Abstract
Two major classes of small noncoding RNAs have emerged as important regulators of gene expression in eukaryotes, the short interfering RNAs (siRNAs) associated with RNA silencing and endogenous micro-RNAs (miRNAs) implicated in regulation of gene expression. Helper component-proteinase (HC-Pro) is a viral protein that blocks RNA silencing in plants. Here we examine the effect of HC-Pro on the accumulation of siRNAs and endogenous miRNAs. siRNAs were analyzed in transgenic tobacco plants silenced in response to three different classes of transgenes: sense-transgenes, inverted-repeat transgenes, and amplicon-transgenes. HC-Pro suppressed silencing in each line, blocking accumulation of the associated siRNAs and allowing accumulation of transcripts from the previously silenced loci. HC-Pro-suppression of silencing in the inverted-repeat- and amplicon-transgenic lines was accompanied by the apparent accumulation of long double-stranded RNAs and proportional amounts of small RNAs that are larger than the siRNAs that accumulate during silencing. Analysis of these results suggests that HC-Pro interferes with silencing either by inhibiting siRNA processing from double-stranded RNA precursors or by destabilizing siRNAs. In contrast to siRNAs, the accumulation of endogenous miRNAs was greatly enhanced in all of the HC-Pro-expressing lines. Thus, our results demonstrate that accumulation of siRNAs and miRNAs in plants can be differentially regulated by a viral protein. The fact that HC-Pro affects the miRNA pathway raises the possibility that this pathway is targeted by plant viruses as a means to control gene expression in the host.
RNA silencing is a eukaryotic surveillance mechanism that detects and eliminates double-stranded RNA (dsRNA) and any homologous single-stranded RNA, providing a defense against invasive nucleic acids such as viruses, transposons, and transgenes. The term refers to a set of related pathways found in plants [posttranscriptional gene silencing (PTGS)], fungi (quelling), and animals [RNA interference (RNAi)] (for recent reviews of RNA silencing in various organisms, see refs. 1–8). Consistent with the antiviral nature of RNA silencing, many viruses use a counterdefensive strategy, encoding proteins that block one or more steps in the RNA-silencing pathway (6, 9, 10). Our work has focused on using helper component-proteinase (HC-Pro), a viral suppressor of RNA silencing encoded by plant potyviruses, as a tool to help elucidate the mechanism of RNA silencing.
A key conserved feature of RNA silencing in different organisms is that the process is triggered by the presence of dsRNA. In Caenorhabditis elegans and Drosphilia, dsRNA is processed into ≈22-nt short interfering RNAs (siRNAs) by an RNase III termed Dicer (11). The siRNAs incorporate into a multicomponent silencing complex termed the RNA-induced silencing complex, where they act as guides to promote sequence-specific destruction of target RNAs (11–16). We and others (17–20) have shown that HC-Pro blocks RNA silencing induced by sense constructs and severely reduces or eliminates siRNA accumulation. However, biogenesis of siRNAs is likely a multistep pathway, and the step where HC-Pro inhibits is unknown.
Three different classes of transgenes, sense, inverted-repeat (IR), and amplicon, have been shown to trigger RNA silencing in plants. These represent separate branches of the silencing pathway that converge at production of dsRNA. IR transgenes are thought to produce dsRNA directly by readthrough transcription, whereas amplicon transgenes encode replication-competent viral RNAs that produce dsRNA by the activity of a virus-encoded RNA-dependent RNA polymerase (RdRp) (21–23). Studies using stable transgenic lines, as well as those using Agrobacterium infiltration, a transient expression technique, suggest that the genetic requirements for silencing and the response to HC-Pro differ depending on the nature of the inducing transgene. Sense transgene silencing depends on a set of cellular genes that are not required for IR transgene silencing including an RdRp (SGS2/SDE1), an RNA helicase (SDE3), a coiled-coil protein (SGS3), and a Piwi/PAZ domain protein (AGO1) (24–28). Therefore, IR transgene silencing bypasses a portion of the RNA-silencing pathway that is specific to sense transgene silencing and provides a valuable model to understand steps of the pathway that are downstream of dsRNA. In transgenic lines as well as in the Agrobacterium-mediated transient expression system, the suppression of sense-transgene silencing by HC-Pro is associated with a block in siRNA accumulation (17–20). In contrast, experiments using IR constructs in the transient system suggest that HC-Pro only partially suppresses this type of silencing, allowing siRNA accumulation to continue (20). Although HC-Pro has been shown to suppress the silencing induced by stably integrated IR and amplicon transgenes (24, 29, 30), the influence of HC-Pro on siRNA accumulation in these IR or amplicon transgenic lines is unknown.
Another class of small regulatory RNAs, the micro-RNAs (miRNAs), exists in a broad range of eukaryotes ranging from humans to plants (31–35). The first examples of miRNAs, lin-4 and let-7, were found in C. elegans, where they arise from genes that do not encode protein and function by binding to the 3′-untranslated region of target mRNAs, blocking their translation. In these two known cases, the miRNA targets encode important regulators of development (36–40). The discovery of miRNAs in many other organisms suggests that the miRNA pathway, like the RNA-silencing pathway, arose early in eukaryotic evolution. Although the functions of most miRNAs are unknown, bioinformatic analysis suggests that many plant miRNAs regulate the expression of transcription factors that function in plant development (41). Although lin-4 and let-7 miRNAs control developmental processes at the level of translation, one plant miRNA has been shown to mediate mRNA cleavage (42). Considering the large number of miRNA genes in diverse species, some miRNAs may regulate gene expression at other levels, such as transcription, mRNA localization, or mRNA processing.
The siRNAs and miRNAs differ in their origin. Whereas siRNAs come from dsRNA that has been processed into fragments, miRNAs come from endogenous transcripts processed such that a single miRNA is produced from one arm of the structured precursor molecule (13, 16, 31–35, 43). Nonetheless, siRNAs and miRNAs share similarities in their biogenesis. In animals, both siRNAs and miRNAs are processed by the activity of Dicer (12, 13, 43–45). A similar situation is expected in plants, where the Dicer homolog CAF/SIN/SUS (46–48) is required for miRNA accumulation (34), although the RNase III that produces siRNAs has not yet been identified. In addition, siRNAs and miRNAs are similar in size (21–24 nt), and both have been found as an integral part of a multicomponent complex containing a member of the Piwi/PAZ domain family, siRNAs in the RNA-induced silencing complex and miRNAs in the microribonucleoprotein (12, 49). In fact, recent evidence suggests that the RNA-induced silencing complex and the microribonucleoprotein might in fact be the same complex (41, 42, 50). The shared features of miRNA and siRNA biogenesis together with the result that HC-Pro decreases siRNA accumulation raises the possibility that HC-Pro may also alter the accumulation of miRNAs and lead to profound effects on gene expression.
To further dissect the RNA-silencing and miRNA pathways, we compared the effect of HC-Pro in a sense-transgene silenced line to that in representative IR- and amplicon-silenced lines. We found that HC-Pro blocks RNA silencing in the stably transformed lines regardless of the class of inducing transgene and that, in each case, HC-Pro-suppression of silencing was accompanied by elimination of detectable levels of siRNAs. Analysis of the accumulation of dsRNA in these lines suggests that HC-Pro blocks RNA silencing by blocking siRNA processing from dsRNA precursors or by destabilizing siRNAs. Surprisingly, HC-Pro dramatically enhances the accumulation of miRNAs. Our results indicate that HC-Pro alters small RNA metabolism in plants, differentially regulating the accumulation of these two classes of small regulatory RNAs.
Materials and Methods
Transgenic Tobacco Lines.
The β-glucuronidase (GUS)-silenced transgenic tobacco lines 6b5 (51), 155 (SA93006) (23), and T4 (52), the GUS-expressing line T19 (52), and P1/HC-Pro (derived from tobacco etch virus) expressing line X-27-8 (18) have been described.
RNA Gel Blot Analysis.
Total nucleic acid was isolated, and RNA gel blot analysis was performed as described (53). For the probe, PCR-amplified fragments representing the entire GUS-coding sequence were labeled with 32P by using the Amersham Pharmacia, Megaprime DNA-labeling system. Ethidium bromide (EtdBr)-stained 25S rRNA is shown in each figure as a loading control.
Low molecular weight RNAs were isolated, separated by denaturing polyacrylamide gel electrophoresis, and blotted to a nylon membrane (Hybond NX, Amersham Pharmacia) as described (18). Sense and antisense RNA probes for small RNAs were generated by transcription from the 3′ 700 nucleotides of the GUS-coding sequence in the appropriate direction by using the Ambion, Maxiscript SP6/T7 kit. MiRNA probes were prepared by end-labeling antisense oligonucleotides using T4 polynucleotide kinase (New England Biolabs). The small RNAs (2 μg per lane) were also separated by agarose gel electrophoresis and the most predominant EtdBr-stained species of RNA on these gels, running at ≈200 nt, was used as a loading control.
RNase A Analysis.
Total RNA was digested at 37°C for 30 min with 2.2 μg/ml RNase A/T1 (Ambion) in RNase buffer containing 0.01 M Tris⋅HCl (pH 7.5), 0.2 M NaCl, 0.1 M LiCl, and 1 mM EDTA in a final reaction volume of 250 μl. After RNase digestion, proteinase K and SDS were added to a final concentration of 1 mg/ml and 0.5%, respectively, and the reaction was incubated at 37°C for 15 min. Samples were purified with phenol/chloroform and ethanol precipitation. As a control, total RNA was denatured by boiling for 5 min before RNase A/T1 digestion. RNA gel blot analysis was performed as described above.
Calf Intestinal Phosphatase (CIP) and Polynucleotide Kinase Reactions.
Approximately 10–15 μg of small RNAs were incubated with 20 units of CIP (New England Biolabs) for 1 h at 37°C in the buffer supplied by the manufacturer. Polynucleotide kinase reactions were performed as suggested by the manufacturer at 37°C for 30 min by using 10 units of T4 kinase (New England Biolabs) and 1 mM ATP (Ambion). Samples were separated by denaturing PAGE and blotted to nylon membrane (Amersham Pharmacia) as described (18).
Results
HC-Pro Suppression of IR- or Amplicon-Induced RNA Silencing Interferes with the Accumulation of siRNAs and Promotes Accumulation of Larger Small RNAs.
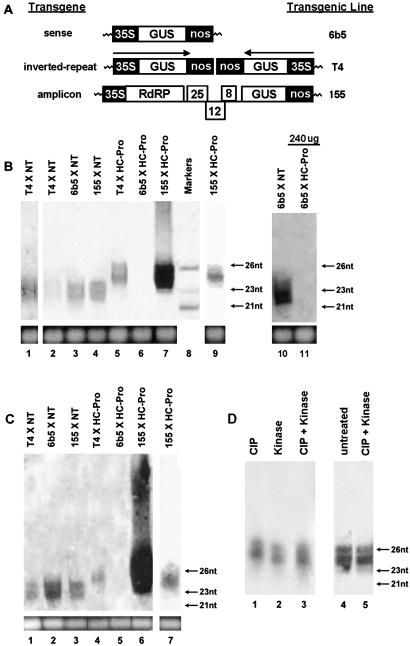
RNA silencing is induced by dsRNA that is cleaved into the siRNAs that mediate sequence-specific RNA degradation. We have previously shown that HC-Pro suppression of RNA silencing induced by a GUS sense transgene eliminates the accumulation of siRNAs (Fig. 1A, line 6b5, and Fig. 1B, lanes 3 and 6; ref. 18). To determine whether HC-Pro suppression of RNA silencing in IR and amplicon transgenic lines T4 and 155 (23, 52) also eliminates siRNA accumulation, we analyzed small RNAs from the offspring of crosses between these lines and either a nontransformed control plant or an HC-Pro expressing line (line X-27-8) (18). Southern analysis demonstrated that the transgene locus in line T4 contains two GUS genes arranged as an IR (Fig. 1A; ref. 52), and this line is therefore thought to produce GUS dsRNA via read-through transcription. The amplicon transgene in line 155 is comprised of complementary DNA encoding the potato virus X (PVX) genomic RNA in which the coat protein gene is replaced by the GUS locus (Fig. 1A). The transcript produced by the amplicon transgene is a replicating PVX vector and thus produces dsRNA replication intermediates with homology to both PVX and GUS sequences.
Fig 1.
HC-Pro suppression of IR- and amplicon-induced RNA silencing prevents the accumulation of siRNAs and results in accumulation of a new size class of small RNAs. (A) Diagram of the transgene loci in lines 6b5, T4, and 155 as predicted by DNA blot analysis. 35S indicates the position of the cauliflower mosaic virus 35S promoter, and nos indicates the position of the nopaline synthase terminator. Arrows indicate the direction of transcription in line T4 based on the predicted arrangement of the 35S promoters (52). The amplicon transgene in line 155 encodes a PVX complementary DNA in which the coat protein gene is replaced by the GUS locus. The PVX viral ORFs encode RdRp and three movement proteins, p25 (25), p12 (12), and p8 (8) (23). (B) RNA gel blot analysis of small RNAs from silenced transgenic lines and lines in which silencing has been suppressed by HC-Pro. Lanes 1–4 and 10 show small RNAs from IR-silenced tobacco line T4 (lanes 1 and 2; lane 1 is a longer exposure of lane 2), sense transgene-silenced tobacco line 6b5 (lanes 3 and 10), and an amplicon-silenced tobacco line 155 (lane 4). Lanes 5–7, 9, and 11 show small RNAs from an IR line expressing HC-Pro (T4 × HC-Pro, lane 5), a sense transgene line expressing HC-Pro (6b5 × HC-Pro, lanes 6 and 11), and an amplicon line expressing HC-Pro (155 × HC-Pro, lane 7; lane 9 is a shorter exposure of lane 7). The probe was 32P-labeled RNA corresponding to the sense strand of the 3′ 700 nt of the GUS-coding sequence and detects anti-sense strand GUS RNAs. The migration of 21-, 23-, and 26-nt DNA oligomers is shown in lane 8. EtdBr staining of the predominant RNA species in the fractionated sample is shown as a loading control. Low molecular weight RNA (20 μg) was loaded in each lane, except for lanes 7 and 9 (155 × HC-Pro), in which 5 μg was loaded, and lane 11, in which 240 μg was loaded. (C) RNA gel blot analysis of small RNAs from the same samples shown in B. The probe was 32P-labeled RNA corresponding to the anti-sense strand of the 3′ 700 nt of the GUS-coding sequence and detects sense-strand, GUS small RNAs. EtdBr staining of the predominant RNA species in the fractionated sample is shown as a loading control. Low molecular weight RNA (20 μg) was loaded, except for lanes 6 and 7 (155 × HC-Pro), in which 7 μg was loaded. (D) The 5′ phosphorylation status of the 25- to 27-nt larger small RNAs. Small RNAs were isolated from the HC-Pro-amplicon transgenic line (155 × HC-Pro) and treated with CIP and polynucleotide kinase (kinase) as indicated, and sizes of the resulting RNAs were analyzed by RNA gel blot analysis. The migration of 21-, 23-, and 26-nt DNA oligomers is indicated.
As expected, the IR- and amplicon-silenced lines, like the sense transgene-silenced line 6b5, accumulated siRNAs. The small RNAs produced in these three different lines were analyzed by high-resolution gel electrophoresis. In each case, the size of the small RNAs was very similar, ranging from 21 to 24 nt (Fig. 1B, lanes 1–4; lane 1 is a longer exposure of lane 2). The small RNAs in each of these silenced lines hybridized to probes specific to both sense-strand GUS RNA (Fig. 1B) and antisense-strand GUS RNA (Fig. 1C), indicating that both polarities are represented as expected for siRNAs. Thus, these small RNAs have the characteristics of bona fide siRNAs.
Examination of small RNAs isolated from sense, IR, and amplicon transgenic lines crossed with a line expressing HC-Pro showed that HC-Pro interfered with accumulation of the 21- to 24-nt siRNAs in all three of these lines (Fig. 1B, lanes 5–7 and 9; lane 9 is a shorter exposure of lane 7). However, surprisingly, HC-Pro suppression of IR- and amplicon-induced RNA silencing resulted in accumulation of a longer species of small RNAs, ≈25–27 nt (Fig. 1B, lanes 5, 7, and 9). These 25- to 27-nt small RNAs accumulated to very high levels during HC-Pro suppression of amplicon-induced RNA silencing (Fig. 1B, lane 7), presumably reflecting the vastly increased amount of replicating PVX-GUS RNA in this line (Fig. 2A, lane 3, vs. Fig. 2B, lane 4; ref. 30). Like the RNA silencing-associated siRNAs, the larger small RNAs also hybridized with both antisense-strand (Fig. 1B) and sense-strand GUS RNA probes (Fig. 1C). Thus, suppression of RNA silencing by HC-Pro eliminates siRNAs for all three types of transgenes, but in the case of IR and amplicon transgenes, a larger class of small RNAs accumulates instead.
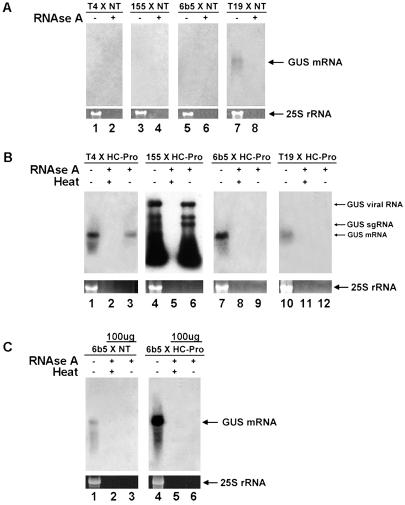
Fig 2.
HC-Pro suppression of both IR and amplicon but not sense transgene-induced RNA silencing results in the accumulation of full-length GUS dsRNA. (A) RNA gel blot showing the level of GUS RNA before and after RNase A digestion in silenced lines T4 (lanes 1 and 2), 155 (lanes 3 and 4), 6b5 (lanes 5 and 6), and a GUS-expressing control line T19 (lanes 7 and 8). Total RNA (25 μg) was digested for each plant line. EtdBr staining of 25S rRNA is shown as a loading control. (B) RNA gel blot showing the level of GUS RNA before and after RNase A digestion in plant lines T4 × HC-Pro (lanes 1–3), 155 × HC-Pro (lanes 4–6), 6b5 × HC-Pro (lanes 7–9), and the GUS expressing line T19 × HC-Pro (lanes 10–12). The position of GUS viral RNA and subgenomic RNAs (sgRNAs) is indicated. Total RNA (25 μg) was digested for each plant line. Heat refers to boiling the samples immediately before RNase A digestion to denature dsRNA. EtdBr staining of 25S rRNA is shown as a loading control. (C) RNA gel blot showing the level of GUS mRNA before and after RNase A digestion in silenced line 6b5 (lanes 1–3) and the unsilenced line 6b5 × HC-Pro (lanes 4–6). Total RNA (100 μg) was digested for each plant line, and 10 μg of total RNA was used for the untreated sample. The heat control is described in B. EtdBr staining of 25S rRNA is shown as a loading control.
The 5′-Phosphorylation Status of the Larger Small RNAs.
Previous experiments indicate that siRNAs must be phosphorylated at the 5′ end to direct silencing (54). To determine whether the larger (25–27 nt) GUS small RNAs have a 5′ phosphate and are therefore silencing competent in this regard, larger small RNAs derived from HC-Pro-amplicon plants were treated with CIP and/or polynucleotide kinase. CIP will remove phosphates regardless of whether they are located at the 3′ or 5′ end, whereas polynucleotide kinase adds a single phosphate to the 5′ end. Fig. 1D shows that CIP treatment retards the migration of the larger small RNAs, as expected by the removal of negatively charged group(s). In addition, larger small RNAs treated with polynucleotide kinase, as well as those treated with CIP followed by polynucleotide kinase, have the same mobility as untreated RNAs (Fig. 1D). The only explanation that is consistent with these data is that the larger small RNAs possess a single phosphate that is located at their 5′ ends and are therefore silencing competent in this regard. Furthermore, the presence of a 5′ phosphate suggests that the larger small RNAs are the products of an RNase III.
HC-Pro Suppression of IR- and Amplicon-Induced RNA Silencing Appears to Promote the Accumulation of Long dsRNA.
HC-Pro may interfere with siRNA accumulation by interfering with the production of the double-stranded siRNA precursors. To investigate whether dsRNA accumulation was altered during HC-Pro suppression of RNA silencing induced by the three different classes of transgenes, we isolated RNA from each line in the presence and the absence of HC-Pro and measured the level of dsRNA. In these experiments, we digested the isolated RNA with RNase A under conditions that eliminate single-stranded but not dsRNA. No RNase A-resistant high molecular weight RNA was detected from any of the silenced lines (Fig. 2A, lanes 2, 4, and 6), presumably because of efficient Dicer-mediated processing of dsRNA into the 21- to 24-nt siRNAs. In contrast, RNase A treatment of total RNA extracted from both T4 × HC-Pro and 155 × HC-Pro plants revealed the presence of dsRNA from the IR and amplicon lines when RNA silencing is suppressed by HC-Pro (Fig. 2B, lanes 3 and 6, respectively). Note that we cannot rule out the possibility that the dsRNA was not present in the cells but instead formed during our RNA extraction procedure from single-stranded sense and antisense strands. Nonetheless, the detection of dsRNA following extraction indicates that HC-Pro increased the accumulation of both RNA strands in the cells, and it is plausible that a large proportion of the GUS RNA accumulating in these lines is double stranded within the cell. Furthermore, HC-Pro expression was insufficient to cause dsRNA accumulation in the GUS-expressing control line T19 (Fig. 2 A and B; ref. 52). The multiple bands found in the 155 × HC-Pro line represent viral genomic and subgenomic dsRNAs. Denaturation of the RNA before RNase A treatment eliminated the signal (Fig. 2B, lanes 2 and 5) indicating that we were, indeed, detecting dsRNA. These results indicate that HC-Pro does not interfere with dsRNA accumulation in the IR and amplicon lines and suggest that the effects on small RNA accumulation in these lines are because of changes in dsRNA processing or small RNA stability.
Surprisingly, double-stranded GUS RNA did not accumulate to detectable levels during HC-Pro suppression of RNA silencing in the sense transgene line 6b5 (Fig. 2B, lane 9). Further attempts were made to detect dsRNA from the 6b5 × HC-Pro transgenic line. We increased the amount of RNA assayed by using four times more RNA than previously (100 μg vs. 25 μg) and still failed to detect dsRNA in the resolution range of these gels (>≈150 nt; Fig. 2C). In addition, we used high specific activity RNA probes and no dsRNA was detected (data not shown). We estimate that we would have detected dsRNA in the 6b5 line if it were present at 1/30 of the level detected in T4 × HC-Pro plants. Perhaps dsRNA is produced in the 6b5 × HC-Pro line, but it is too small or too heterogeneous to be easily detected. Therefore, we analyzed samples enriched for RNA <200 nt. We did not detect any GUS sequences in the range of 30–150 nt, even when using large amounts of these RNAs (240 μg; Fig. 1B, lane 11). Even though dsRNA was not detected in the silenced 6b5 plants, this line accumulates siRNAs of both polarities (Fig. 1 B and C; ref. 18), suggesting that dsRNA is produced in this line. Thus, we cannot determine whether HC-Pro affects dsRNA accumulation in the sense transgene line 6b5 because we cannot detect dsRNA from these plants. However, it is likely that HC-Pro inhibits dsRNA processing or destabilizes siRNAs in the sense transgene line, because it acts at one of these steps in the IR and amplicon lines.
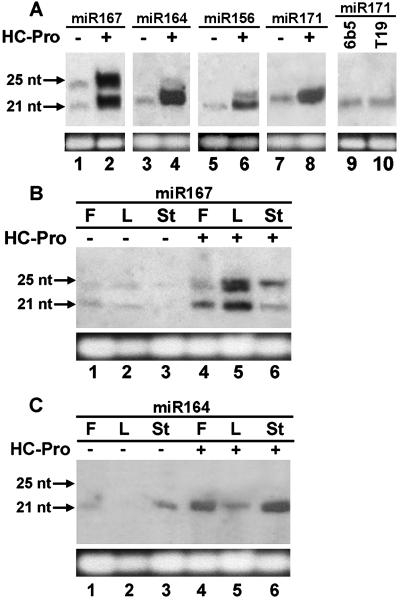
HC-Pro Increases the Level of miRNA Accumulation in Tobacco.
HC-Pro inhibits siRNA accumulation (Fig. 1), suggesting that HC-Pro may alter the activity of the Dicer-like RNase III responsible for siRNA production. Because miRNAs are also products of a Dicer-like RNase III in plants (34), we examined the effect of HC-Pro on miRNA accumulation (Fig. 3). Four miRNAs were chosen for these experiments. Each was identified in Arabidopsis but predicted, on the basis of sequence similarity, to be highly conserved throughout the flowering plants (34). The expression of all four miRNAs was detected in tobacco, supporting the phylogenetic conservation of the miRNAs. Strikingly, we found that HC-Pro increased the accumulation of these miRNAs 5- to 10-fold in leaves (Fig. 3A). A similar increase in miR164 and miR167 accumulation was also detected in flowers and stems of HC-Pro-expressing plants (Fig. 3 B and C). No miRNA precursors were detected in HC-Pro-expressing or nonexpressing lines, suggesting that they are rapidly processed, degraded, or too large to be observed on these gels. The increase in miRNA accumulation was independent of the silencing locus present in the plants because there was little difference in miRNA levels in 155 × HC-Pro, 6b5 × HC-Pro, and T4 × HC-Pro plant lines (data not shown). Furthermore, the level of miRNA accumulation was similar in a GUS-expressing control line (T19) and a GUS-silenced line (6b5) (Fig. 3A, lanes 9 and 10), suggesting that transgene-induced RNA silencing does not reduce miRNA accumulation by competing for components shared by the two pathways.
Fig 3.
HC-Pro expression leads to increased miRNA accumulation in tobacco. (A) RNA gel blot analysis of 20 μg of small RNAs from leaf tissue of the silenced line 6b5 (lanes 1, 3, 5, 7, and 9), the HC-Pro-expressing line 6b5 × HC-Pro (lanes 2, 4, 6, and 8), and the GUS-expressing control line T19 (lane 10). The specific probe used to detect each miRNA is noted (miR167, miR164, miR156, and miR171). The migration of 21- and 25-nt DNA oligomers is shown on the left, and EtdBr staining of the predominant RNA species in the fractionated sample is shown as a loading control. (B and C) RNA gel blot analysis of miR167 and miR164 miRNAs, extracted from flower (F), leaf (L), and stem (St) tissue of a control line Xanthi (lanes 1–3) and the HC-Pro-expressing line X-27-8 (lanes 4–6). The migration of 21- and 25-nt DNA oligomers is shown on the left, and EtdBr staining of the predominant RNA species in the fractionated sample is shown as a loading control.
Discussion
Our results show that HC-Pro alters the accumulation of multiple classes of small RNAs in plants: the siRNAs that mediate RNA silencing, the miRNAs thought to regulate endogenous gene expression, and a novel class of larger small RNAs of unknown function. siRNAs are eliminated, whereas miRNAs and the larger small RNAs accumulate to higher levels in HC-Pro-expressing plants. In addition, HC-Pro has been shown to enhance the accumulation of small RNAs associated with dsRNA-mediated transcriptional gene silencing (55). These results raise the possibility that HC-Pro affects a shared step in the biogenesis or turnover of different classes of small RNAs.
We examined the effect of HC-Pro on siRNA accumulation in three transgenic lines, each carrying a different class of transgene. As expected, silencing of the transgene in each of these lines was associated with accumulation of the previously reported siRNAs, ≈22 nt in length, representing both polarities of the silenced locus (14). Expression of HC-Pro in each of these transgenic lines suppressed the silencing as assayed by appearance of transcripts from the previously silenced loci, and in each case, the HC-Pro suppression of silencing was correlated with elimination of siRNAs. These results confirm previous observations when using a sense-transgene silenced line (17, 18), extend these findings to IR and amplicon transgenes, and suggest that HC-Pro suppresses silencing by preventing the accumulation of siRNAs.
The accumulation of dsRNA was examined during HC-Pro suppression of silencing induced by the three classes of transgene to determine whether the effect of HC-Pro on siRNA accumulation might reflect changes in the production or the processing of dsRNA. Although dsRNA has previously been detected during the silencing of IR transgenes in Petunia (56), dsRNA was detected in our experiments only after HC-Pro suppression of IR- and amplicon-induced silencing. dsRNA likely accumulated in the IR and amplicon lines because silencing was suppressed, and dsRNA was formed either directly in the case of the IR lines or during viral replication in the case of amplicon lines. However, formation of dsRNA in the sense transgene line requires a cellular RdRp, SGS2/SDE1 (26, 28). If this RdRp requires siRNAs as primers for dsRNA production, dsRNA would not be expected to accumulate during HC-Pro suppression of sense transgene silencing because the siRNAs are eliminated. The fact that dsRNA accumulates in the IR and amplicon lines that are suppressed for silencing suggests that HC-Pro blocks the accumulation of siRNAs by acting downstream of dsRNA in these lines. HC-Pro could inhibit the processing of dsRNA into siRNAs by interfering with a plant Dicer. However, the results are also consistent with other mechanisms. For example, siRNAs could be produced at similar rates in the silenced and the HC-Pro-expressing plants but be rapidly degraded in the HC-Pro-expressing plants and thus fail to accumulate.
A population of larger small RNAs with homology to the previously silenced locus accumulates in HC-Pro-expressing IR and amplicon lines. However, the biogenesis of these RNAs and their functions are not yet understood. The larger small RNAs in our experiments are of both polarities, possess 5′-terminal phosphates, and accumulate to levels proportional to the amount of long dsRNA. These features suggest that they are derived from dsRNA and are most likely produced by an RNase III. Plants encode a minimum of seven RNase IIIs, at least four of which are putative Dicers; therefore, the larger small RNAs and the siRNAs could be products of different RNase IIIs. Alternatively, they could be products of the Dicer(s) that has been altered by HC-Pro to produce 25- to 27-nt small RNAs instead of siRNAs. Even though the larger small RNAs have some characteristics of siRNAs, they are not correlated with RNA degradation and only appear in plants where silencing has been suppressed by HC-Pro. Larger small RNAs derived from reporter genes also have been detected in Agrobacterium-infiltration transient assay experiments, and larger small RNAs derived from endogenous retroelements have been detected in nontransgenic plants (19). As in our experiments, the presence of these larger small RNAs in the transient assays is not correlated with degradation of the corresponding mRNA. Rather, the larger small RNAs detected in the transient systems are correlated with systemic silencing and those from retroelements with methylation of homologous DNA (19). However, neither of these correlations is observed in the case of a sense-transgene silenced line expressing HC-Pro (6b5 × HC-Pro; ref. 18). This line is competent for systemic silencing and transgene methylation in the absence of detectable levels of either siRNAs or the larger small RNAs (18).
Our results using transgenic lines differ in several respects from those reported using the Agrobacterium-mediated transient expression assay, likely reflecting basic differences in these two model systems. For example, larger small RNAs are detected during the silencing of sense constructs in the Agrobacterium-mediated system (19), whereas they are only detected during suppression of silencing by HC-Pro in the IR and amplicon transgenic lines (Fig. 1). It is not clear whether the larger small RNAs have the same origin in the two systems. Furthermore, HC-Pro only partially suppresses the mRNA degradation of IR transcripts in the Agrobacterium-mediated system (20), whereas it completely suppresses mRNA degradation in the IR transgene line. However, siRNA accumulation correlates with sequence-specific RNA degradation in both systems (17–20). Thus, the two assays are complementary, each providing valuable, but not completely overlapping information.
In contrast to siRNAs, the accumulation of endogenous miRNAs is dramatically enhanced in HC-Pro expressing plants. How does HC-Pro differentially regulate siRNA and miRNA accumulation? One possibility is that HC-Pro either redirects or differentially affects the Dicer(s) that processes these different classes of small RNAs. A simple competition between miRNA and siRNA biogenesis might not account for the increase in miRNA accumulation in HC-Pro-expressing plants, because this increase occurs even in the absence of a silenced transgene (compare Fig. 3A to Fig. 3 B and C). However, in considering the idea of direct competition, it is important to consider the possibility of numerous endogenous siRNAs (34, 35), which would also be competing for the processing machinery, possibly overwhelming any competition from the transgene RNAs. Another possibility is that HC-Pro differentially affects the assembly of the RNA-inducing silencing complex/microribonucleoprotein. For example, HC-Pro might affect the stability of small RNAs by inhibiting or enhancing their incorporation into these RNA–protein complexes. It has been reported that HC-Pro alters the stability of the small RNAs formed during transcriptional silencing (55).
The impact of HC-Pro on accumulation of endogenous miRNAs has implications for viral pathogenesis. The symptoms elicited by many viruses include developmental abnormalities. Given the role of at least some miRNAs in regulating development, such symptoms may reflect viral counterdefensive strategies that target host small regulatory RNA metabolism. The developmental abnormalities exhibited by transgenic plants that express high levels of HC-Pro or that overexpress rgs-CaM, a cellular suppressor of RNA silencing that interacts with HC-Pro (57), might also reflect changes in miRNA accumulation. HC-Pro suppression of siRNA accumulation serves as an obvious counterdefensive strategy to thwart RNA silencing. However, this strategy may represent just part of a double-edged, global tactic that also uses the miRNA pathway to influence expression of proteins involved in other host defense pathways. In light of these findings, it will be interesting to examine the effect of other viral proteins on miRNA accumulation.
Acknowledgments
We thank David Baulcombe for amplicon line SA93006, IR line T4, and line T19 and Herve Vaucheret for line 6b5. We also thank Gail Pruss and Erin Connolly for critical review of the manuscript. This work was supported by grants to V.B.V. from the U.S. Department of Agriculture, Competitive Grants Program, and by a grant to V.B.V. and L.H.B. from the National Institutes of Health. A.C.M. was supported in part by a National Science Foundation Industry/Graduate Research Traineeship.
Abbreviations
dsRNA, double-stranded RNA
GUS, β-glucuronidase
miRNA, micro-RNA
siRNA, short interfering RNA
IR, inverted repeat
EtdBr, ethidium bromide
RdRp, RNA-dependent RNA polymerase
HC-Pro, helper component-proteinase
PVX, potato virus X
CIP, calf intestinal phosphatase
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Zamore P. D. (2002) Science 296, 1265-1269. [DOI] [PubMed] [Google Scholar]
- 2.Ahlquist P. (2002) Science 296, 1270-1273. [DOI] [PubMed] [Google Scholar]
- 3.Hannon G. J. (2002) Nature 418, 244-251. [DOI] [PubMed] [Google Scholar]
- 4.Grishok A. & Mello, C. C. (2002) Adv. Genet. 46, 339-360. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein E., Denli, A. M. & Hannon, G. J. (2001) RNA 7, 1509-1521. [PMC free article] [PubMed] [Google Scholar]
- 6.Vance V. & Vaucheret, H. (2001) Science 292, 2277-2280. [DOI] [PubMed] [Google Scholar]
- 7.Waterhouse P. M., Wang, M. B. & Lough, T. (2001) Nature 411, 834-842. [DOI] [PubMed] [Google Scholar]
- 8.Pickford A. S., Catalanotto, C., Cogoni, C. & Macino, G. (2002) Adv. Genet. 46, 277-303. [DOI] [PubMed] [Google Scholar]
- 9.Li H., Li, W. X. & Ding, S. W. (2002) Science 296, 1319-1321. [DOI] [PubMed] [Google Scholar]
- 10.Li W. X. & Ding, S. W. (2001) Curr. Opin. Biotechnol. 12, 150-154. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363-366. [DOI] [PubMed] [Google Scholar]
- 12.Hammond S. M., Bernstein, E., Beach, D. & Hannon, G. J. (2000) Nature 404, 293-296. [DOI] [PubMed] [Google Scholar]
- 13.Zamore P. D., Tuschl, T., Sharp, P. A. & Bartel, D. P. (2000) Cell 101, 25-33. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton A. J. & Baulcombe, D. C. (1999) Science 286, 950-952. [DOI] [PubMed] [Google Scholar]
- 15.Parrish S., Fleenor, J., Xu, S., Mello, C. & Fire, A. (2000) Mol. Cell 6, 1077-1087. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir S. M., Lendeckel, W. & Tuschl, T. (2001) Genes Dev. 15, 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Llave C., Kasschau, K. D. & Carrington, J. C. (2000) Proc. Natl. Acad. Sci. USA 97, 13401-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallory A. C., Ely, L., Smith, T. H., Marathe, R., Anandalakshmi, R., Fagard, M., Vaucheret, H., Pruss, G., Bowman, L. & Vance, V. B. (2001) Plant Cell 13, 571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton A., Voinnet, O., Chappell, L. & Baulcombe, D. (2002) EMBO J. 21, 4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansen L. K. & Carrington, J. C. (2001) Plant Physiol. 126, 930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith N. A., Singh, S. P., Wang, M. B., Stoutjesdijk, P. A., Green, A. G. & Waterhouse, P. M. (2000) Nature 407, 319-320. [DOI] [PubMed] [Google Scholar]
- 22.Wang M. B. & Waterhouse, P. M. (2000) Plant Mol. Biol. 43, 67-82. [DOI] [PubMed] [Google Scholar]
- 23.Angell S. M. & Baulcombe, D. C. (1997) EMBO J. 16, 3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beclin C., Boutet, S., Waterhouse, P. & Vaucheret, H. (2002) Curr. Biol. 12, 684-688. [DOI] [PubMed] [Google Scholar]
- 25.Dalmay T., Horsefield, R., Braunstein, T. H. & Baulcombe, D. C. (2001) EMBO J. 20, 2069-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalmay T., Hamilton, A., Rudd, S., Angell, S. & Baulcombe, D. C. (2000) Cell 101, 543-553. [DOI] [PubMed] [Google Scholar]
- 27.Fagard M., Boutet, S., Morel, J. B., Bellini, C. & Vaucheret, H. (2000) Proc. Natl. Acad. Sci. USA 97, 11650-11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mourrain P., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J. B., Jouette, D., Lacombe, A. M., Nikic, S., Picault, N., et al. (2000) Cell 101, 533-542. [DOI] [PubMed] [Google Scholar]
- 29.Anandalakshmi R., Pruss, G. J., Ge, X., Marathe, R., Mallory, A. C., Smith, T. H. & Vance, V. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mallory A. C., Parks, G., Endres, M. W., Baulcombe, D., Bowman, L. H., Pruss, G. J. & Vance, V. B. (2002) Nat. Biotechnol. 20, 622-625. [DOI] [PubMed] [Google Scholar]
- 31.Lee R. C. & Ambros, V. (2001) Science 294, 862-864. [DOI] [PubMed] [Google Scholar]
- 32.Lau N. C., Lim, L. P., Weinstein, E. G. & Bartel, D. P. (2001) Science 294, 858-862. [DOI] [PubMed] [Google Scholar]
- 33.Lagos-Quintana M., Rauhut, R., Lendeckel, W. & Tuschl, T. (2001) Science 294, 853-858. [DOI] [PubMed] [Google Scholar]
- 34.Reinhart B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B. & Bartel, D. P. (2002) Genes Dev. 16, 1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llave C., Kasschau, K. D., Rector, M. A. & Carrington, J. C. (2002) Plant Cell 14, 1605-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee R. C., Feinbaum, R. L. & Ambros, V. (1993) Cell 75, 843-854. [DOI] [PubMed] [Google Scholar]
- 37.Moss E. G., Lee, R. C. & Ambros, V. (1997) Cell 88, 637-646. [DOI] [PubMed] [Google Scholar]
- 38.Olsen P. H. & Ambros, V. (1999) Dev. Biol. 216, 671-680. [DOI] [PubMed] [Google Scholar]
- 39.Reinhart B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., Horvitz, H. R. & Ruvkun, G. (2000) Nature 403, 901-906. [DOI] [PubMed] [Google Scholar]
- 40.Slack F. J., Basson, M., Liu, Z., Ambros, V., Horvitz, H. R. & Ruvkun, G. (2000) Mol. Cell 5, 659-669. [DOI] [PubMed] [Google Scholar]
- 41.Rhoades M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B. & Bartel, D. P. (2002) Cell 110, 513-520. [DOI] [PubMed] [Google Scholar]
- 42.Llave C., Xie, Z., Kasschau, K. D. & Carrington, J. C. (2002) Science 297, 2053-2056. [DOI] [PubMed] [Google Scholar]
- 43.Hutvagner G., McLachlan, J., Pasquinelli, A. E., Balint, E., Tuschl, T. & Zamore, P. D. (2001) Science 293, 834-838. [DOI] [PubMed] [Google Scholar]
- 44.Grishok A., Pasquinelli, A. E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D. L., Fire, A., Ruvkun, G. & Mello, C. C. (2001) Cell 106, 23-34. [DOI] [PubMed] [Google Scholar]
- 45.Ketting R. F., Fischer, S. E., Bernstein, E., Sijen, T., Hannon, G. J. & Plasterk, R. H. (2001) Genes Dev. 15, 2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobsen S. E., Running, M. P. & Meyerowitz, E. M. (1999) Development (Cambridge, U.K.) 126, 5231-5243. [DOI] [PubMed] [Google Scholar]
- 47.Ray A., Lang, J. D., Golden, T. & Ray, S. (1996) Development (Cambridge, U.K.) 122, 2631-2638. [DOI] [PubMed] [Google Scholar]
- 48.Castle L. A., Errampalli, D., Atherton, T. L., Franzmann, L. H., Yoon, E. S. & Meinke, D. W. (1993) Mol. Gen. Genet. 241, 504-514. [DOI] [PubMed] [Google Scholar]
- 49.Mourelatos Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M. & Dreyfuss, G. (2002) Genes Dev. 16, 720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hutvagner G. & Zamore, P. D. (2002) Science 297, 2056-2060. [DOI] [PubMed] [Google Scholar]
- 51.Elmayan T. & Vaucheret, H. (1996) Plant J. 9, 787-797. [Google Scholar]
- 52.Hobbs S. L., Kpodar, P. & DeLong, C. M. (1990) Plant Mol. Biol. 15, 851-864. [DOI] [PubMed] [Google Scholar]
- 53.Vance V. B. (1991) Virology 182, 486-494. [DOI] [PubMed] [Google Scholar]
- 54.Nykanen A., Haley, B. & Zamore, P. D. (2001) Cell 107, 309-321. [DOI] [PubMed] [Google Scholar]
- 55.Mette M. F., Matzke, A. J. & Matzke, M. A. (2001) Curr. Biol. 11, 1119-1123. [DOI] [PubMed] [Google Scholar]
- 56.Sijen T., Vijn, I., Rebocho, A., van Blokland, R., Roelofs, D., Mol, J. N. & Kooter, J. M. (2001) Curr. Biol. 11, 436-440. [DOI] [PubMed] [Google Scholar]
- 57.Anandalakshmi R., Marathe, R., Ge, X., Herr, J. M., Jr., Mau, C., Mallory, A., Pruss, G., Bowman, L. & Vance, V. B. (2000) Science 290, 142-144. [DOI] [PubMed] [Google Scholar]