219-5228. [DOI] [PubMed] [Google Scholar]
- 42.Mongiat M., Otto, J., Oldershaw, R., Ferrer, F., Sato, J. D. & Iozzo, R. V. (2001) J. Biol. Chem. 276, 10263-10271. [DOI] [PubMed] [Google Scholar]
- 43.Wiberg C., Hedbom, E., Khairullina, A., Lamande, S. R., Oldberg, A., Timpl, R., Morgelin, M. & Heinegard, D. (2001) J. Biol. Chem. 276, 18947-18952. [DOI] [PubMed] [Google Scholar]
- 44.Bowe M. A., Mendis, D. B. & Fallon, J. R. (2000) J. Cell Biol. 148, 801-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo I. K., Marchuk, L. L., Hart, D. A. & Frank, C. B. (1998) J. Orthop. Res. 16, 421-428. [DOI] [PubMed] [Google Scholar]
- 46.Hildebrand A., Romaris, M., Rasmussen, L. M., Heinegard, D., Twardzik, D. R., Border, W. A. & Ruoslahti, E. (1994) Biochem. J. 302, 527-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bradshaw A. D. & Sage, E. H. (2001) J. Clin. Invest. 107, 1049-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brekken R. A. & Sage, E. H. (2000) Matrix Biol. 19, 569-580. [DOI] [PubMed] [Google Scholar]
- 49.Yan Q. & Sage, E. H. (1999) J. Histochem. Cytochem. 47, 1495-1506. [DOI] [PubMed] [Google Scholar]
- 50.Cho W. J., Kim, E. J., Lee, S. J., Kim, H. D., Shin, H. J. & Lim, W. K. (2000) Biochem. Biophys. Res. Commun. 271, 630-634. [DOI] [PubMed] [Google Scholar]
- 51.Liaw L., Birk, D. E., Ballas, C. B., Whitsitt, J. S., Davidson, J. M. & Hogan, B. L. (1998) J. Clin. Invest. 101, 1468-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trueblood N. A., Xie, Z., Communal, C., Sam, F., Ngoy, S., Liaw, L., Jenkins, A. W., Wang, J., Sawyer, D. B., Bing, O. H., et al. (2001) Circ. Res. 88, 1080-1087. [DOI] [PubMed] [Google Scholar]
- 53.Beck G. R., Zerler, B. & Moran, E. (2000) Proc. Natl. Acad. Sci. USA 97, 8352-8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ashkar S., Weber, G. F., Panoutsakopoulou, V., Sanchirico, M. E., Jansson, M., Zawaideh, S., Rittling, S. R., Denhardt, D. T., Glimcher, M. J. & Cantor, H. (2000) Science 287, 860-864. [DOI] [PubMed] [Google Scholar]
- 55.Brenman J. E., Chao, D. S., Xia, H., Aldape, K. & Bredt, D. S. (1995) Cell 82, 743-752. [DOI] [PubMed] [Google Scholar]
- 56.Grozdanovic Z., Gosztonyi, G. & Gossrau, R. (1996) Acta Histochem. 98, 61-69. [DOI] [PubMed] [Google Scholar]
- 57.Wehling M., Spencer, M. J. & Tidball, J. G. (2001) J. Cell Biol. 155, 123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leiper J. & Vallance, P. (1999) Cardiovasc. Res. 43, 542-548. [DOI] [PubMed] [Google Scholar]
- 59.Greenberg S. A., Sanoudou, D., Haslett, J. N., Kohane, I. S., Kunkel, L. K., Beggs, A. H. & Amato, A. A. (2002) Neurology 59, 1170-1182. [DOI] [PubMed] [Google Scholar]
- 60.Bakay M., Zhao, P., Chen, J. & Hoffman, F. (2002) Neuromuscul. Disord. 12, S125-S142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.