Abstract
By using dendritic cells (DCs) transduced with retroviruses encoding covalent Abβ/peptide fusion proteins tagged with fluorescent proteins, we followed the relocation of class II MHC molecules loaded with agonist or null peptides during the onset of activation of naive and effector CD4+ T cells. Clusters of T cell receptor (TCR)/CD3 complex formed in parallel with clusters of agonist class II MHC/peptide complexes on the surface of DCs. However, activation of naive but not effector T cells was accompanied by expulsion of the null class II MHC/peptide complexes from the T cell–DC interface. These effects were perturbed in the presence of exogenously supplied antagonist peptide. These results suggest that interference with selective relocation of agonist and null MHC/peptide complexes in the immunological synapse contributes to the inhibitory effect of antagonist peptides on the response of naive CD4+ T cells to agonist ligands.
Antigenic peptides with affinity sufficient to activate T cell response, so-called agonists, are not the only peptides encountered by the T cell receptor (TCR) on the surface of antigen-presenting cells (APCs). Usually, TCR faces a myriad of different peptides bound to MHC molecules, which are detained in the interface between the APC and the T cell, most of which have undetectable (i.e., null) affinity for the TCR. The rules and mechanisms that govern the trafficking of MHC/peptide complexes on the membrane of the APC, which are involved in the regulation of the T cell response, are not well understood. Some recent evidence suggests that APCs can autonomously organize the distribution of MHC/peptide complexes on their surface. Colocalization of a diverse set of these complexes within lipid rafts may improve the presentation of less frequent peptides to T cells, whereas the accumulation of class II MHC molecules bound with a few abundant peptides in tetraspan microdomains may be a favorable target for particular subpopulations of T cells (1, 2). On the other hand, the qualitative sorting of MHC/peptide complexes on APCs is also actively driven by TCRs during the onset of T cell activation (3). Initially, it was reported that on activation, CD4+ T cells selectively cluster agonist class II MHC/peptide complexes, but in subsequent studies it was found that null class II MHC/peptide complexes with weak or no affinity for TCRs are also present within the cell–cell contact site (3, 4). It was also shown that TCR engagement by MHC molecules loaded with so-called antagonist peptide may inhibit T cell response (5). It is not clear whether this effect is because of the direct interference with TCR binding to agonist MHC/peptide complexes (6–8) or because of the interference with the formation of functional MHC/peptide clusters at the T cell–APC interface, as observed for other receptor/ligand interactions (9). To investigate the latter possibility we analyzed the effect of exogenously added antagonist peptide on the redistribution of agonist and null class II MHC/peptide complexes in the interface area of dendritic cells (DCs) interacting with naive CD4+ T cells expressing transgenic antigen-specific TCR (TCRTg). Our results demonstrate that in contrast to the interaction with effector CD4+ T cells, interaction with naive CD4+ T cells results in expulsion of null MHC/peptide complexes from the T cell–DC interface and that this process in inhibited in the presence of antagonist peptides.
Materials and Methods
Mice.
Mice deficient for invariant chain (AbIi−) were kindly provided by R. Germain (National Institutes of Health, Bethesda). Mice transgenic for the Vα4JVβ8.1 TCR (TCRTg) were generated at the Medical College of Georgia (Augusta) (10). All mice were housed under specific pathogen-free conditions in the animal care facility at the Medical College of Georgia.
DNA Constructs.
DNA encoding the β chain of the Ab molecule covalently bound with the class II MHC Eα chain (Eα) (52–68) peptide, Ep (ASFEAQGALANIAVDKA) (AbβEp), or covalently bound with the analog of the pigeon cytochrome c (PCC)-(43–58) peptide, PCC50V54A (AEGFSYTVANKAKGIT) (AbβPCCVA), was amplified as described (11). The DNA inserts encoding AbβEp or AbβPCCVA were cloned into pEYFP-N1 and pECFP-N1 vectors (CLONTECH) and DNA fragments encoding AbβEp-cyan fluorescent protein (CFP) [or yellow fluorescent protein (YFP)] and AbβPCCVA-CFP (or YFP) were recloned into Moloney murine leukemia virus (MoMLV)-based LZRS-pBMN-Z retroviral vector (kindly provided by G. Nolan, Stanford University School of Medicine, Stanford, CA).
DNA encoding CD3ζ (kindly provided by Makio Iwashima, Medical College of Georgia) was cloned into pECFP-N1 and pEYFP-N1 vectors. DNA fragments encoding CD3ζ-CFP or CD3ζ-YFP were recloned into the LZRS-pBMN-Z retroviral vector.
Production of Retroviral Supernatants.
Phoenix Eco 293T packaging cell line (provided by G. Nolan) was transfected with the appropriate DNA construct by using calcium phosphate (12). On days 2, 3, and 4 after transfection, the supernatant was collected and viral titer was determined by transducing NIH 3T3 cells. The expression of covalent Ab/peptide complexes was checked by flow cytometry (for the YFP constructs) or fluorescence microscopy (for the CFP constructs). The average viral titer used was 1.5–2.5 × 107 viral particles per ml. Viral supernatants were snap frozen in liquid nitrogen and stored at −70°C.
Isolation of Naive T Cells.
Lymph node cells of the TCRTg mouse were used as a source of naive CD4+ T cells. Cells, resuspended at the concentration of 1 × 107 per ml, were incubated for 30 min at 4°C with anti-CD4 magnetic beads (CD4 MicroBeads, Miltenyi Biotec, Auburn, CA). TCRTg CD4+ T cells were isolated by positive selection with the autoMACS sorter (Miltenyi Biotec). The purity of cells was 95–99% as tested by flow cytometry.
Generation of Activated CD4+ T Cells.
Lymph node cells from a TCRTg mouse (2 × 106 per ml) were cultured in the presence of 50 μM agonist PCCVA peptide. After 48 h, cells were spun on the Lymphocyte Separation Medium (Cellgro, Herndon, VA) and blasts were further expanded in the presence of IL-2. Activated TCRTg CD4+ T cells were used for microscopy experiments on days 6–8 after isolation. The purity of cells was >95% as tested by flow cytometry.
Transduction of Activated T Cells.
At day 3 of activation, TCRTg CD4+ T cells were transferred into 6-well plates and resuspended in 1 ml per well of IL-2-supplemented medium at a concentration 2 × 106 cells per ml. Viral supernatants (300 μl) and Polybrene (8 μg/ml, Sigma) were added to each well. Transduction was enhanced by the centrifugation of six-well plates enclosed in CO2-supplemented zip-lock bags at 1,000 × g for 1 h at ambient temperature (28–32°C). Sixteen hours after transduction, Polybrene and viral supernatants were washed off and the cells were resuspended in medium with IL-2. On day 5, transduced activated TCRTg CD4+ T cells were analyzed by flow cytometry and used for microscopy experiments on days 6–8 after isolation.
Transduction of Bone Marrow (BM)-Derived DCs.
To isolate DCs, a modified protocol of Lutz et al. (13) was used. Depleted of erythrocytes, BM cells from AbIi− mice were incubated in medium supplemented with granulocyte/macrophage colony-stimulating factor (GM-CSF)-containing supernatant (15%) and IL-4 (50 units/ml, PeproTech, Rocky Hill, NJ) and plated onto 10 mm Petri dishes at ≈3 × 106 cells per ml. On day 2 cells were transferred into six-well plates and resuspended in 1 ml per well of cytokine-supplemented medium at a concentration of 3–4 × 106 cells per ml. Viral supernatants and Polybrene (8 μg/ml) were added to each well. Transduction was enhanced by centrifugation of six-well plates enclosed in CO2-supplemented zip-lock bags at 1,000 × g for 1 h at ambient temperature. The transduction procedure was repeated on days 3 and 4. Eight to 10 hours after transduction, Polybrene and viral supernatants were washed off and the cells were resuspended in medium containing GM-CSF and IL-4 (14). On day 8, transduced DCs were analyzed by flow cytometry to determine the efficiency of transduction, and they were used for microscopy experiments on days 8–10 after isolation.
Flow Cytometry Analysis.
The reagents used for flow cytometry analyses were as follows: anti-CD11c-PE (PharMingen), anti-AbEp-biotin (YAe, self-prepared), anti-Ab-biotin (Y3P, self-prepared), and streptavidin-CyChrome (PharMingen). Staining was done on ice in BSS buffer containing 2% FBS and 0.1% NaN3. All fluorescence-activated cell sorter (FACS) analyses were performed by using a FACSCalibur flow cytometer (Becton Dickinson) and cellquest software (Becton Dickinson).
Detection of Ca2+ Influx.
Calcium influx was detected by using fluo-3 AM (Molecular Probes). Fluo-3 was loaded into cells as described (15) with minor modifications. Cells (1 × 107 per ml) were incubated in PBS with 2 μM fluo-3 AM and 0.02% pluronic acid F-127 (Molecular Probes) for 20 min at room temperature. Next, the cells were diluted to a final concentration of 2 × 106 per ml with PBS containing 1% FCS, and incubated for an additional 40 min in a 37.5 ± 0.1°C water bath (16). Cells were then washed twice with PBS and maintained in RPMI medium 1640 (Cellgro), buffered with 25 mM Hepes and supplemented with 10% FBS/2 mM l-glutamine (Cellgro). An increase in the intensity of fluorescence of at least 50% above the resting level was considered significant for calcium influx.
Microscopy.
Pictures were taken with an Axiovert S100 2TV microscope (Zeiss) equipped with the DeltaVision system (Applied Precision, Issaquah, WA) by using a CFP/YFP set of narrow-band filters (Chroma Technology, Brattleboro, VT). An oil-immersion ×100/1.40 numerical aperture objective warmed to 37°C by the Objective Heater System (Bioptechs, Butler, PA) was used. All images were deconvoluted by using a constrained-iterative mode of deconvolution, applying 10 iterations. Three-dimensional images were reconstructed from Z-stacks of 2D/two-wavelength images. For image analysis LSM 510 software (Zeiss) was used. Live T cells and DCs were resuspended in Hepes-buffered medium, mixed together, and placed in a temperature-controlled chamber (Bioptechs) for immediate imaging (for calcium influx experiments with fluo-3). Alternatively, to increase the number of conjugates, T cells and DCs were mixed and spun at 50 × g for 5 min and incubated at 37°C for 15 min. Next, pellets were gently resuspended and cells were placed in a temperature-controlled chamber. After 15 min, T cell–DC conjugates were subjected to imaging.
Analysis of Cell–Cell Contacts.
The area of contact in the T cell–DC conjugate, showing clear cell–cell contact, was defined based on the differential interference contrast (DIC) images. Accumulation of Ab/peptide complexes was considered as clustering when the ratio of fluorescence intensity at the interface over the side of the cell (Figs. 6 and 7, which are published as supporting information on the PNAS web site, www.pnas.org) was greater than 1.5 (50% increase in fluorescence intensity).
For 3D visualization of intercellular contacts only those complexes were taken into consideration whose contact areas were flat enough to be contained in a rectangular volume for a facing projection. Conjugates in which the plane of the interface was not orthogonal to the Z-stacks were disregarded during analysis. Exclusion of a particular complex was defined by at least a 50% decrease in the fluorescence intensity in comparison with the intensity of complexes in the proximity of the place of contact (Figs. 6 and 7, which are published as supporting information on the PNAS web site).
In Vitro Proliferation and Antagonism Assay.
BM-derived DCs from AbIi− mice were transduced with recombinant retroviruses encoding AbPCCVA-CFP (or YFP) and/or AbEp-CFP (or YFP) complexes as described above, and irradiated at day 8 with a dose of 3,000 rad. These DCs (104 per well) were mixed with TCRTg CD4+ T cells (2 × 104 per well) and were incubated in triplicate for 3 days in U-bottom 96-well plates (Costar, Cambridge, MA) at 37°C. During the last 12 to 16 h of the culture, the incorporation of [3H]thymidine [0.5 μCi (1 Ci = 37 GBq) per well] by proliferating lymphocytes was measured.
For in vitro antagonism assay, transduced DCs were pulsed for 3 h with different concentrations of either antagonist (PCC50E) or null (PCC50Q) peptides (10). DCs were then cocultured with TCRTg CD4+ T cells as described above. For microscopy experiments transduced DCs were pulsed for 3 h with 50 μM antagonist peptide PCC50E as indicated.
Results
Covalent Ab/Peptide Complexes Expressed by Transduced AbIi− DCs Retain the Specificity of Noncovalent Complexes and Form Functional Clusters on Engagement by TCRs.
To study the relocation of class II MHC/peptide complexes on the surface of DCs during interaction with naive and effector CD4+ T cells, DCs were transduced with recombinant retroviruses encoding the Abβ chain tethered by means of a flexible linker to agonist (PCCVA) or null (Ep) peptides. To prevent the cleavage of such covalent Ab/peptide complexes in transduced cells, DCs were derived from AbIi− mice, which have an impaired mechanism of peptide processing (17). To follow the distribution of covalent AbPCCVA and AbEp complexes separately, we tagged them on their intracellular ends with different fluorescent proteins (CFP or YFP).
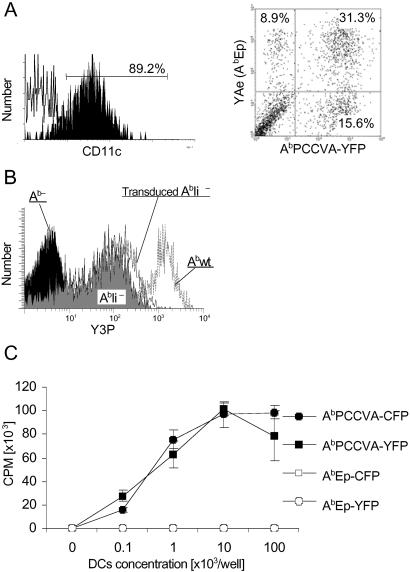
As shown in Fig. 1A, substantial proportion of DCs transduced simultaneously with both types of retroviruses expressed both complexes on the cell surface at comparable levels. Importantly, transduction did not result in a significant increase of the expression level of peptide/MHC II complexes on BM-derived DCs (Fig. 1B). When transduced separately with one type of vector, only DCs expressing the covalent AbPCCVA complexes, but not DCs expressing covalent AbEp complexes, were able to activate transgenic CD4+ T cells, demonstrating that engineered complexes retained their original specificity (Fig. 1C). The uniform distribution of the covalent Ab/peptide complexes on the surface of DCs indicates that the spontaneous clustering of these complexes (2) does not occur (data not shown).
Fig 1.
Covalent Ab/peptide complexes are expressed on the surface of DCs and retain the same T cell specificity as noncovalent complexes. (A) BM-derived AbIi− DCs, transduced with AbβPCCVA-YFP and AbβEp-CFP complexes, were stained with anti-CD11c-PE or YAe-biotin/SA-CyChrome (specific to AbEp) mAbs and analyzed by FACS. YAe mAb was used to identify AbEp because CFP is not detectable by FACS. A dot plot represents results obtained on CD11c-positive cells, gated as indicated. (B) Comparison of expression levels on transduced BM-derived AbIi− DCs vs. BM-derived DCs from AbIi− and Abwt mice. Cells were stained with Y3P-biotin/SA-CyChrome mAb (specific to Ab) and analyzed by FACS. (C) TCRTg CD4+ T cells were incubated with irradiated BM-derived DCs expressing indicated covalent complexes. Incorporation of [3H]thymidine by proliferating T cells ± SD is shown. Data are representative of three independent experiments.
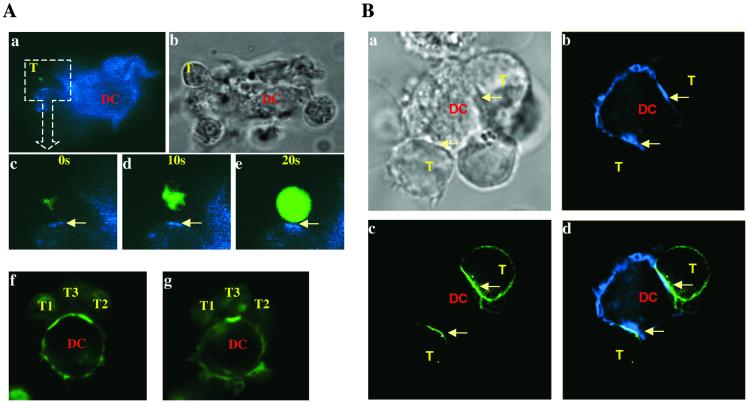
To test whether class II MHC molecules covalently bound with agonist peptides can be clustered in the T cell–APC interface as observed during formation of the immunological synapse (4, 18), DCs expressing AbPCCVA complexes tagged with CFP were mixed either with naive or with effector TCRTg CD4+ T cells loaded with the calcium influx indicator fluo-3 or transfected with CD3ζ-YFP, respectively (Fig. 2). The transduced T cells expressed a physiological level of TCR, probably because remaining endogenous CD3 chains limit expression of TCR/CD3 complexes. The monitoring of fluctuations in fluo-3 fluorescence as well as clustering of TCR/CD3 complexes allowed us to ensure that the observed T cell–DC contacts were functional, i.e., led to the T cell activation. The TCR/CD3 complex tagged with YFP also allowed us to follow the spatial relationship between formation of TCR and AbPCCVA clusters. As shown in Fig. 2 (see also Table 1), clusters of agonist covalent AbPCCVA complexes and TCRTg/CD3 complexes were formed at the T cell–DC interface, which was associated with T cell activation. The clustering occurs when both naive and effector T cells become activated. We also noticed that simultaneous interaction of several naive (Fig. 2A f and g) or effector (Fig. 2B a and d) TCRTg CD4+ T cells with one DC formed separate clusters of AbPCCVA complexes at each individual contact site. This observation provides strong additional evidence that one DC can activate several T cells at the same time (see review, ref. 19).
Fig 2.
Visualization of functional clusters of covalent agonist MHC class II/peptide complexes and TCRTg/CD3 complexes (arrows) at the T cell–DC interface. Naive TCRTg CD4+ T cells loaded with fluo-3 (A) or effector TCRTg CD4+ T cells transduced with CD3ζ-YFP (B) were mixed with AbIi− DCs expressing agonist AbPCCVA-CFP (blue, A a–e and B b–d) or AbPCCVA-YFP complexes (green, A f and g). Photo recording started 20 min after initial contact of a T cell with a DC (A c–e). T cell activation, detected by calcium flux (note the increase in green fluorescence), was recorded every 10 s. (A f and g) Two different Z-sections of one DC interacting with three naive TCRTg CD4+ T cells. (B b–d) One DC interacting with two effector T cells. (Bb) Clusters of AbPCCVA-CFP on DCs. (Bc) Clusters of TCR/CD3 on T cells. (Bd) An overlay of Bb and Bc. The AbPCCVA complexes were clustered in 82% of recorded contacts between DCs and naive T cells, in contrast to null AbEp complexes, which were clustered in 9.2% of contacts (not shown). Four independent experiments were done and at least 30 contacts per experiment were analyzed (Table 1). (Ab and Ba) DIC images; T, T cell; DC, dendritic cell.
Table 1.
Percentage of T cell–DC conjugates with clustering or expulsion of MHC/peptide complexes
| T cells
|
DCs transduced with: | AbEp | AbPCCVA | AbEp and AbPCCVA | ||
|---|---|---|---|---|---|---|
| Exogenously added peptide: | None | None | None | PCC50E (antagonist) | PCC50Q (null) | |
| Naive | ||||||
| AbPCCVA clustering | — | 82 ± 15% | 76 ± 13% | 61 ± 21% | 70 ± 9.8% | |
| (n = 118) | (n = 88) | (n = 103) | (n = 60) | |||
| AbEp clustering | 9.2 ± 4.5% | — | 4.0 ± 3.8% | 62 ± 12% | 7.0 ± 6.2% | |
| (n = 116) | (n = 67) | (n = 63) | (n = 42) | |||
| AbEp expulsion | 0% | — | 61 ± 7.5% | 11 ± 5.2% | 55 ± 8.6% | |
| (n = 116) | (n = 67) | (n = 63) | (n = 42) | |||
| Activated | ||||||
| AbPCCVA clustering | — | 59 ± 19% | 52 ± 12% | 66 ± 14% | 57 ± 8.9% | |
| (n = 44) | (n = 75) | (n = 54) | (n = 65) | |||
| AbEp clustering | 0% | — | 92 ± 6.9% | 89 ± 4.7% | 94 ± 3.1% | |
| (n = 38) | (n = 39) | (n = 36) | (n = 37) | |||
| AbEp expulsion | 0% | — | 0% | 0% | 2.1 ± 3.6% | |
| (n = 38) | (n = 39) | (n = 36) | (n = 37) | |||
n = number of analyzed T cell–DC conjugates.
Conjugates with AbPCCVA clustering.
, P = 0.001;
, P = 0.024;
, P = 0.025 (t test, 95% confidence interval of the difference, significant when P < 0.05).
Null MHC/Peptide Complexes Are Expelled from the Site of Contact Between DC and Naive T Cells but Not Between DC and Effector T Cells.
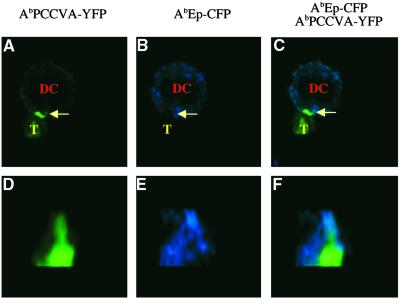
To examine the bilateral relocation of agonist and null Ab/peptide complexes on the interaction with naive and effector TCRTg CD4+ T cells, DCs coexpressing both complexes were used. As shown in Fig. 3 A–C, the activation of naive TCRTg CD4+ T cells (visualized by calcium flux) correlated with the visible expulsion of the null AbEp complexes and the increased concentration of the agonist AbPCCVA complexes in the cell–cell contact site. This phenomenon of preferential relocation of agonist complexes inward, and of null complexes outward of the DC–T cell junction, observed in the majority of contact sites (see Table 1), was confirmed by the 3D reconstruction of the interface of DC contacting the naive TCRTg CD4+ T cell (Fig. 3 D–F).
Fig 3.
Naive CD4+ T cells expel null MHC class II/peptide complexes from the center of the contact sites with DCs. Naive TCRTg CD4+ T cells (T) were loaded with calcium indicator fluo-3 and mixed with AbIi− DCs expressing agonist AbPCCVA-YFP and neutral AbEp-CFP complexes. (A–C) A single 2D optical section in the center of the cell conjugate. (D–F) A 3D view of the T cell–DC contact. (A and D) AbPCCVA-YFP only (green). (B and E) AbEp-CFP only (blue). (C and F) AbPCCVA-YFP and AbEp-CFP overlay. Agonist AbPCCVA complexes were clustered in 76% of recorded contacts between DCs and T cells. Of the clusters, 61% were characterized by the expulsion of neutral AbEp-CFP complexes. Exclusion of a particular complex was defined as at least a 50% decrease in the intensity of fluorescence in comparison with the intensity in the proximity of the place of contact (Fig. 6, which is published as supporting information on the PNAS web site). Three independent experiments were done, and a total of 88 contacts were analyzed (Table 1). This phenomenon was also observed in the opposite experimental setup, where TCR-specific Ab/PCCVA complex was labeled with CFP and neutral Ab/Ep complex was labeled with YFP (data not shown; Fig. 7, which is published as supporting information on the PNAS web site).
In contrast to naive CD4+ T cells, the above described selective reorganization of agonist and null Ab/peptide complexes at the DC–T cell interface was not observed during the interaction of effector CD4+ T cells with DCs (Fig. 4 and Table 1). In the latter case, both types of complexes are clustered at the DC–T cell interface as indicated by the formation of “overlaid patches” of agonist AbPCCVA-YFP (green) and null AbEp-CFP (blue) complexes (compare Figs. 3 and 4). These results suggest that to become activated, naive CD4+ T cells, unlike effector T cells, tend to exclude irrelevant class II MHC/peptide complexes from the site of TCR-mediated interaction with DCs.
Fig 4.
Effector TCRTg CD4+ T cells do not expel irrelevant MHC class II/peptide complexes from the center of the contact sites with DCs. In vitro activated TCRTg CD4+ T cells (T) were mixed with AbIi− DCs (DC) expressing agonist AbPCCVA-YFP and null AbEp-CFP complexes. (A–D) A single 2D optical section in the center of the cell conjugate. (E–G) A 3D view of the cell–cell contact. (A) DIC image. (B and E) AbPCCVA-YFP only (green). (C and F) AbEp-CFP only (blue). (D and G) AbPCCVA-YFP and AbEp-CFP overlay. This image is representative for 39 contacts analyzed in three independent experiments (Table 1).
Antagonist Peptides Abrogate the Reorganization of Agonist and Null MHC Class II/Peptide Complexes at the Site of Contact Between DC and Naive TCRTg CD4+ T Cells.
Above, we have provided evidence that agonist and null Ab/peptide complexes are often selectively relocated in the site of contact with naive but not with effector CD4+ T cells, which could reflect the requirement of naive T cells for higher concentration of agonist ligands to become activated. If this selective relocation indeed plays a role in activation of naive CD4+ T cells, then factors interfering with this process should inhibit the response of these cells to the agonist ligands.
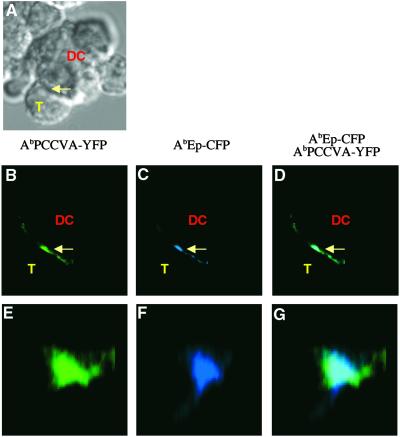
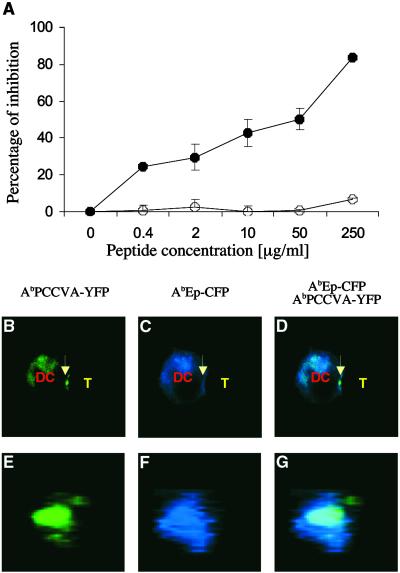
To test whether antagonist peptides are able to interfere with the described relocation of agonist and null MHC/peptide complexes, DCs coexpressing covalent AbPCCVA and AbEp molecules were preincubated with soluble antagonist peptide (PCC50E) and mixed with naive CD4+ TCRTg T cells. The DCs coexpressing covalent AbPCCVA-YFP and AbEp-CFP molecules, which were preincubated with irrelevant (null) peptide (PCC50Q) for the transgenic TCR, served as a control. The covalent binding of peptides to Ab molecules excluded the possibility of peptide exchange, which was essential for the demonstration that exposure to an excess of antagonist peptide disturbs the reorganization of MHC complexes bound to agonist and null peptides. As shown in Fig. 5A, copresentation of the antagonist PCC50E peptide, but not of the null PCC50Q peptide, by endogenous class II MHC molecules together with covalent AbPCCVA and AbEp complexes, inhibited the activation of naive CD4+ TCRTg T cells. Fig. 5 B–G clearly demonstrates that the inhibition of the T cell activation by antagonist PCC50E peptide is associated with poor expulsion of the null AbEp complexes from the DC–T cell interface (compare with Fig. 3). The presentation of the null PCC50Q peptide had no such effect (Table 1). We conclude that interference with selective relocation of agonist and null MHC/peptide complexes on the surface of DCs contributes to the inhibitory effect of antagonist peptides on the response of naive CD4+ T cells to agonist ligands.
Fig 5.
Antagonist peptide, which inhibits the proliferation of naive TCRTg CD4+ T cells to agonist peptide, also inhibits expulsion of null MHC class II/peptide complexes from the center of the T cell–DC contact site. AbIi− DCs expressing agonist AbPCCVA-YFP and null AbEp-CFP complexes were preincubated with the indicated concentration of antagonist PCC50E (•) or null PCC52Q (○) peptides (A) or with 50 μM antagonist PCC50E peptide (B–G) and cultured with naive TCRTg CD4+ T cells. (A) Inhibition of [3H]thymidine incorporation ± SD into proliferating T cells (representative of three independent experiments) is shown. (B–D) A single 2D optical section in the center of the T cell–DC conjugate. (E–G) A 3D view of the T cell–DC contact. (B and E) AbPCCVA-YFP only (green). (C and F) AbEp-CFP only (blue). (D and G) an AbPCCVA-YFP and AbEp-CFP overlay. Agonist AbPCCVA complexes were clustered in 61% of recorded contacts between DCs and T cells. Only 11% of the clusters were characterized by the exclusion of neutral AbEp-CFP complexes (Table 1). Expulsion of a particular complex was defined by at least a 50% decrease in the intensity of fluorescence in comparison with the intensity in the proximity of the place of contact. T cell–DC contacts (103 in total) were analyzed in three independent experiments.
Discussion
We have shown that DCs coexpressing recombinant complexes of class II MHC molecules covalently bound to agonist and null peptides represent a suitable experimental model for the study of the reorganization of TCR-specific and TCR-nonspecific MHC/peptide complexes, which occur at the intercellular contact site during interaction with naive or effector T cells. In agreement with previous experiments, by using noncovalent TCR ligands, we observed that contact with DCs expressing covalent agonist Ab/peptide complexes resulted in TCR/CD3 clustering and detectable calcium flux in both types of T cells, which was accompanied by simultaneous clustering of the MHC/peptide complexes at the T cell–DC interface (4, 18). We found, however, a striking difference between the behavior of null MHC/peptide complexes at the contact site of DCs interacting with naive or effector CD4+ T cells. In contrast to effector T cells, naive T cells tended to exclude the MHC molecules bound to null peptides from the contact area with DCs, suggesting that a higher local concentration of agonist ligand is required for activation of “antigen-inexperienced” than “antigen-experienced” T cells. This observation is intriguing in view of the recent report that not only agonist, but also null MHC/peptide complexes, accumulate in immune synapses and contribute to T cell recognition (3). Our findings imply that the enhancing role of nonagonist MHC/peptide may be restricted to effector T cells only, whereas the presence of such complexes in the “synapse” formed by naive T cell and DC would have an opposite effect. At present, one can only speculate about the mechanism enabling naive T cells to selectively expel the nonagonist complexes from the immune synapse, and therefore more experiments will be needed to propose the explanation for this phenomenon. As far as the “T cell side” is concerned, it was reported (20) that activation-induced membrane changes increasing TCR avidity may contribute to the increased sensitivity of activated T cells to antigen. One could imagine that the lack of activation-induced membrane changes in naive T cells could be compensated by local increase in antigen concentration on DCs brought about by expulsion of null MHC/peptide complexes from the contact area. However, irrespective of the mechanism involved, we made another interesting observation with regard to the possible mechanism contributing to the inhibitory effect of antagonist peptides on the antigen-specific response of naive CD4+ T cells. Several mechanisms have been postulated to account for the inhibition of T cell activation by antagonist peptides. Most of them are concerned with the effects on TCR-mediated signaling pathways in T cells (21–24), and largely ignored the possible influence of antagonist peptides on the relocation of MHC/peptide complexes in T cell–DC interface. It was therefore very interesting to find that exposure to antagonist peptides obstructs exclusion of the null MHC/peptide complexes from the immunological synapse formed by DCs interacting with naive CD4+ T cells. In this context, it is worth mentioning that according to a recent study (25) TCR-mediated signaling in naive T cells peaks before the mature synapse is formed, suggesting another, as-yet-unrecognized, role of TCR/MHC/peptide clusters in activation of naive T cells. One favored possibility is that cluster formation is important for TCR down-modulation and endocytosis. If this is the case, our results may indicate that antagonist peptides act by causing the retention of irrelevant MHC/peptide complexes in the synapse and prevent sufficient contact between TCR and agonist MHC/peptide ligands required for TCR internalization. The fact that antagonist peptides were shown to inhibit TCR endocytosis (26) is consistent with this idea. Thus, the reorganization of agonist and nonagonist MHC/peptide complexes on the surface of DCs during the activation of naive CD4+ T cells may be an important part of the mechanism that controls the initiation of the adaptive immune response.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Basic Research Grants AI41145-01A1 and HD36302-02 (to L.I.). P.K. is supported by the Howard Hughes Medical Institute.
Abbreviations
TCR, T cell receptor
APCs, antigen-presenting cells
FACS, fluorescence-activated cell sorter
CFP, cyan fluorescent protein
YFP, yellow fluorescent protein
DC, dendritic cell
PCC, pigeon cytochrome c
DIC, differential interference contrast
BM, bone marrow
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kropshofer H., Spindeldreher, S., Rohn, T. A., Platania, N., Grygar, C., Daniel, N., Wolpl, A., Langen, H., Horejsi, V. & Vogt, A. B. (2002) Nat. Immunol. 3, 61-68. [DOI] [PubMed] [Google Scholar]
- 2.Anderson H. A., Hiltbold, E. M. & Roche, P. A. (2000) Nat. Immunol. 1, 156-162. [DOI] [PubMed] [Google Scholar]
- 3.Wulfing C., Sumen, C., Sjaastad, M. D., Wu, L. C., Dustin, M. L. & Davis, M. M. (2002) Nat. Immunol. 3, 42-47. [DOI] [PubMed] [Google Scholar]
- 4.Grakoui A., Bromley, S. K., Sumen, C., Davis, M. M., Shaw, A. S., Allen, P. M. & Dustin, M. L. (1999) Science 285, 221-227. [DOI] [PubMed] [Google Scholar]
- 5.Sette A., Alexander, J., Ruppert, J., Snoke, K., Franco, A., Ishioka, G. & Grey, H. M. (1994) Annu. Rev. Immunol. 12, 413-431. [DOI] [PubMed] [Google Scholar]
- 6.Kersh B. E., Kersh, G. J. & Allen, P. M. (1999) J. Exp. Med. 190, 1627-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stotz S. H., Bolliger, L., Carbone, F. R. & Palmer, E. (1999) J. Exp. Med. 189, 253-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachmann M. F., Speiser, D. E., Zakarian, A. & Ohashi, P. S. (1998) Eur. J. Immunol. 28, 3110-3119. [DOI] [PubMed] [Google Scholar]
- 9.Torigoe C., Inman, J. K. & Metzger, H. (1998) Science 281, 568-572. [DOI] [PubMed] [Google Scholar]
- 10.Kraj P., Pacholczyk, R., Ignatowicz, H., Kisielow, P., Jensen, P. & Ignatowicz, L. (2001) J. Exp. Med. 194, 407-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozono H., White, J., Clements, J., Marrack, P. & Kappler, J. (1994) Nature 369, 151-154. [DOI] [PubMed] [Google Scholar]
- 12.Kinsella T. M. & Nolan, G. P. (1996) Hum. Gene Ther. 7, 1405-1413. [DOI] [PubMed] [Google Scholar]
- 13.Lutz M. B., Kukutsch, N., Ogilvie, A. L., Rossner, S., Koch, F., Romani, N. & Schuler, G. (1999) J. Immunol. Methods 223, 77-92. [DOI] [PubMed] [Google Scholar]
- 14.De Veerman M., Heirman, C., Van Meirvenne, S., Devos, S., Corthals, J., Moser, M. & Thielemans, K. (1999) J. Immunol. 162, 144-151. [PubMed] [Google Scholar]
- 15.Vandenberghe P. A. & Ceuppens, J. L. (1990) J. Immunol. Methods 127, 197-205. [DOI] [PubMed] [Google Scholar]
- 16.Greimers R., Trebak, M., Moutschen, M., Jacobs, N. & Boniver, J. (1996) Cytometry 23, 205-217. [DOI] [PubMed] [Google Scholar]
- 17.Ignatowicz L., Winslow, G., Bill, J., Kappler, J. & Marrack, P. (1995) J. Immunol. 154, 3852-3862. [PubMed] [Google Scholar]
- 18.Monks C. R., Freiberg, B. A., Kupfer, H., Sciaky, N. & Kupfer, A. (1998) Nature 395, 82-86. [DOI] [PubMed] [Google Scholar]
- 19.Banchereau J. & Steinman, R. M. (1998) Nature 392, 245-252. [DOI] [PubMed] [Google Scholar]
- 20.Fahmy T. M., Bieler, J. G., Edidin, M. & Schneck, J. P. (2001) Immunity 14, 135-143. [PubMed] [Google Scholar]
- 21.Dittel B. N., Stefanova, I., Germain, R. N. & Janeway, C. A. (1999) Immunity 11, 289-298. [DOI] [PubMed] [Google Scholar]
- 22.Hemmer B., Stefanova, I., Vergelli, M., Germain, R. N. & Martin, R. (1998) J. Immunol. 160, 5807-5814. [PubMed] [Google Scholar]
- 23.Kersh G. J., Kersh, E. N., Fremont, D. H. & Allen, P. M. (1998) Immunity 9, 817-826. [DOI] [PubMed] [Google Scholar]
- 24.Wulfing C., Bauch, A., Crabtree, G. R. & Davis, M. M. (2000) Proc. Natl. Acad. Sci. USA 97, 10150-10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K. H., Holdorf, A. D., Dustin, M. L., Chan, A. C., Allen, P. M. & Shaw, A. S. (2002) Science 295, 1539-1542. [DOI] [PubMed] [Google Scholar]
- 26.Preckel T., Grimm, R., Martin, S. & Weltzien, H. U. (1997) J. Exp. Med. 185, 1803-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.