Abstract
Feedback de-excitation (qE) regulates light harvesting in plants to prevent inhibition of photosynthesis when light absorption exceeds photosynthetic capacity. Although the mechanism of qE is not completely understood, it is known to require a low thylakoid lumen pH, de-epoxidized xanthophylls, and the photosystem II protein PsbS. During a short-term 4-h exposure to excess light, three PsbS- and qE-deficient Arabidopsis thaliana mutants that differed in xanthophyll composition were more photoinhibited than the wild type. The extent of photoinhibition was the same in all of the mutants, suggesting that qE capacity rather than xanthophyll composition is critical for photoprotection in short-term high light, in contrast to longer-term high light conditions (days) when additional antioxidant roles of specific xanthophylls are evident. Plants with a 2-fold increase in qE capacity were generated by overexpression of PsbS, demonstrating that the level of PsbS limits the qE capacity in wild-type Arabidopsis. These results are consistent with the idea that variations in PsbS expression are responsible for species-specific and environmentally induced differences in qE capacity observed in nature. Furthermore, plants with higher qE capacity were more resistant to photoinhibition than the wild type. Increased qE was associated with decreased photosystem II excitation pressure and changes in the fractional areas of chlorophyll a fluorescence lifetime distributions, but not the lifetime centers, suggesting that qE protects from photoinhibition by preventing overreduction of photosystem II electron acceptors. Engineering of qE capacity by PsbS overexpression could potentially yield crop plants that are more resistant to environmental stress.
Plants experience fluctuations in light intensity in nature, with quantitative changes occurring over several orders of magnitude and on time scales ranging from seconds to seasons. Because of these fluctuations, imbalances between light harvesting and utilization of absorbed light energy in photosynthesis occur frequently for most plants. Absorption of light in excess of a plant's capacity for CO2 fixation has the potential to increase production of reactive intermediates and reactive oxygen species that can cause an inhibition of photosynthesis. To prevent this photoinhibition, plants and algae have evolved several regulatory mechanisms and antioxidant systems (1).
In excess light, photosynthetic light harvesting is regulated by feedback de-excitation of singlet excited chlorophyll molecules in photosystem (PS) II. Feedback de-excitation, which can be measured as the qE component of nonphotochemical quenching of chlorophyll fluorescence (NPQ), is a nearly ubiquitous phenomenon in plants and eukaryotic algae that results in harmless thermal dissipation of excess absorbed light energy (reviewed in refs. 2 and 3). Induction and relaxation of qE occur rapidly (time scale of seconds to minutes), and qE has been considered to represent a protective mechanism that prevents PS II photoinhibition during short-term fluctuations in light intensity (3). Three elements are necessary for qE: de-epoxidized xanthophylls with a 3-hydroxy β-ring endgroup, the PsbS protein, and a low thylakoid lumen pH.
The xanthophylls involved in qE in plants are generated primarily by the xanthophyll cycle, the enzyme-catalyzed pigment conversion from violaxanthin to zeaxanthin via antheraxanthin in excess light by violaxanthin de-epoxidase (VDE) and back to violaxanthin in low light by zeaxanthin epoxidase (4–6). Zeaxanthin and antheraxanthin are the active molecules that are involved in qE (5, 7). Lutein, because of its structural similarity to antheraxanthin, might also function in qE (8, 9). Direct evidence for the involvement of these pigments in qE has come from studies of mutants of the green alga Chlamydomonas reinhardtii (8) and the laboratory weed Arabidopsis thaliana (9–11) and from experiments with transgenic tobacco plants with reduced levels of VDE (12). For example, the Arabidopsis npq1 mutant, which is defective in the gene encoding VDE, has very low levels of qE (10). The Arabidopsis lut2 mutant is defective in the lycopene ɛ-cyclase, is unable to synthesize α-carotene and lutein, and exhibits slower induction and a lower extent of qE (9). Essentially all qE is abolished in the npq1 lut2 double mutant (11).
PsbS is a 22-kDa PS II subunit that belongs to the light-harvesting complex (LHC) protein superfamily (13, 14). Unlike most LHC proteins, which have three transmembrane helices, PsbS has four transmembrane helices (15), and the pigment-binding characteristics of PsbS also differ from those of typical LHC proteins (16, 17). PsbS has been shown to be specifically involved in qE, because qE is absent in an Arabidopsis psbS deletion mutant, npq4–1 (hereafter called npq4), whereas light harvesting and other photosynthetic parameters are not affected (18). Analysis of the genetic semidominance of npq4 in heterozygous plants has demonstrated a stoichiometric dependence of qE capacity on the amount of PsbS protein (19).
The third element that is necessary for qE is a low thylakoid lumen pH, which occurs when the ΔpH across the thylakoid membrane increases in magnitude in excess light. The resulting decrease in lumen pH activates VDE (6) and causes the protonation of PsbS (17, 19, 20) and possibly other PS II proteins (2). This protonation is thought to cause some conformational change of these proteins (2, 3) that appears to involve a change in the energetics of one or two bound zeaxanthin molecules per PS II (21).
Photoprotective functions of qE and de-epoxidized xanthophylls have been strongly supported by studies of plants growing in natural conditions. In these investigations, growth at high photon flux densities (PFDs) was closely associated with increases in qE capacity, xanthophyll cycle de-epoxidation state, and total xanthophyll cycle pool size (reviewed in refs. 5 and 22). Other evidence has come from experiments in which VDE activity and qE in leaves were blocked by using DTT as an inhibitor of VDE (23–25), by mutation of the VDE gene in npq1 mutants (10), or by antisense inhibition of VDE expression (26). These experiments indicated a photoprotective role for qE but did not distinguish the contribution of qE from protection by de-epoxidized xanthophylls themselves. This is especially important, because in addition to their role in qE, xanthophylls are important lipid-soluble antioxidants that can quench excited triplet chlorophyll and singlet oxygen species, inhibit lipid peroxidation, and stabilize membranes (27). Studies of the Arabidopsis mutants npq1, npq4, and the double mutant npq4 npq1 have revealed that zeaxanthin, besides its role in qE, is involved in protection against photo-oxidative membrane damage during long-term high light stress (28). An additional role for lutein has also been suggested by long-term light stress experiments with the npq1 lut2 double mutant (11). Recent experiments with npq4 mutants have provided evidence for the protective function of qE under short-term high light stress (29) or a combination of high light and chilling stress (30). Furthermore, both npq4 and npq1 mutants exhibit a fitness defect when grown either under natural sunlight in the field or in rapidly fluctuating moderate light in the laboratory (31), demonstrating the important role of qE in a variable light environment.
To dissect further the photoprotective role of qE from that of xanthophyll pigments and to address in particular the physiological significance of qE, we constructed and characterized a series of Arabidopsis mutants with stepwise impairments of qE and xanthophyll metabolism, as well as Arabidopsis transgenic plants with enhanced qE but normal xanthophyll composition. Pigment composition and photosynthetic parameters were tested before, during, and after a short-term high light treatment. The results confirm that qE protects PS II from photoinhibition during short-term light stress and show that the level of PsbS limits qE capacity in wild-type Arabidopsis, demonstrating the feasibility of engineering plants with significantly enhanced photoprotective qE.
Materials and Methods
Plant Material.
All A. thaliana plants were of the ecotype Columbia (Col-0). The npq4 npq1 lut2 triple mutant was constructed by crossing npq4 npq1 (18, 28) with npq1 lut2 (11). PsbS-overexpressing plants were generated by transforming Col-0 with a psbS genomic clone. A 3,198-bp fragment of BAC clone F9J23 containing the psbS gene with its endogenous promoter was subcloned into the pPZP121 vector (32) to generate pXPL7. Plants were transformed (33) by using Agrobacterium tumefaciens GV3101 containing pXPL7. DNA gel blotting was conducted on the T1 generation to select transgenic lines with single insertions. Two independent homozygous lines, L5 and L17, were identified in the T3 generation.
Growth, Treatment, and Recovery Conditions.
Plants were grown in a growth chamber with high intensity discharge lighting (E15, Conviron, Winnipeg, Canada) at 150 μmol photons m−2⋅s−1 (low light, LL) and a 10-h light (22°C)/14-h dark (18°C) cycle for ≈7 weeks (just before bolting). For RNA extraction and immunoblot analysis, rosette leaves were harvested 3 h into the light period. For the high-light treatment of wild-type and mutants, plants were first illuminated with LL for 2 h in the morning and then exposed to 1,700 μmol photons m−2⋅s−1 (high light, HL) for 4 h followed by a recovery period in LL for another 4 h and then overnight in the dark. The experiment was repeated 4–6 times with independently grown batches of plants. For the transgenic lines, leaves were cut from plants and floated on a large volume of water. Leaves were exposed to LL for 2 h and then to HL for 6 h followed by a recovery period in LL for 1 h and then overnight in the dark. Five leaves were analyzed for each time point, and this experiment was repeated three times with independently grown batches of plants.
Pigment Measurements.
Determination of carotenoids and chlorophylls by HPLC was performed as described (34).
Chlorophyll Fluorescence and Oxygen Evolution Measurements.
Standard chlorophyll fluorescence parameters (35) were measured by using a commercial fluorometer (FMS2, Hansatech, King's Lynn, U. K.). NPQ was calculated as (Fm − Fm′)/Fm′; 1 − qP was calculated as (Fs − Fo′)/(Fm′ − Fo′); and Fv/Fm was calculated as (Fm − Fo)/Fm. In Figs. 1 and 5, qE was calculated as (Fm − Fm′)/Fm′, where the value of Fm was determined after relaxation of qE for 15 min in the dark. Oxygen evolution was measured in detached leaves with a leaf disk electrode system (LD2/2, Hansatech) in response to a series of increasing PFDs. The apparent quantum yield of oxygen evolution [Φ(O2)] was calculated from the initial slope of the light response curve. The fluorescence lifetimes were measured with an ISS K2–004 phase and modulation fluorometer (ISS Inc., Urbana, IL). The basic instrumental setup, including the temperature-controlled sample compartment, fiber-optic probe, data acquisition protocol, and data analysis software and procedures are described elsewhere (36, 37). Plants were dark-adapted for 12 h and kept in darkness before leaf pieces (2 cm2) were vacuum infiltrated with 0.35 M glucose for 30 min. After the lifetime determination, leaf pieces were frozen in liquid nitrogen and stored at −80°C before pigment analysis.
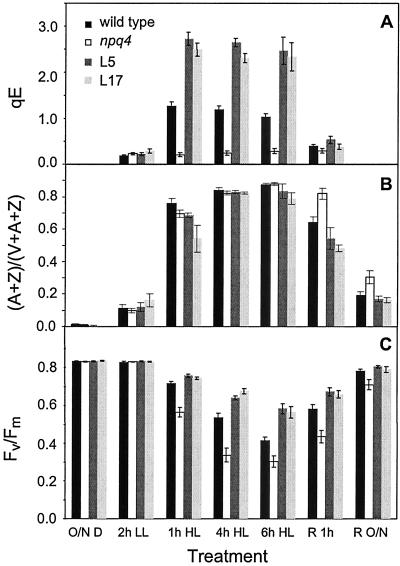
Fig 1.
Characteristics of LL (150 μmol photons m−2⋅s−1)-grown wild-type and mutant Arabidopsis plants before, during, and after treatment with HL (1,700 μmol photons m−2⋅s−1). (A) qE. Fm′ was measured at the indicated time points in LL or HL, and Fv and Fm were measured after 15 min in the dark. qE was calculated as (Fm − Fm′)/Fm′. Data are shown as the means ± SE (n ≥ 12). (B) Xanthophyll cycle de-epoxidation state ([A+Z]/[V+A+Z]). Data are shown as the means ± SE (n = 4). (C) PS II photochemical efficiency (Fv/Fm). Data are shown as the means ± SE (n ≥ 12).
Fig 5.
Characteristics of LL-grown wild type, L5, L17, and npq4 plants before, during, and after HL treatment. Data are shown as the means ± SE (n = 15). (A) qE. Fm′ was measured at the indicated time points in LL or HL, and Fv and Fm were measured after 15 min in the dark. qE was calculated as (Fm − Fm′)/Fm′. Fm′ could not be measured after the overnight dark periods, so no qE values were calculated for the O/N D and R O/N time points. (B) Xanthophyll cycle de-epoxidation state ([A+Z]/[V+A+Z]). (C) PS II photochemical efficiency (Fv/Fm). O/N D, overnight dark; R, recovery in LL; R O/N, recovery overnight in the dark.
DNA and RNA Blotting.
Genomic DNA was extracted from young flower buds (38), and RNA was extracted from leaves (39). For DNA gel blot analysis, a 1.1-kb PCR product generated from pXPL7 with the primers XPL14 (5′-CTTTTTCCCCCATGTAAGC-3′) and PZP1 (5′-CAGCGGAGGGGTTGGATC-3′) was used as a hybridization probe using the AlkPhos Direct kit and CDP-Star detection (Amersham Pharmacia). A 0.9-kb fragment of the psbS cDNA (EST clone 137M5T7) was amplified by PCR with the primers KN118 (5′-TCCTTCTCTCATCCTCAGAAA-3′) and KN119 (5′-CAACATGAAGAGAAGGTCACA-3′) and used as a hybridization probe for RNA blots. RNA gel blot analysis was done using the DIG-High Prime DNA Labeling and Detection kit (Roche Molecular Biochemicals).
Immunoblot Analysis.
Thylakoid isolation and PsbS immunoblotting were performed as described (20). The D1 antibody was kindly provided by Anastasios Melis (University of California, Berkeley), and the Lhcb1, Lhcb2, Lhcb3, Lhcb5, and Lhcb6 antibodies were kindly provided by Stefan Jansson (University of Umeå, Umeå, Sweden). The MLH2 monoclonal antibody (40) recognizing Lhcb4 was prepared from supernatants of hybridoma cells obtained from the American Type Culture Collection (CRL-1779).
Results
Characteristics of Wild-Type Plants and npq Mutants Before HL Treatment.
To differentiate the photoprotection caused by qE from additional protective functions of xanthophyll pigments, we compared the npq4 single mutant, npq4 npq1 double mutant, and npq4 npq1 lut2 triple mutant with wild-type Arabidopsis plants. The npq4 mutant specifically lacks qE (18), the npq4 npq1 double mutant lacks both qE and zeaxanthin in high light, and the npq4 npq1 lut2 triple mutant additionally lacks lutein. Under LL conditions (150 μmol photons m−2⋅s−1), wild-type and mutant plants had the same total chlorophyll content and chlorophyll a/b ratio, with the exception of the triple mutant that had a slightly lower chlorophyll content and slightly higher chlorophyll a/b ratio (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). Xanthophyll cycle pigment pool size was the same in the wild type, npq4, and npq4 npq1, and as expected it was nearly four times higher in the npq4 npq1 lut2 triple mutant due to the lack of lutein (see Table 2). Lutein concentrations in the npq4 and npq4 npq1 mutants were the same as that of the wild type (see Table 2). None of the mutants had any PsbS protein (because of the npq4 mutation), but there were no differences in the level of the PS II reaction center protein, D1 (data not shown).
qE, Xanthophyll Cycle Pigment Conversion, and Increased Photoinhibition During HL Treatment of Mutants.
Changes in qE and pigment composition were measured during a 4-h treatment of LL-grown plants with HL (1,700 μmol photons m−2⋅s−1). As shown in Fig. 1A, qE increased in wild-type plants during the HL treatment, but qE stayed at a very low level in all three mutants because of the npq4 mutation (18). The xanthophyll de-epoxidation state ([A + Z]/[V + A + Z]) increased dramatically in wild type and npq4 under HL and then decreased gradually after the plants were shifted back to LL (Fig. 1B). In the double and triple mutants, the xanthophyll cycle was blocked by the npq1 mutation, and thus the de-epoxidation state did not change substantially in the double and triple mutants (Fig. 1B).
Photoinhibition during the HL treatment and recovery period was measured by chlorophyll fluorescence and oxygen evolution. The maximal PS II photochemical efficiency, represented by the chlorophyll fluorescence parameter Fv/Fm, decreased gradually during exposure to HL and then increased during the LL recovery period (Fig. 1C). The HL-induced decrease in Fv/Fm was significantly more pronounced in the mutants than in the wild type; however, the extent of photoinhibition of Fv/Fm was similar in all three mutants, despite their differences in xanthophyll composition. During the LL recovery period, Fv/Fm in the three mutants did not recover to a level as high as the wild type. The apparent quantum yield of oxygen evolution [Φ(O2)], which also reflects the quantum efficiency of PS II, was compared between wild type and npq4. Corroborating the Fv/Fm data, Φ(O2) decreased more in npq4 than in the wild type during the HL treatment (Fig. 2).
Fig 2.
Apparent quantum yield of oxygen evolution [Φ(O2)] before, during, and after HL treatment of wild type and npq4. Data are shown as the means ± SE (n = 4). At the 1- and 4-h HL time points, the wild type and npq4 differ significantly (P < 0.05, t test).
Increased qE Capacity in Plants That Overexpress PsbS.
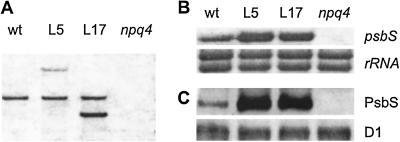
The results obtained with the npq4, npq4 npq1, and npq4 npq1 lut2 mutants suggested that qE capacity rather than differences in xanthophyll composition was critical for photoprotection in short-term HL, so we attempted to increase the qE capacity of Arabidopsis plants. Previous studies of npq4 have suggested that qE capacity is determined by PsbS protein level (18, 19). To investigate the effect of increasing expression of the PsbS protein above the wild-type level, wild-type plants were transformed with the psbS gene under the control of its own promoter. Two independent homozygous transgenic lines, L5 and L17, were selected for analysis. DNA gel blotting on the T3 generation confirmed that L5 and L17 each had only one additional band that hybridized with a psbS probe (Fig. 3A). Higher psbS mRNA levels were detected in both L5 and L17 (Fig. 3B).
Fig 3.
DNA, RNA, and protein analyses in wild-type plants (wt), psbS-overexpressing lines (L5 and L17), and the npq4 mutant. (A) DNA gel blot analysis. Genomic DNA (8 μg) was digested with XhoI and hybridized with a psbS probe. (B) RNA gel blot analysis. Total RNA (10 μg) was hybridized with a psbS probe. rRNA was visualized by staining of the blot with methylene blue. (C) Immunoblot analysis of PsbS and D1.
The levels of PsbS and other Lhcb proteins in L5 and L17 were determined by immunoblotting using specific antibodies. Compared with the wild type, L5 and L17 had several times more PsbS protein relative to the PS II reaction center protein D1, and as expected npq4 totally lacked any PsbS protein (Fig. 3C). Despite the differences in the PsbS levels between wild type, L5, L17, and npq4, the amounts of all other Lhcb proteins (Lhcb1–6) (see Fig. 7, which is published as supporting information on the PNAS web site) and the D1 protein (Fig. 3C) were the same.
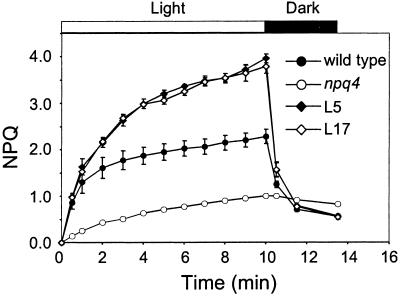
The level of total NPQ induced by exposure to a PFD equivalent to full sunlight (2,000 μmol m−2⋅s−1) was substantially higher in L5 and L17 compared with the wild type (Fig. 4). The total NPQ reached values of 2.3, 4.0, 3.8, and 1.0 in the wild type, L5, L17, and npq4, respectively. By subtracting the residual NPQ in each genotype after relaxation in the dark, we estimated the capacity of the qE component after 10-min light induction in the wild type to be 1.7, whereas it was 3.4 and 3.3 in L5 and L17, respectively, and only 0.2 in npq4.
Fig 4.
NPQ induction curves in wild type, L5, L17, and npq4. Plants were dark-adapted overnight before exposure to actinic light of 2,000 μmol photons m−2⋅s−1. Data are shown as the means ± SE (n = 3).
Resistance to Photoinhibition and Lower PS II Excitation Pressure in Leaves with Higher qE Capacity.
To assess the effects of HL on L5 and L17, leaves of plants grown in LL were exposed to HL for 6 h. Consistent with the results of Fig. 4, L5 and L17 exhibited qE levels in HL that were twice as high as that of the wild type (Fig. 5A). All four genotypes had a similar xanthophyll cycle pool size (data not shown) and a similar xanthophyll cycle de-epoxidation state during the HL treatment (Fig. 5B). During recovery, a slower re-epoxidation of zeaxanthin to violaxanthin was observed in npq4, resulting in a sustained higher de-epoxidation state similar to that observed after severe photoinhibition of other plants (41, 42). Photoinhibition of PS II during the HL treatment and recovery was again estimated by measuring the PS II photochemical efficiency as Fv/Fm. All genotypes had the same Fv/Fm before exposure to HL, but the two lines with higher qE capacity (L5 and L17) were significantly more resistant to photoinhibition (Fig. 5C). During this more severe HL treatment, npq4 was even more sensitive to photoinhibition than in the earlier experiment (Fig. 1).
NPQ and PS II excitation pressure (estimated as 1-qP) were measured in wild type, L5, L17, and npq4 as a function of PFD. During exposure to a series of increasing PFDs as shown in Fig. 6A, increases in NPQ occurred at all PFDs higher than the growth light. L5 and L17 plants again exhibited much higher levels of NPQ, especially at higher PFDs (Fig. 6A). After exposure to the highest PFD in the series (2,400 μmol m−2⋅s−1), leaves were darkened to allow for relaxation of the qE component of NPQ (data not shown). By subtracting the residual NPQ in the dark from the NPQ measured at 2,400 μmol m−2⋅s−1, the capacity of the qE component was estimated as 1.9, 4.4, 4.0, and 0.2 in the wild type, L5, L17, and npq4, respectively. Compared with Fig. 4, the higher qE capacities in this experiment were due to both the higher maximum PFD and longer total exposure time to excess light. The higher levels of qE in L5 and L17 were accompanied by significantly lower levels of PS II excitation pressure than in the wild type, whereas npq4 showed the highest excitation pressure (Fig. 6B).
Fig 6.
Light response curves for NPQ and PS II excitation pressure. Chlorophyll fluorescence was measured during exposure of attached rosette leaves to an increasing series of PFDs for 5 min each. Data are shown as the means ± SE (n = 3). (A) NPQ. (B) PS II excitation pressure (1-qP).
Global Fluorescence Lifetime Distribution Analysis of Leaves with Higher qE Capacity.
The influence of increased qE capacity on PS II chlorophyll a fluorescence lifetime distributions under conditions of steady-state photosynthesis was determined in L5 and L17 and compared with the wild type and npq4. After induction of qE by illumination with HL (1,200 μmol m−2⋅s−1, white light) at 25°C and attenuation of the temperature to 2°C, the laser diode illumination (120 μmol m−2⋅s−1, red light) was sufficient to maintain qE, with subsaturating reduction of PS II electron acceptors, during the lifetime data acquisition. Table 1 shows that the steady-state fluorescence intensity (Fs) for all four genotypes could be modeled by using six Lorentzian lifetime distributions with the lifetime centers and widths globally linked and fractional areas free-floating. Although PsbS overexpression in L5 and L17 did not change the lifetime centers, the relative fractions of faster lifetime distributions were increased, thereby decreasing the average lifetimes. Three PsbS-dependent lifetime components (c1, c3, and c5) were found in the wild type, L5, and L17, whereas the npq4 fluorescence decay could be described with only two positive components (c4 and c6). The rapid c1 and c3 components were consistent with enhanced energy dissipation in open PS II reaction centers, and the c5 component was consistent with energy dissipation reported in closed PS II reaction centers (19). The c4 and c6 components in npq4 likely reflected the dissipation rates of open and closed PS II reaction centers, respectively, in the absence of PsbS. There was also a significant negative amplitude component (c2) in all lines that was consistent with antenna to PS II core energy transfer processes (43). Consistent with measurements of qE based on fluorescence intensity (i.e., Fig. 5A), the average lifetimes were most rapid in L5 and L17 (≈330 ps), which were almost twice as fast as the wild type (568 ps) and more than three times faster than npq4 (1,046 ps). Compared with the wild type, the lower PS II excitation pressure in L5 and L17 was evident mainly as a fractional increase in the c1 component and decreases in the closed PSII center fractions, c5 and c6.
Table 1.
PS II chlorophyll fluorescence lifetime distribution parameters obtained by global analysis of multifrequency phase and modulation data from Arabidopsis leaves with varying levels of PsbS expression
| Component
|
Lifetime distribution | Fractional intensity area | ||||
|---|---|---|---|---|---|---|
| Center, ps | Width, ps | Wild type | L5 | L17 | npq4 | |
| c1 | <20 | 16 | 0.158 | 0.447 | 0.438 | 0.000 |
| c2 | 144 | 78 | NA | NA | NA | NA |
| c3 | 203 | 22 | 0.129 | 0.127 | 0.131 | 0.000 |
| c4 | 274 | 22 | 0.000 | 0.004 | 0.001 | 0.410 |
| c5 | 515 | 10 | 0.633 | 0.416 | 0.422 | 0.000 |
| c6 | 1,168 | 194 | 0.081 | 0.007 | 0.008 | 0.590 |
Experimental conditions correspond to the steady-state intensity of PS II fluorescence (Fs) with saturating levels of qE and subsaturating reduction of PS II electron acceptors. The computed global statistic (36) was χ2 = 1.773, which was calculated using the number of free-fitting parameters (m = 45), total frequency points (2N = 184), and standard frequency-independent values for the errors in the phase angle shift (0.2°) and demodulation ratios (0.004).
Values were obtained by normalizing the sum of the integrated positive fractional intensity area components and excluding the negative fraction of c2; the calculated average lifetimes including the c2 fraction are described in the text.
Discussion
The Photoprotective Function of qE.
Although npq4, npq4 npq1, and npq4 npq1 lut2 plants did not exhibit any major differences in photosynthetic parameters when grown in LL, the three mutants were clearly more sensitive to photoinhibition than the wild type during a short-term exposure to HL. The decline in the fluorescence parameter Fv/Fm was the same for all three mutants (Fig. 1C), suggesting that the enhanced susceptibility to photoinhibition is due specifically to the lack of qE. The use of fluorescence data to measure photoinhibition was validated by measurements of apparent quantum yields of oxygen evolution in npq4 (Fig. 2). In contrast to the importance of qE, the presence or absence of zeaxanthin or lutein or the size of the xanthophyll cycle pool in the mutants did not appear to make a significant difference during the short-term HL treatment, despite the fact that zeaxanthin in particular has been shown to have a protective role in longer-term HL (28). The role of zeaxanthin in preventing photooxidative damage (e.g., lipid peroxidation) might become evident only during more prolonged light stress, perhaps when the capacity of other antioxidant systems is overwhelmed (28).
The protective function of qE is further strongly supported by the psbS overexpression experiments. Transgenic PsbS-overexpressing Arabidopsis plants (lines L5 and L17) had a qE capacity that was approximately twice as high as that of the wild type. Remarkably, the higher qE capacity conferred a pronounced resistance to photoinhibition induced by HL (Fig. 5C).
Thus, photoprotection due to qE is not only evident in an environment in which variation in PFD is occurring on a time scale of seconds (31), but also on exposure to excess light over a period of minutes to hours. This type of sudden HL exposure of plants that are acclimated to LL mimics canopy gap formation in nature. Under these conditions, plants experience a sudden and prolonged imbalance between light absorption and utilization, and qE could provide photoprotection while longer-term acclimation responses, such as increases in antioxidant enzymes and small molecules and increases in photosynthetic capacity, are being induced.
An inverse relationship was observed between qE capacity and PS II excitation pressure (Fig. 6), with npq4 plants that lack qE having the highest excitation pressure and lines L5 and L17 having the lowest excitation pressure over a wide range of PFDs. The lower excitation pressure in L5 and L17 was evident in measurements of chlorophyll fluorescence lifetime distributions (Table 1), where the fractional increase in the rapid c1 and c3 components indicated a higher proportion of open PS II reaction centers with more rapid energy dissipation. In various plant species, it has been shown that photoinhibition is enhanced when the PS II excitation pressure, measured as the chlorophyll fluorescence parameter 1-qP, exceeds a value of 0.4 (44). This threshold value was reached at a PFD of ≈300 μmol photons m−2⋅s−1 in npq4, 500 μmol photons m−2⋅s−1 in the wild type, and 700–800 μmol photons m−2⋅s−1 in L5 and L17 (Fig. 6). Our results are consistent with the idea that qE protects PS II from photoinhibition by dissipating excess absorbed light energy as heat in the PS II antenna, thereby preventing overexcitation of PS II, decreasing the rate of charge separation in the PS II reaction centers, and preventing overreduction of PS II electron acceptors.
PsbS Expression Level and qE Capacity.
The semidominance of the npq4 deletion mutant first suggested that psbS gene dosage might affect qE capacity (18). Heterozygous (npq4/NPQ4) Arabidopsis plants containing only a single dose of the psbS gene exhibit a level of qE that is intermediate between that of the wild type and homozygous npq4 mutant (18, 19). Expression of both psbS mRNA and PsbS protein is ≈60% of the wild-type level in npq4 heterozygotes, establishing a correlation between qE capacity and the amount of PsbS protein (19).
By increasing the psbS gene dosage using transformation, we achieved a several-fold increase in PsbS protein level (Fig. 3C) and a doubling of qE capacity in the L5 and L17 lines (Figs. 4 and 5A), demonstrating that PsbS levels are limiting for qE in LL-grown wild-type Arabidopsis. PsbS overexpression in L5 and L17 affected the relative fractions of chlorophyll fluorescence lifetime distributions, but not the lifetimes themselves (Table 1), resulting in overall decreases in the average lifetime that agreed with the qE measurements. It is evident from characterization of the npq4 mutant and the L5 and L17 transgenic lines that the average stoichiometry of PsbS per PS II can vary enormously in Arabidopsis, ranging from the complete absence of PsbS to several times the wild-type level, without affecting pigment composition, accumulation of other PS II subunits, or PS II photosynthetic parameters.
This clear demonstration of the relationship between PsbS protein level and qE capacity strengthens the suggestion that natural variations in qE capacity in different species and in response to different growth PFDs might be caused by differences in PsbS expression (18, 19). Many plants respond to HL by increasing qE capacity (5, 22), which presumably confers enhanced resistance to photoinhibition. However, if more PsbS and more qE are advantageous, then one might ask why Arabidopsis does not constitutively express a level of PsbS protein that saturates qE capacity. As observed for herbicide resistance in Arabidopsis (45), there might also be a cost involved in constitutive expression of high levels of PsbS. It will be interesting to examine the fitness of PsbS-overexpressing transgenic plants like L5 and L17 under natural conditions. To what extent would potential benefits of increased qE capacity be offset by the cost of high constitutive expression of PsbS? Depending on the outcome of such experiments, in the future it might be possible to generate stress-resistant crops by overexpressing PsbS and increasing qE capacity.
Supplementary Material
Acknowledgments
We thank Talila Golan for comments on the manuscript, Nancy Holt for helpful discussions, and Stan Grell of the University of California, Berkeley, Hybridoma Facility for the MLH2 antibody preparation. This work was supported by the Director, Office of Science, Office of Basic Energy Sciences, Chemical Sciences Division, of the U.S. Department of Energy under Contract no. DE-AC03-76SF00098, by the U.S. Department of Agriculture National Research Initiative Grant 98-35306-6600, and by the Searle Scholars Program/The Chicago Community Trust.
Abbreviations
HL, high light
LL, low light
NPQ, nonphotochemical quenching
qE, pH- and xanthophyll-dependent component of nonphotochemical quenching (feedback de-excitation)
PFD, photon flux density
PS II, photosystem II
VDE, violaxanthin de-epoxidase
LHC, light harvesting complex
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Niyogi K. K. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333-359. [DOI] [PubMed] [Google Scholar]
- 2.Horton P., Ruban, A. V. & Walters, R. G. (1996) Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 655-684. [DOI] [PubMed] [Google Scholar]
- 3.Müller P., Li, X.-P. & Niyogi, K. K. (2001) Plant Physiol. 125, 1558-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto H. Y., Nakayama, T. O. M. & Chichester, C. O. (1962) Arch. Biochem. Biophys. 97, 168-173. [DOI] [PubMed] [Google Scholar]
- 5.Demmig-Adams B. (1990) Biochim. Biophys. Acta 1020, 1-24. [Google Scholar]
- 6.Yamamoto H. Y., Bugos, R. C. & Hieber, A. D. (1999) in The Photochemistry of Carotenoids, eds. Frank, H. A., Young, A. J., Britton, G. & Cogdell, R. J. (Kluwer Academic, Dordrecht, The Netherlands), pp. 293–303.
- 7.Gilmore A. M. & Yamamoto, H. Y. (1993) Photosynth. Res. 35, 67-78. [DOI] [PubMed] [Google Scholar]
- 8.Niyogi K. K., Björkman, O. & Grossman, A. R. (1997) Proc. Natl. Acad. Sci. USA 94, 14162-14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pogson B. J., Niyogi, K. K., Björkman, O. & DellaPenna, D. (1998) Proc. Natl. Acad. Sci. USA 95, 13324-13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niyogi K. K., Grossman, A. R. & Björkman, O. (1998) Plant Cell 10, 1121-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niyogi K. K., Shih, C., Chow, W. S., Pogson, B. J., DellaPenna, D. & Björkman, O. (2001) Photosynth. Res. 67, 139-145. [DOI] [PubMed] [Google Scholar]
- 12.Chang S.-H., Bugos, R. C., Sun, W.-H. & Yamamoto, H. Y. (2000) Photosynth. Res. 64, 95-103. [DOI] [PubMed] [Google Scholar]
- 13.Kim S., Sandusky, P., Bowlby, N. R., Aebersold, R., Green, B. R., Vlahakis, S., Yocum, C. F. & Pichersky, E. (1992) FEBS Lett. 314, 67-71. [DOI] [PubMed] [Google Scholar]
- 14.Wedel N., Klein, R., Ljungberg, U., Andersson, B. & Herrmann, R. G. (1992) FEBS Lett. 314, 61-66. [DOI] [PubMed] [Google Scholar]
- 15.Kim S., Pichersky, E. & Yocum, C. F. (1994) Biochim. Biophys. Acta 1188, 339-348. [DOI] [PubMed] [Google Scholar]
- 16.Funk C., Schröder, W. P., Napiwotzki, A., Tjus, S. E., Renger, G. & Andersson, B. (1995) Biochemistry 34, 11133-11141. [DOI] [PubMed] [Google Scholar]
- 17.Dominici P., Caffarri, S., Armenante, F., Ceoldo, S., Crimi, M. & Bassi, R. (2002) J. Biol. Chem. 277, 22750-22758. [DOI] [PubMed] [Google Scholar]
- 18.Li X.-P., Björkman, O., Shih, C., Grossman, A. R., Rosenquist, M., Jansson, S. & Niyogi, K. K. (2000) Nature 403, 391-395. [DOI] [PubMed] [Google Scholar]
- 19.Li X.-P., Gilmore, A. M. & Niyogi, K. K. (2002) J. Biol. Chem. 277, 33590-33597. [DOI] [PubMed] [Google Scholar]
- 20.Li X.-P., Phippard, A., Pasari, J. & Niyogi, K. K. (2002) Funct. Plant Biol. 29, 1131-1139. [DOI] [PubMed] [Google Scholar]
- 21.Ruban A. V., Pascal, A. A., Robert, B. & Horton, P. (2002) J. Biol. Chem. 277, 7785-7789. [DOI] [PubMed] [Google Scholar]
- 22.Demmig-Adams B. & Adams, W. W., III (1992) Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 599-626. [Google Scholar]
- 23.Winter K. & Königer, M. (1989) Planta 180, 24-31. [DOI] [PubMed] [Google Scholar]
- 24.Bilger W. & Björkman, O. (1990) Photosynth. Res. 25, 173-185. [DOI] [PubMed] [Google Scholar]
- 25.Demmig-Adams B., Adams, W. W., III, Heber, U., Neimanis, S., Winter, K., Krüger, A., Czygan, F.-C., Bilger, W. & Björkman, O. (1990) Plant Physiol. 92, 293-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verhoeven A. S., Bugos, R. C. & Yamamoto, H. Y. (2001) Photosynth. Res. 67, 27-39. [DOI] [PubMed] [Google Scholar]
- 27.Havaux M. (1998) Trends Plant. Sci. 3, 147-151. [Google Scholar]
- 28.Havaux M. & Niyogi, K. K. (1999) Proc. Natl. Acad. Sci. USA 96, 8762-8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graßes T., Pesaresi, P., Schiavon, F., Varotto, C., Salamini, F., Jahns, P. & Leister, D. (2002) Plant Physiol. Biochem. 40, 41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Havaux M. & Kloppstech, K. (2001) Planta 213, 953-966. [DOI] [PubMed] [Google Scholar]
- 31.Külheim C., Ågren, J. & Jansson, S. (2002) Science 297, 91-93. [DOI] [PubMed] [Google Scholar]
- 32.Hajdukiewicz P., Svab, Z. & Maliga, P. (1994) Plant Mol. Biol. 25, 989-994. [DOI] [PubMed] [Google Scholar]
- 33.Clough S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 34.Müller-Moulé P., Conklin, P. L. & Niyogi, K. K. (2002) Plant Physiol. 128, 970-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Kooten O. & Snel, J. F. H. (1990) Photosynth. Res. 25, 147-150. [DOI] [PubMed] [Google Scholar]
- 36.Gilmore A. M. (2003) in Chlorophyll Fluorescence: The Signature of Photosynthetic Efficiency, eds. Papageorgiou, G. & Govindjee, (Kluwer Academic, Dordrecht, The Netherlands), in press.
- 37.Matsubara, S., Gilmore, A. M., Ball, M. C., Anderson, J. M. & Osmond, C. B. (2002) Funct. Plant Biol., in press. [DOI] [PubMed]
- 38.Shure M., Wessler, S. & Federoff, N. (1983) Cell 35, 225-233. [DOI] [PubMed] [Google Scholar]
- 39.Bugos R. C. & Yamamoto, H. Y. (1996) Proc. Natl. Acad. Sci. USA 93, 6320-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darr S. C., Somerville, S. C. & Arntzen, C. J. (1986) J. Cell Biol. 103, 733-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jahns P. & Miehe, B. (1996) Planta 198, 202-210. [Google Scholar]
- 42.Demmig-Adams B., Moeller, D. L., Logan, B. A. & Adams, W. W., III (1998) Planta 205, 367-374. [Google Scholar]
- 43.Mimuro M., Tamai, N., Yamazaki, T. & Yamazaki, I. (1987) in Primary Processes in Photobiology, ed. Kobayashi, T. (Springer, Berlin), pp. 22–32.
- 44.Öquist G., Chow, W. S. & Anderson, J. M. (1992) Planta 186, 450-460. [DOI] [PubMed] [Google Scholar]
- 45.Purrington C. B. & Bergelson, J. (1997) Genetics 145, 807-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.