Abstract
Phylogenies of indigenous microbes have been used as surrogates for the origins of the hosts that carry them. Conversely, polymorphisms may be used to date the spread of a microbial species when information about their host populations is available. Therefore, we examined polymorphisms in Helicobacter pylori, which persistently colonize the human stomach, to test the hypothesis that they have been ancient inhabitants of humans. Three H. pylori loci that previously have been shown to have phylogeographic affinity have been analyzed for two populations with different ethnic origins from Venezuela. In a group of Amerindian subjects from Amazonia, East Asian H. pylori genotypes were present for each of the loci examined but were absent in a mestizo population from Caracas. These findings provide evidence that H. pylori has been present in humans at least since ancestors of Amerindians migrated from Asia more than 11,000 years ago.
The peopling of the Americas occurred at least 11,000 years and up to 43,000 years before the present with origins in east Central Asia (1, 2). In addition to using host mitochondrial and Y-chromosome polymorphisms to reconstruct the population history of the indigenous Amerindians, inquiries also have focused on the demogeographic polymorphisms present in persistent viruses (e.g., JC virus and human T-lymphotrophic virus types I and II) carried by humans (3–9). The generally consistent findings of these studies indicate that the phylogenies of indigenous microbes can be used as surrogates for the origins of the hosts that carry them. Conversely, here we examine polymorphisms in Helicobacter pylori, which persistently colonize the human stomach, to determine whether evidence supports its presence in humans at least since ancestors of Amerindians migrated from Asia (10).
H. pylori, spiral, microaerophilic, Gram-negative bacteria that colonize the human stomach, are associated with increased risk of developing gastric cancer and peptic ulcer disease (11). Although common in developing country populations, H. pylori has been disappearing gradually from industrialized countries (12, 13). H. pylori strains are highly diverse, and within all loci examined, both extensive polymorphism and multiple informative sites are present that can be used to construct phylogenies (14, 15). In particular, specific genotypes associated with either Europe or East Asia have been recognized (14–16). Extensive studies have shown that the vacA s allelic types have a specific geographic distribution (15, 16). In particular, the vacA s1c allele is present exclusively in persons originating from East Asia (15, 17–19). Two other genes, babB and HP0638, also have been shown to have strong phylogeographic affinity and can be used to distinguish between strains from hosts of Eastern or Western origin (14, 20). Thus, in Europe and North America, individuals carrying H. pylori strains with East Asian markers either themselves immigrated from East Asia, or their parents or grandparents had done so (21).
However, the period during which humans have been colonized with H. pylori is controversial (22, 23). Studies examining polymorphisms in H. pylori isolates from Latin American patients showed European genotypes, and Kersulyte et al. have suggested that South America may have been H. pylori-free before the European colonization and Europeans might have introduced H. pylori to the New World after Columbus (15, 23). However, the patients in that study and in our previous work were from metropolitan areas and are mestizo, with mixed European, African, and aboriginal (Amerindian) origins (15, 23). Therefore, finding strains with markers of Western origin in this population of mestizo patients might be expected and is consistent with the hypothesis of a “selective sweep” in which the European genotypes could have replaced preexisting genotypes. To differentiate among these possibilities, it is ideal to examine individuals who more closely reflect the pre-Columbian peoples. To accomplish this aim, we examined gastric biopsy samples from Amerindians in the Venezuelan Amazon, because their ancestors migrated from East Central Asia no less than 11,000 years ago (1). Finding markers of East Asian strains in this relatively isolated population of true Amerindian descent would provide evidence that H. pylori arrived in the New World with their ancestors and has been indigenously transmitted since.
Materials and Methods
Biopsy Samples.
DNA was obtained from antrum biopsies from patients from Caracas, the urban capital of Venezuela, and Puerto Ayacucho, a regional center in Venezuelan Amazonia, which provides medical services to people from surrounding villages. All 103 persons from Caracas (mean age 42 yr) were of European or mixed ancestry. The 102 persons studied from the Puerto Ayacucho area (mean age 37 yr), were of Amerindian ancestry (including 96 Piaroas, 5 Guajibos, and 1 Yanomami). All patients were undergoing upper gastrointestinal endoscopy for dyspeptic symptoms and signed a consent form to participate in this study. Biopsy samples were obtained from patients in 1999. DNA was extracted from each biopsy as described (23).
PCR Analysis.
Primers used for H. pylori PCR amplification were: (i) GlmM1 and GlmM2 for a 296-bp glmM fragment (14); (ii) R1 and R2 for a 100-bp cagA fragment (24); (iii) VAIF and VAIXR for the vacA s region (15); (iv) 181 and 214 for a 401-bp fragment including parts of HP0637 and HP0638; and (v) C9655 and C5733 for a 496-bp region of babB (25). Human mitochondrial DNA primers mt L15905 and mt R16498 were used to amplify a 593-bp region of the hypervariable region I (26). In each case, PCR was performed with 100 ng of each primer and 100 ng of template DNA as described (23) with appropriate negative and positive controls.
Sequence Analysis.
Purified PCR products (QIAquick PCR purification kit, Qiagen, Valencia, CA) were sequenced directly on both strands at the New York University Skirball Institute of Biomedical Sciences core facility. Resulting sequences were assembled by using SEQUENCHER 3.1.1. (Gene Codes, Ann Arbor, MI), and edited alignments were created by using CLUSTALW (GenBank accession nos. are AF485664–AF485758) (27). Phylograms based on nucleotide alignments were generated by using Kimura two-parameter distance matrices (28) using paup 4.0b8 (Sinauer Associates, Sunderland, MA). Analysis of synonymous and nonsynonymous substitution rates was performed by using the method of Nei and Gojobori with SWAAP 1.0.0 (29, 30). Likelihood analysis was performed to determine the probability that the phylogenies created at the three loci could be reproduced by chance alone (31). Briefly, for each locus 1,000 trees were generated with random topology and then compared with the observed tree to determine the log likelihood (Δ − lnL) that each tree could be reproduced given the sequence data at each locus. Log-likelihood values were calculated for each random tree, and the 99th percentile bounding the random distribution was determined. For each observed tree, the calculated likelihood value was compared with the 99th percentile boundary of the random distribution; for values beyond the boundary, the hypothesis that the observed phylogenies could be reproduced by chance alone is rejected (31). Random trees and likelihood values were determined by using PAUP 4.0b8. Analysis of the number of polymorphic and informative sites that support the East Asian–Amerindian association in all the phylogenies was determined by using MACCLADE 4.04 (Sinauer Associates).
Results
Mitochondrial DNA Variation Between Two Populations of Venezuela.
We examined samples obtained from two groups of Venezuelan patients as described above: from Puerto Ayacucho, representing an Amerindian population, and from Caracas, representing the mestizo population. To confirm the ethnicity of the study populations, we examined a subset of the study subjects using sequence analysis of the mitochondrial DNA type1 hypervariable region (Table 1; refs. 4–6). Of 12 Caracas patients, 8 had phylogeographic affinity with Native American sequences and 4 with either West Eurasian or Sub-Saharan African sequences. In contrast, all 18 sequences from Puerto Ayacucho patients had Native American phylogeographic affinities (4–6). Thus, the Puerto Ayacucho population studied was suitable for exploration of the competing hypotheses.
Table 1.
Mitochondrial DNA lineages in gastric biopsy specimens from Venezuela
| HVR-I haplotype | Haplogroup | Phylogeographic affinity | Sample designation |
|---|---|---|---|
| 111 223 290 319 362 | A | Native American | VC 17, 22, 77, 93 |
| 111 145 223 290 319 362 | A | Native American | VP 26 |
| 111 223 290 293 304 319 362 | A | Native American | VP 29, 33, 35, 36, 41, 43, 66 |
| 86 182C 183C 189 217 | B | Native American | VC 62 |
| 51 223 298 325 327 | C | Native American | VP 52, 78 |
| 51 129 223 325 | C | Native American | VC 95 |
| 129 223 298 325 327 | C | Native American | VP 24 |
| 223 239 298 325 327 | D | Native American | VP 25 |
| 142 147 223 325 327 | D | Native American | VP 03 |
| 142 147 223 235 325 362 | D | Native American | VP 49 |
| 142 147 223 325 362 | D | Native American | VP 08, 55, 84 |
| 142 179 223 295 325 362 | D | Native American | VC 14, 35 |
| 223 311 312 325 362 | D | Native American | VP 19 |
| 0 | CRS | West Eurasian | VC 94 |
| 189 192 223 265 278 294 309 390 | L2 | Sub-Sahara African | VC 100 |
| 38 129 187 189 223 256 278 293 294 360 | L1c | Sub-Sahara African | VC 72 |
| 111 126 187 189 223 239 270 278 293 311 | L1b | Sub-Sahara African | VC 88 |
Sequence types for hypervariable region I (HVR-I) between base pairs 16,021 and 16,400 are shown as variant positions minus 16,000 relative to Cambridge reference sequence (26). Polymorphic positions are transitions unless the change is specified.
Haplogroup assignment of each sequence type is based on diagnostic motifs as described in refs. 4–6.
VP, Venezuela Puerto Ayacucho; VC, Venezuela Caracas.
CRS, Cambridge reference sequence. This sequence seems associated with several Eurasian haplogroups and is the most frequent type in populations of Europe and the Middle East (47).
Prevalence of H. pylori and Specific Genotypes in the Two Studied Populations of Venezuela.
For the total of 205 patients studied in both locales, DNA could be extracted from 195 (95%) of the biopsies in sufficient quantity and quality to permit PCR amplification. By glmM PCR of these biopsy samples, 78.4 and 80.6% of the 97 Puerto Ayacucho and 98 Caracas patients, respectively, were found to carry H. pylori (23). cagA is a marker for a 35- to 40-kb pathogenicity island in H. pylori (32–34), the presence of which is significantly correlated with clinical outcome in persons carrying the organisms (35, 36). The prevalence of cagA varies in different host populations, but it is present in nearly all strains of East Asian origin (37) and only half of the strains in North America (32). In a subset analysis, the rate of PCR positivity for cagA was nearly identical in H. pylori-positive patients from Puerto Ayacucho (n = 35, 62%) and Caracas (n = 31, 58%). Thus, the rate of H. pylori colonization and overall strain type was present to similar extents in these two study groups.
Analysis of vacA s Allelic Type and HP0638 and babB Types in the Two Populations of Venezuela.
Next we examined three polymorphic H. pylori loci with distinctive demogeographic features: (i) the s region of the vacA-encoded signal sequence (15); (ii) the 5′ 167 bp of HP0638 and the upstream 188 bp (20, 38); and (iii) a 5′ 378-bp region of babB (14). Due to small biopsy sizes and difficult specimen transport from Amazonia, sequence information was determined from representative samples, which gave sufficient PCR yields. Comparing the sequences obtained from the subjects of this study with those of the controls in other defined geographic regions showed extensive polymorphism in each locus, as expected.
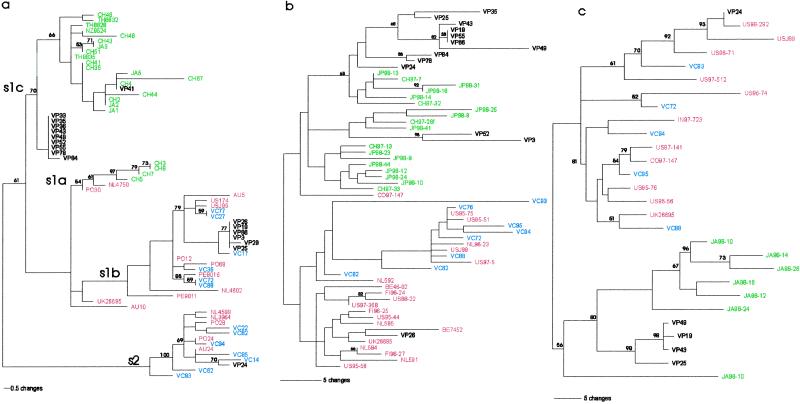
For vacA, all samples from Caracas had s1b or s2 sequences (39), consistent with earlier studies of mestizo populations (Table 2; refs. 15 and 23). In contrast, from Puerto Ayacucho, 58% were s1c, a strong marker for East Asian origin (15); by phylogenetic analysis, most of these s1c sequences represent a subpopulation distinct from the East Asian/Pacific sequences (Fig. 1a). For the 5′ HP0638 locus (20), the sequences from 12 of 13 Amazonian patients studied clustered with East Asian strains (Fig. 1b). For the HP0638 CT-repeat number, which also shows distinctive demogeographic features (20), all samples from the Caracas patients showed Western patterns, whereas those from the Amazonian patients showed either East Asian (54%) or Western (46%) patterns (Table 3; ref. 20). In analysis of the babB demogeographic locus, 4 of 5 Amazonian sequences were on a distinct branch related to the East Asian sequences, whereas all 5 Caracas patient sequences were on the branch with Western sequences (Fig. 1c). Although strain VP25 and VP19 babB sequences were on the same branch as for VP49 and VP43, the former strains were vacA s1b and the latter were s1c. These findings are indicative of intrahost recombination between Western and East Asian strains, as would be predicted from earlier analysis of H. pylori recombination (40, 41). Likelihood analysis was performed to test the hypothesis that the phylogenies created for the three loci could be reproduced by chance alone (31). Analyses using 1,000 random trees for each locus showed that the likelihood values for the observed phylogenies were far beyond the 99th percentile of the null distribution (P < 0.0001 in each case), indicating that chance alone would not explain their structure (data not shown).
Table 2.
Prevalence of vacA s-region allotypes in samples from East Asia, Western countries, or Venezuela
|
s allotype
|
Percent of sample showing specified allotypes | |||
|---|---|---|---|---|
| East Asia (n = 15)
|
Western countries (n = 15)
|
Venezuela | ||
| Puerto Ayacucho (n = 17) | Caracas (n = 13) | |||
| s1 | 100 | 67 | 94 | 46 |
| s1a | 27 | 20 | 0 | 0 |
| s1b | 0 | 47 | 36 | 46 |
| s1c | 73 | 0 | 58 | 0 |
| s2 | 0 | 33 | 6 | 53 |
s allotype was determined by sequence of PCR products using primers VAIF and VAIX as described.
Fig 1.
Neighbor-joining trees of H. pylori based on sequences at three loci. Sequences are from H. pylori strains from East Asia/Pacific (green), Western countries (red), Venezuela/Puerto Ayacucho (VP, black), Venezuela/Caracas (VC, blue). (a) the vacA s region has four major allotypes: s1a, s1b, s1c, and s2. The majority of Venezuela/Puerto Ayacucho sequences are on the s1c branch on a relatively conserved subbranch or amidst the East Asian/Pacific isolates. (b) HP0638 sequences divide into two major groupings, with sequences from East Asia, the majority of Venezuela/Caracas, and one from Colombia together (Top), and the Venezuela/Caracas sequences and most Western sequences (Bottom). (c) babB sequences divide into two major groupings, with East Asia and most of Venezuela/Puerto Ayacucho (Bottom) together, and Venezuela/Caracas and Western together (Top). CH, China; TH, Thailand; J, Japan; NL, The Netherlands; PO, Portugal; NZ, Maori; AU, Australia; US, United States; PE, Peru; UK, United Kingdom; CO, Colombia; BE, Belgium; FI, Finland; IN, India. Bootstrap values >50 and branch-length indices are shown.
Table 3.
Variation in the CT-repeat number in H. pylori HP0638 in samples from East Asia, Western countries, or Venezuela
| No. of CT repeats
|
Percent of individuals with H. pylori strain with specified CT-repeat number | |||
|---|---|---|---|---|
| East Asia (n = 47)
|
Western countries (n = 37)
|
Venezuela | ||
| Caracas (n = 8) | Puerto Ayacucho (n = 13) | |||
| ≤4 | 100 | 0 | 0 | 53 |
| 6–7 | 0 | 68 | 50 | 38 |
| ≥8 | 0 | 32 | 50 | 8 |
Sequences included from ref. 20.
Analysis of Polymorphic and Informative Sites.
In each locus, multiple polymorphic sites were present (Table 4). Analysis of the vacA s region phylogeny shows 40 informative sites, of which 10 (25%) directly support the hypothesis that the East Asian and Amerindian vacA s1c sequences are present on the same branch. Despite high levels of homoplasy, there are 97 informative sites in the babB phylogeny, of which 10 directly support the association, and in the HP0638 phylogeny 10 of the 72 informative sites also directly support the hypothesis that H. pylori strains from present-day Amerindians have a common ancestry with present-day East Asian H. pylori strains. The other informative sites support other phylogenetic divisions and are neutral for this hypothesis.
Table 4.
Informative sites in vacA, babB, and HP0638 sequences in H. pylori isolates
| Gene
|
No. of isolates included in analysis
|
Total sites
|
No. of sites | ||
|---|---|---|---|---|---|
| Polymorphic | Noninformative | Informative | |||
| vacA | 69 | 132 | 58 | 18 | 40 |
| babB | 28 | 378 | 130 | 33 | 97 |
| HP0638 | 59 | 399 | 138 | 57 | 72 |
Includes sequences from China (14), Thailand (3), Japan (4), New Zealand (1), Puerto Ayacucho (17), Caracas (13), The Netherlands (4), Portugal (5), Australia (3), the United States (2), Peru (2), and the United Kingdom (1).
Analysis of Synonymous and Nonsynonymous Substitutions.
Comparison of synonymous (Ks) and nonsynonymous (Ka) nucleotide substitutions within the vacA s region confirms both extensive diversity and functional variation; within each allele, the overall diversity and functional differences were reduced (Table 5 and data not shown). Within the vacA s1c allele, the level of synonymous substitutions within the sequences from Puerto Ayacucho and East Asia (Ks = 0.25) was higher than for either group alone, which suggests a substantial time interval since the divergence of these two groups; that the level of nonsynonymous substitution between these two groups was zero (Ka = 0.00) indicates the absence of functional differences (Table 5). For babB and HP0638 as well, sequences from Puerto Ayacucho and East Asia, when analyzed together, have higher rates of synonymous substitutions than when examined separately.
Table 5.
Synonymous and nonsynonymous substitutions within vacA s region, babB, and HP0638
| Genetic locus | Ks | Ka | Ka/Ks |
|---|---|---|---|
| vacA s-region | |||
| Total (n = 69) | 0.36 ± 0.18 | 0.13 ± 0.09 | 0.38 ± 0.32 |
| PA s1c (n = 10) | 0.05 ± 0.10 | 0.00 ± 0.01 | 0.06 ± 0.06 |
| East Asian s1c (n = 18) | 0.13 ± 0.08 | 0.01 ± 0.02 | 0.07 ± 0.11 |
| PA vs East Asian s1c | 0.25 ± 0.09 | 0.00 ± 0.01 | 0.02 ± 0.05 |
| babB | |||
| Total (n = 28) | 0.37 ± 0.14 | 0.07 ± 0.03 | 0.19 ± 0.08 |
| PA (n = 5) | 0.24 ± 0.25 | 0.06 ± 0.05 | 0.23 ± 0.16 |
| East Asian (n = 7) | 0.23 ± 0.05 | 0.03 ± 0.01 | 0.13 ± 0.04 |
| PA vs. East Asian | 0.36 ± 0.12 | 0.08 ± 0.02 | 0.22 ± 0.05 |
| HP0638 | |||
| Total (n = 59) | 0.17 ± 0.09 | 0.05 ± 0.02 | 0.26 ± 0.22 |
| PA (n = 13) | 0.12 ± 0.11 | 0.03 ± 0.02 | 0.24 ± 0.21 |
| East Asian (n = 18) | 0.15 ± 0.04 | 0.06 ± 0.05 | 0.25 ± 0.20 |
| PA vs. East Asian | 0.16 ± 0.07 | 0.05 ± 0.02 | 0.41 ± 0.26 |
Discussion
Although the mixture of Amerindian, European, and African human and microbial genes has been occurring in South America for >500 years, examination of relatively isolated populations provides a means to assess for genes that predate the European conquest (1, 7–9). That transmission of H. pylori occurs primarily in families (42) and that strains from geographically separated human populations can be distinguished indicate that such differences can be used to assess strain origins (15, 23, 25, 38, 43). However, even in Puerto Ayacucho nonindigenous genes clearly have been introduced as indicated by the Western H. pylori genotypes observed in a proportion of the patients. Despite intermixing, the prominence of East Asian genotypes in H. pylori populations from patients in Puerto Ayacucho and their absence from the control Caracas (mestizo) population provide evidence that these genotgypes are indigenous in the Amerindian population. Generally parallel findings in each of the three loci examined increase confidence that the polymorphisms observed represent clonal descent from East Asian ancestors rather than isolated homoplasies. The substantial level of synonymous substitutions between the Puerto Ayacucho and East Asian s1c alleles indicates that the divergence of these two groups is not recent and virtually eliminates as a possibility a recent transfer of the s1c allele to Puerto Ayacucho from East Asian strains. This level of synonymous substitutions, combined with the lack of nonsynonymous substitutions, is indicative of substantial random drift that occurred since the last common ancestral strain that carried this allele.
These findings imply that the ancestors of present-day Amerindians carried H. pylori (in the stomach) when they migrated from Asia and thus that H. pylori has been present in humans for a minimum of 11,000 years (2), which is consistent with recent findings involving isolates from Amerindian people in Colombia (16). A more precise temporal assessment of the duration of H. pylori in human populations could not be done because of the lack of samples collected over an extended time from the same individual. Based on examining paired isolates obtained ≈2 yr apart from individual hosts, Falush et al. estimated that the presence of H. pylori in humans is at least 2,500 years (41). The distance of the Puerto Ayacucho vacA s1c sequences from the East Asian s1c sequences is consistent with long-standing separation of these two gene pools, and their relative homogeneity is consistent with an evolutionary bottleneck such as may have occurred during passage across the Bering Strait (1) or as a consequence of relative genetic isolation within Amazonia. Although data from each of the three different genetic loci provide evidence for East Asian origins of Amerindian H. pylori strains, their phylogenies are not strictly clonal, consistent with frequent recombination among H. pylori strains (40, 41) and the selection of strains with mixed East Asian and European characteristics after the European conquest.
Whereas for the populations examined, the mitochondrial haplotypes predominantly reflect Amerindian origins, the dominance in the mestizo population of H. pylori genotypes introduced after Columbus, as well as their presence in a more indigenous Amerindian population, suggest strong fitness advantages of these alleles. However, the factors permitting a selective sweep by these alleles have not been determined, and as shown in this report, recombination may have occurred frequently. Evidence that H. pylori has been present in the human stomach for at least thousands of years, causing little disease during reproductive age (11), suggests that colonization may be beneficial or of low biological cost to humans (44–46).
Acknowledgments
We acknowledge the collaboration of the Gastroenterology service at Hospital General del Oeste (Caracas, Venezuela) and Dr. Euclides Gonzalez from the Hospital Central de Puerto Ayacucho. This work was supported by Grant R01GM63270 and the Medical Scientist Training Program from the National Institutes of Health, the Medical Research Service of the Department of Veterans Affairs, the Institute for Urban and Global Health (New York University), and the Instituto Venezolano de Investigaciones Cientificas.
References
- 1.Hoffecker J. F., Powers, W. R. & Goebel, T. (1993) Science 259, 46-53. [DOI] [PubMed] [Google Scholar]
- 2.Bonatto S. L. & Salzano, F. M. (1997) Proc. Natl. Acad. Sci. USA 94, 1866-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santos F. R., Pandya, A., Tyler-Smith, C., Pena, S. D., Schanfield, M., Leonard, W. R., Osipova, L., Crawford, M. H. & Mitchell, R. J. (1999) Am. J. Hum. Genet. 64, 619-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Rienzo A. & Wilson, A. C. (1991) Ann. Hum. Genet. 88, 1597-1601. [Google Scholar]
- 5.Macaulay V., Richards, M., Hickey, E., Vega, E., Cruciani, F., Guida, V., Scozzari, R., Bonne-Tamir, B., Sykes, B. & Torroni, A. (1999) Am. J. Hum. Genet. 64, 232-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torroni A., Rengo, C., Guida, V., Cruciani, F., Sellitto, D., Coppa, A., Calderon, F. L., Simionati, B., Valle, G., Richards, M., Macaulay, V. & Scozzari, R. (2001) Am. J. Hum. Genet. 69, 1348-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agostini H. T., Yanagihara, R., Davis, V., Ryschkewitsch, C. F. & Stoner, G. L. (1997) Proc. Natl. Acad. Sci. USA 94, 1452-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandamme A. M., Hall, W. W., Lewis, M. J., Goubau, P. & Salemi, M. (1999) Nat. Med. 5, 1428-1432. [DOI] [PubMed] [Google Scholar]
- 9.Neel J. V., Biggar, R. J. & Sukernik, R. I. (1994) Proc. Natl. Acad. Sci. USA 91, 10737-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuipers E. J, Israel, D. A., Kusters, J. G., Gerrits, M. M., Weel, J., van Der Ende, A., van Der Hulst, R., Wirth, H. P., Hook-Nikanne, J, Thompson, S. A. & Blaser, M. J. (2000) J. Infect. Dis. 181, 273-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peek R. M. & Blaser, M. J. (2002) Nat. Rev. Cancer 2, 28-37. [DOI] [PubMed] [Google Scholar]
- 12.Rehnberg-Laiho L., Rautelin, H., Koskela, P., Sarna, S., Pukkala, E., Aromaa, A., Knekt, P. & Kosunen, T. U. (2001) Epidemiol. Infect. 126, 37-42. [PMC free article] [PubMed] [Google Scholar]
- 13.Perez-Perez G. I., Salomaa, A., Kosunen, T. U., Daverman, B., Rautelin, H., Aromaa, A., Knekt, P. & Blaser, M. J. (2002) Gut 50, 295-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pride D. T. & Blaser, M. J. (2002) J. Mol. Biol. 316, 627-640. [DOI] [PubMed] [Google Scholar]
- 15.van Doorn L.-J., Figueiredo, C., Mégraud, F., Pena, S., Midolo, P., Queiroz, D. M. D. M., Carneiro, F., Vanderborght, B., Pegado, M. D. G. F., Sanna, R., et al. (1999) Gastroenterology 116, 823-830. [DOI] [PubMed] [Google Scholar]
- 16.Yamaoka Y., Orito, E., Mizokami, M., Gutierrez, O., Saitou, N., Kodama, T., Osato, M. S., Kim, J. G., Ramirez, F. C., Mahachai, V. & Graham, D. Y. (2002) FEBS Lett. 517, 180-184. [DOI] [PubMed] [Google Scholar]
- 17.Wong B. C., Yin, Y., Berg, D. E., Xia, H. H., Zhang, J. Z., Wang, W. H., Wong, W. M., Huang, X. R., Tang, V. S. & Lam, S. K. (2001) Helicobacter 6, 317-324. [DOI] [PubMed] [Google Scholar]
- 18.Kim S. Y., Woo, C. W., Lee, Y. M., Son, B. R., Kim, J. W., Chae, H. B., Youn, S. J. & Park, S. M. (2001) J. Korean Med. Sci. 16, 579-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park I. S., Lee, Y. C., Park, H. J., Kim, T. I., Lee, S. I., Kim, H., Chung, K. S. & Lee-Kim, Y. C. (2001) Yonsei Med. J. 42, 457-470. [DOI] [PubMed] [Google Scholar]
- 20.Ando T., Peek, R. M., Pride, D., Levine, S. M., Takata, T., Lee, Y.-C., Kusugami, K., van der Ende, A., Kuipers, E. J., Kusters, J. G. & Blaser, M. J. (2002) J. Clin. Microbiol. 40, 239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gold B., van Doorn, L.-J., Guarner, J., Owens, M., Pierce-Smith, D., Song, Q., Hutwagner, L., Sherman, P. M., de Mola, O. L. & Czinn, S. J. (2001) J. Clin. Microbiol. 39, 1348-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaser M. J. (1998) Gut 43, 721-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kersulyte D., Mukhopadhyay, A. K., Velapatiño, B., Su, W. W., Pan, Z. J., Garcia, C., Hernandez, V., Valdez, Y., Mistry, R. S., Gilman, R. H., et al. (2000) J. Bacteriol. 182, 3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rugge M., Busatto, G., Cassaro, M., Shiao, Y. H., Russo, V., Leandro, G., Avellini, C., Fabiano, A., Sidoni, A. & Covacci, A. (1999) Cancer 85, 2506-2511. [PubMed] [Google Scholar]
- 25.Pride D. T. & Blaser, M. J. (2001) Infect. Immun. 69, 1160-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson S., Bankier, A. T., Barrell, B. G., de Bruijn, M. H., Coulson, A. R., Drouin, J., Eperon, I. C., Nierlich, D. P., Roe, B. A., Sanger, F., et al. (1981) Nature 290, 457-465. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura M. (1980) J. Mol. Evol. 16, 111-120. [DOI] [PubMed] [Google Scholar]
- 29.Pride D. T. & Blaser, M. J. (2002) Genome Lett. 1, 2-15. [Google Scholar]
- 30.Nei M. & Gojobori, T. (1986) Mol. Biol. Evol. 3, 418-426. [DOI] [PubMed] [Google Scholar]
- 31.Feil E. J., Holmes, E. C., Bessen, D. E., Chan, M. S., Day, N. P., Enright, M. C., Goldstein, R., Hood, D. W., Kalia, A., Moore, C. E., Zhou, J. & Spratt, B. G. (2001) Proc. Natl. Acad. Sci. USA 98, 182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tummuru M., Cover, T. L. & Blaser, M. J. (1993) Infect. Immun. 61, 1799-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Censini S., Lange, C., Xiang, Z., Crabtree, J. E., Ghiara, P., Borodovsky, M., Rappuoli, R. & Covacci, A. (1996) Proc. Natl. Acad. Sci. USA 93, 14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akopyants N. S., Clifton, S. W., Kersulyte, D., Crabtree, J. E., Youree, B. E., Reece, C. A., Bukanov, N. O., Drazek, E. S., Roe, B. A. & Berg, D. E. (1998) Mol. Microbiol. 28, 37-53. [DOI] [PubMed] [Google Scholar]
- 35.Blaser M. J., Perez-Perez, G. I., Kleanthous, H., Cover, T. L., Peek, R. M., Chyou, P. H., Stemmermann, G. N. & Nomura, A. (1995) Cancer Res. 55, 2111-2115. [PubMed] [Google Scholar]
- 36.Parsonnet J., Friedman, G. D., Orentreich, N. & Vogelman, H. (1997) Gut 40, 297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan Z. J., van der Hulst, R. W., Feller, M., Xiao, S. D., Tytgat, G. N., Dankert, J. & van der Ende, A. (1997) J. Clin. Microbiol. 35, 1344-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaoka Y., Kwon, D. H. & Graham, D. Y. (1995) Proc. Natl. Acad. Sci. USA 97, 7533-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atherton J. C., Cao, P., Peek, R. M., Jr., Tummuru, M. K., Blaser, M. J. & Cover, T. L. (1995) J. Biol. Chem. 270, 17771-17777. [DOI] [PubMed] [Google Scholar]
- 40.Suerbaum S., Smith, J. M., Bapumia, K., Morelli, G., Smith, N. H., Kunstmann, E., Dyrek, I. & Achtman, M. (1998) Proc. Natl. Acad. Sci. USA 95, 12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falush D., Kraft, C., Taylor, N. S., Correa, P., Fox, J. G., Achtman, M. & Suerbaum, S. (2001) Proc. Natl. Acad. Sci. USA 98, 15056-15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodman K. J. & Correa, P. (2000) Lancet 355, 358-362. [DOI] [PubMed] [Google Scholar]
- 43.Drumm B., Perez-Perez, G. I., Blaser, M. J. & Sherman, P. M. (1990) N. Engl. J. Med. 322, 359-363. [DOI] [PubMed] [Google Scholar]
- 44.Putsep K., Branden, K., Boman, H. G. & Normark, S. (1999) Nature 398, 671-672. [DOI] [PubMed] [Google Scholar]
- 45.Rothenbacher D., Blaser, M. J., Bode, G. & Brenner, H. (2000) J. Infect. Dis. 182, 1446-1449. [DOI] [PubMed] [Google Scholar]
- 46.Mattsson A., Lönroth, H., Quiding-Järbrink, M. & Svennerholm, A. (1998) J. Clin. Invest. 102, 51-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richards M., Macaulay, V., Hickey, E., Vega, E., Sykes, B., Guida, V., Rengo, C., Sellitto, D., Cruciani, F., Kivisild, T., et al. (2000) Am. J. Hum. Genet. 67, 1251-1276. [PMC free article] [PubMed] [Google Scholar]