Abstract
We present biochemical evidence for the functional similarity of Escherichia coli RecO protein and bacteriophage T4 UvsY protein to eukaryotic Rad52 protein. Although Rad52 protein is conserved in eukaryotes, no sequence homologue has been found in prokaryotes or archeabacteria. Rad52 protein has two unique activities: facilitation of replication protein-A (RPA) displacement by Rad51 protein and annealing of RPA–single-stranded DNA (ssDNA) complexes. Both activities require species-specific interaction between Rad52 protein and RPA. Both RecO and UvsY proteins also possess the former property with regard to their cognate ssDNA-binding protein. Here, we report that RecO protein anneals ssDNA that is complexed with only its cognate ssDNA-binding protein, suggesting the involvement of species-specific interactions. Optimal activity for RecO protein occurs after formation of a 1:1 complex with SSB protein. RecR protein, which is known to stimulate RecO protein to facilitate SSB protein displacement by RecA protein, inhibits annealing by RecO protein, suggesting that RecR protein may regulate the choice between the DNA strand invasion versus annealing pathways. In addition, we show that UvsY protein anneals ssDNA; furthermore, ssDNA, which is complexed only with its cognate ssDNA-binding protein, is annealed in the presence of UvsY protein. These results indicate that RecO and possibly UvsY proteins are functional counterparts of Rad52 protein. Based on the conservation of these functions, we propose a modified double-strand break repair model that includes DNA annealing as an important intermediate step.
Both biochemical and genetic studies in bacteriophage, Bacteria, Eukarya, and Archaea have established many parallels in genetic recombination. Structural and functional homologues of the eubacterial RecA protein are found in T4 phage and in all eukaryotes and archaea (UvsX, Rad51, and RadA proteins, respectively), as well as functional homologues of single-stranded DNA (ssDNA)-binding protein, SSB protein [gene 32 protein (gp32), replication protein-A (RPA), and RPA/SSB, respectively; for review, see refs. 1 and 2]. Despite these and other common features, many unique proteins exist. For example, homologous recombination in the budding yeast Saccharomyces cerevisiae requires the Rad52 protein. Lack of RAD52 function results in total loss of homologous recombination as manifest by defects in double-stranded DNA (dsDNA) break repair, mating type switching, and ssDNA annealing (3, 4). Consistent with the importance of this protein, homologues are present in the Eukarya (5). Yet, despite the importance of Rad52 protein to eukaryotic recombination function, primary sequence homologues have not been identified outside of the Eukarya. However, previous biochemical analyses, and those to be reported here, suggest that Escherichia coli RecO protein (6, 7) and the bacteriophage T4 UvsY protein (8, 9) are functional homologues of the eukaryotic Rad52 protein. The rationale for this hypothesis follows.
Rad52 protein has two distinctive activities. One is the stimulation of DNA-strand exchange mediated by Rad51 protein and RPA (10–12). The second is the annealing of complementary ssDNA (13). A unique feature of DNA annealing by Rad52 protein is that it not only anneals free ssDNA, but it can also anneal ssDNA that is complexed with RPA, acting more effectively with its cognate RPA than with human RPA (14, 15). This biochemical activity is consistent with the essential role of RAD52 in the ssDNA annealing pathway of recombination (16, 17). Rad52 protein physically interacts with Rad51 protein and RPA (15, 18–20), and these species-specific interactions are required for the stimulation of both DNA strand exchange (11) and ssDNA annealing (14, 15).
The E. coli recO gene was first isolated as a mutation that eliminated conjugal recombination in a background that lacked the RecBC pathway of recombination (recBC sbcB) (21). A recO mutation confers UV sensitivity and severely defective plasmid recombination in an otherwise rec+ background and, in an recBC sbcB background, the recO mutation results in a complete loss of both plasmid and conjugal recombination. Genetic analyses showed that recO belongs to the same epistasis group as recF and that recO acts at the earliest stage of the RecF pathway (which includes recA), together with recF and recR (22, 23). Purified RecF, RecO, and RecR proteins can form a three-protein complex by physical interactions between RecO and RecF proteins, and between RecO and RecR proteins (24), supporting the genetic observations that they act at the same stage. In the presence of ATP and dsDNA, RecF and RecR proteins physically interact, even in the absence of RecO protein (25). This RecFR complex has a higher affinity for an ssDNA–dsDNA junction, thus preventing polymerization of RecA protein from ssDNA beyond the junction into the dsDNA (26). On the other hand, the RecOR complex prevents dissociation of RecA protein from linear ssDNA, presumably by stabilizing the RecA–ssDNA complex (27).
One biochemical similarity of RecO and Rad52 proteins is that RecO protein stimulates the activity of RecA protein (6) in the same way that Rad52 protein stimulates Rad51 protein. When ssDNA is bound with SSB protein before RecA protein, SSB protein inhibits DNA strand exchange by preventing the binding of RecA protein to the ssDNA. RecO protein helps RecA protein to overcome this inhibition by SSB protein; this behavior is identical to the mediator function of Rad52 protein (10–12). An additional biochemical similarity of RecO protein to Rad52 protein is that both proteins have ssDNA annealing activity (7, 13); a unique attribute of this activity is that Rad52 protein can also anneal ssDNA that is complexed with its cognate RPA (14). The final parallel to Rad52 protein and RPA is that RecO protein interacts with SSB protein (24, 28); presumably, this interaction mediates the exchange of RecA protein for SSB protein.
The bacteriophage T4 protein, UvsY, also bears some functional similarity to both Rad52 and RecO proteins, despite sharing little, if any, primary sequence similarity. Mutation of the uvsY gene results in T4 phage that displays UV and γ-ray sensitivity, low burst size, and low levels of recombination (29, 30). UvsY interacts with UvsX protein and with gp32 (31, 32). UvsY protein also assists UvsX protein in the displacement of gp32 from ssDNA to facilitate DNA-strand exchange (8, 9).
Because there are many similarities in the genetic features and biochemical activities of RecO, UvsY, and Rad52 proteins, it seems that these very different proteins are functional counterparts of Rad52 protein (6, 33). Yet if they are genuine homologues, then they should also possess the unique ability to specifically anneal ssDNA that is complexed with its cognate ssDNA-binding protein. Here, we report that this is indeed the case for RecO protein. The annealing of complementary ssDNA by RecO protein is supported by E. coli SSB protein; this stimulation is species-specific: that is, neither T4 gp32 nor yeast RPA supports the reaction. In addition, we show that UvsY protein also has annealing activity. The conservation of the annealing function of Rad52, RecO, and UvsY proteins highlights the importance of this common activity in homologous recombination. The role of this protein-dependent annealing of SSB–ssDNA complexes in recombination is discussed.
Materials and Methods
Reagents.
Chemical reagents were purchased from Sigma unless otherwise noted. PstI and SssI were purchased from New England Biolabs. S-[methyl-3H]Adenosyl-L-methionine ([3H]SAM) was purchased from NEN. Oligonucleotides were purchased from Operon Technologies (Alameda, CA). 4′,6-Diamidino-2-phenyllindole (DAPI) was purchased from Molecular Probes. It was dissolved in water and filtered; the concentration was determined by using an extinction coefficient of 3.3 × 104 M−1·cm−1 at 345 nm.
DNA and Proteins.
Sequences of oligonucleotides are as follows: oligo-25, 5′-GCAATTAAGCTCTAAGCCATCCGCAAAAATGACCTCTTATCAAAAGGA-3′; and oligo-26, 5′-TCCTTTTGATAAGAGGTCATTTTTGCGGATGGCTTAGAGCTTAATTGC-3′. The concentrations of oligo-25 and oligo-26 were determined by using extinction coefficients of 1.0 × 104 and 9.6 × 103 M−1·cm−1 at 260 nm, respectively. pBluescript II SK-dsDNA (Stratagene) was digested by PstI, resulting in a 2.9-kb linear dsDNA. To prepare 3H-labeled DNA, 100 μg of linear pBluescript II SK-dsDNA was methylated at 37°C for 3 h with 500 units of SssI methylase by using 1 mCi (1 Ci = 37 GBq) of [3H]SAM in a buffer containing 200 mM Tris⋅HCl, pH 8.0/50 mM NaCl/1 mM DTT. The unincorporated [3H]SAM was removed by Elutip minicolumns (Schleicher & Schuell). Then, 10 μg of tritiated DNA and 90 μg of unlabeled DNA were mixed and alkaline denatured as described (34). The concentration of the denatured tritiated DNA was determined by a Malachite green-ammonium molybdate assay (35). After alkaline denaturation, DNA was heat-denatured before use by incubation at 100°C for 3 min to ensure complete denaturation. All DNA concentrations are expressed as moles of nucleotides.
RecO, Rad52, RecR, and SSB proteins were purified as described (7, 11, 28, 36), respectively. RPA was purified by using a strain kindly provided by R. Kolodner (University of California at San Diego) as described (37) with minor modifications: the Affigel-Blue column chromatography was omitted, and a Resource Q (Amersham Pharmacia) column was used in place of the DEAE-cellulose column. Bacteriophage T4 gene 32 protein was purified as described (38, 39) followed by chromatography on a Mono Q column (Amersham Pharmacia) to separate gp32 from a nuclease contaminant by Dr. Piero Bianco (University of California, Davis). UvsY protein was kindly provided by S. Morrical (University of Vermont). Concentrations of SSB, RPA, RecO, RecR, Rad52, and gp32 proteins were determined by using extinction coefficient of 3.0 × 104, 8.8 × 104, 2.3 × 104, 5.4 × 103, 2.4 × 104, and 4.13 × 104 M−1·cm−1 at 280 nm, respectively.
Complementary ssDNA Annealing Assays.
Annealing of complementary oligonucleotides was monitored by using the increase in DAPI fluorescence after binding to the annealed dsDNA, using an SLM 8000 spectrofluorometer with excitation and emission wavelengths set to 345 and 467 nm, using the bandwidths of 2 and 8 nm, respectively (14). Oligonucleotides (48-mers, oligo-25 and oligo-26) were reacted with appropriate proteins in 400 μl of reaction buffer at 30°C. The reaction buffer contained 25 mM Tris⋅HCl, pH 7.5/8 mM magnesium acetate/1 mM DTT/0.2 μM DAPI. Reactions were started by the addition of oligo-26, except in experiments containing an ssDNA-binding protein and RecO, Rad52, or UvsY protein, which were started by addition of the annealing protein. Concentrations of proteins are listed in each figure legend. Annealing of denatured plasmid 3H-DNA was assayed by using S1 nuclease as described (34), except that our annealing reaction buffer and temperature (30°C) were used.
Results
RecO Protein Mediates the Annealing of Complementary ssDNA That Is Complexed with SSB Protein.
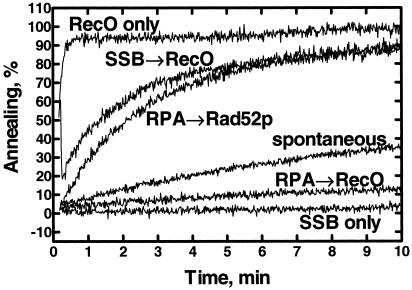
It was previously reported that RecO protein can renature complementary ssDNA (7); however, the effect of SSB protein on the annealing reaction was not examined. To determine whether RecO protein could anneal complementary SSB protein–ssDNA complexes, we monitored ssDNA renaturation using the fluorometric assay that was used previously to analyze annealing by Rad52 protein (14). The substrates that we chose to study first were complementary oligonucleotides that were 48 nucleotides in length (Fig. 1). As reported, in the absence of SSB protein, RecO protein accelerates renaturation ≈50-fold relative to the protein-free reaction (compare RecO only curve versus the spontaneous curve), whereas SSB protein inhibits spontaneous annealing (SSB only). However, RecO protein promotes the annealing of the SSB protein-coated oligonucleotides (SSB → RecO). This ability of RecO protein to mediate the annealing of SSB protein–ssDNA complexes is similar to that of Rad52 protein and RPA: RPA inhibits annealing of ssDNA, whereas Rad52 protein promotes the annealing of the RPA–ssDNA complexes (14). In the case of Rad52 protein, annealing of the RPA–ssDNA complex was species-specific. To determine whether annealing of the SSB–ssDNA complexes was also species-specific, the annealing of RPA–ssDNA complexes by RecO protein was tested. When the oligonucleotides are coated first with RPA, the rate of annealing is less than 10% of the rate obtained with SSB (compare SSB → RecO curve versus RPA → RecO curve).
Fig 1.
RecO protein can anneal complementary oligonucleotides that are complexed with SSB protein. Annealing of complementary 48-mer oligonucleotides (oligo-25 and oligo-26, 200 nM each) is shown. The oligonucleotides were incubated without protein (spontaneous) or in the presence of SSB (SSB only), RecO (RecO only), RPA and RecO (RPA → RecO), RPA and Rad52 (RPA → Rad52p), or SSB and RecO (SSB → RecO) proteins. The final concentrations of SSB, RPA, Rad52, and RecO were 50, 40, 40, and 40 nM, respectively. The extent of DNA annealing is expressed as a percentage of the observed DAPI fluorescence relative to the fluorescence dsDNA 48-mer oligonucleotides (400 nM nucleotides).
To characterize this annealing activity further, reactions were performed at various RecO protein concentrations (Fig. 2). The optimum concentration of RecO protein for this reaction depends on the presence of SSB protein. In the absence of SSB protein, the optimum RecO concentration for annealing is low, requiring about 1 RecO protein per 40 nucleotides (Fig. 2A). In the presence of SSB protein, however, more RecO protein is needed, about 1 RecO protein per 13 nucleotides for optimum annealing, which is approximately the same stoichiometry of SSB protein binding. This suggested that the mode of annealing is different in each case: in the absence of SSB protein, RecO protein acts via the ssDNA, whereas in the presence of SSB protein, it may bind the SSB protein rather than the ssDNA, explaining the different stoichiometry. To determine whether optimal annealing paralleled the concentration of the SSB protein–DNA complex, the protein–DNA complex concentration was varied (Fig. 2B). When the substrate concentration was changed from 200 to 800 nM but the SSB:ssDNA ratio was kept constant at 1:8, the optimum RecO concentration changed accordingly, yielding an approximate value of ≈0.9 RecO monomer per SSB monomer. This result, together with the species specificity demonstrated above, suggests that the active species in this reaction is a RecO–SSB–ssDNA cocomplex, which is analogous to the conclusion derived for RPA and Rad52 protein (14).
Fig 2.
Optimal annealing by RecO protein occurs at stoichiometric amounts of RecO protein, relative to the SSB–ssDNA complex. (A) Annealing of 48-mer oligonucleotides (200 nM each) at various RecO protein concentrations in the absence of SSB protein. (B) Annealing of SSB protein–oligonucleotide complexes. Concentrations of 48-mer oligonucleotides and SSB protein are 100 nM each and 25 nM (▵), 200 nM each and 50 nM (•), and 400 nM each and 100 nM (○), respectively. Data for the 200 nM oligonucleotide reactions (•) are the mean of three replicates, whereas data for other reactions are obtained from a single experiment.
SSB Protein Specifically Permits RecO Protein-Mediated Annealing of Plasmid-Sized ssDNA.
The fluorometric annealing assay using DAPI is a convenient, real-time assay that yields results comparable to gel assays when oligonucleotide substrates are used (14) (data not shown). However, when longer (i.e., plasmid-sized), denatured DNA is the substrate, greater than stoichiometric amounts of an ssDNA-binding protein are needed to remove DNA secondary structure to eliminate background fluorescence arising from DAPI binding to the secondary structure. Therefore, accurate measurement of annealing in the absence or presence of less than stoichiometric amounts of an ssDNA-binding protein is difficult. To overcome this limitation, the S1 nuclease assay was used to follow annealing (34).
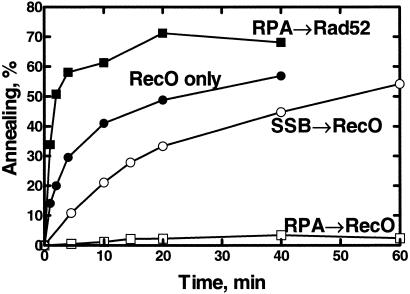
It was reported that RecO protein annealed naked heat-denatured plasmid-sized ssDNA (7). This result was reproduced (Fig. 3, RecO only). In addition, we tested the ability of RecO protein to anneal ssDNA that was complexed with ssDNA-binding proteins. Rad52 protein can anneal heat-denatured plasmid DNA as well as oligonucleotide-length ssDNA (14, 15); however, the annealing of plasmid-sized DNA is unique in its requirement for the yeast RPA. Therefore, if RecO protein is a prokaryotic counterpart of Rad52 protein, then RecO protein should display the same species-specific dependency for a cognate ssDNA-binding protein in the annealing of complex ssDNA. To test this hypothesis, the annealing of denatured plasmid DNA (2.9 kb) that was first coated with an ssDNA-binding protein was examined (Fig. 3).
Fig 3.
RecO protein anneals plasmid-sized ssDNA that is complexed with its cognate ssDNA-binding protein. Annealing was initiated by addition of RecO or Rad52 protein. Either RecO protein (250 nM) or Rad52 protein (200 nM) was added to a complex of denatured, plasmid 3H-DNA (3 μM) and RPA (300 nM), SSB (1.5 μM), or no ssDNA-binding protein. Reactions were started by addition of RecO or Rad52 protein, and annealing was measured by using the S1 nuclease assay.
RecO protein could also anneal SSB-coated, plasmid-length, denatured DNA (SSB → RecO). To test for species specificity, yeast RPA was used instead of SSB protein. RecO protein annealed the RPA–ssDNA complexes very poorly (Fig. 3, RPA → RecO; see also Fig. 1). For comparison, the annealing of RPA-coated plasmid-sized DNA by Rad52 protein is shown (RPA → Rad52); the annealing of naked plasmid-sized DNA can also be detected by this assay (Y. Wu, T.S., and S.C.K., unpublished observations). Therefore, annealing by RecO protein is specific for the SSB protein–ssDNA complex; yeast RPA cannot substitute effectively.
To further address the species-specificity issue, the ability of RecO protein to anneal bacteriophage T4 gp32–ssDNA complexes was also tested (data not shown). As previously reported (40), gp32 annealed plasmid-sized ssDNA; however, when RecO protein was added to the complex of gp32 and ssDNA, annealing was inhibited (data not shown). This was unexpected because gp32 alone can anneal free ssDNA. Nevertheless, this result showed that RecO protein cannot use the gp32–ssDNA complex as a substrate, further arguing that RecO protein acts efficiently only in conjunction with its cognate ssDNA-binding protein. This result is consistent with our inability to detect an interaction between RecO protein and gp32 in biosensor experiments (N.K. and S.C.K., unpublished observation), whereas the same experiments revealed an interaction between RecO and SSB proteins (24, 28).
One characteristic of the annealing of RPA-coated plasmid-sized DNA by Rad52 protein is the inhibition resulting from Rad52 protein concentrations that exceed the optimum concentration (14). To determine whether RecO protein exhibits the same behavior, annealing of SSB-coated DNA was examined at various RecO protein concentrations (data not shown; see also Fig. 2). As for Rad52 protein, annealing is slowed when excess RecO protein was present. This observation further adds to the similarities between Rad52 and RecO proteins.
RecR Protein Inhibits Annealing by RecO Protein.
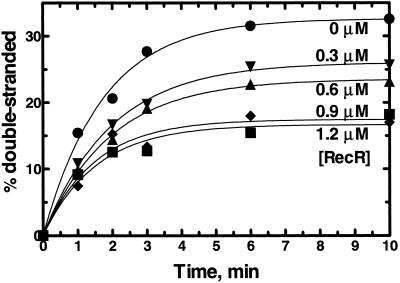
RecR protein is known to interact with RecO protein (24, 28), and RecR protein enhances RecO protein's ability to stimulate displacement of SSB protein from ssDNA by RecA protein (6). Therefore, it was possible that RecR protein could affect annealing by RecO protein. Annealing of plasmid-sized ssDNA in the presence of various concentrations of RecR protein shows that RecR protein inhibits annealing by RecO protein in the absence of SSB protein (Fig. 4). Similar results were obtained when 48-mer oligonucleotides were used as substrate in the presence and absence of SSB protein (data not shown). However, when plasmid-sized ssDNA was used as a substrate in the presence of SSB protein, we were unable to detect inhibition of annealing by RecR protein because annealing of plasmid-sized ssDNA complexed with SSB protein is very slow (data not shown). Fitting the time course data in Fig. 4 to a single-exponential function showed that RecR protein inhibits annealing by RecO protein by reducing the extent of annealing, in direct proportion to the RecR protein concentration, without affecting the rate constant. Thus, combined with the fact that RecR protein does not bind ssDNA (28), RecR protein may modulate the functions of RecO protein.
Fig 4.
RecR protein inhibits annealing of plasmid-sized ssDNA by reducing the extent of annealing without affecting the half-time. 3H-ssDNA (3 μM) was incubated with various concentrations of RecR protein for 2 min at 30°C, and then 250 nM RecO protein was added to start annealing. RecR protein concentrations: 0 μM (•), 0.3 μM (▾), 0.6 μM (▴), 0.9 μM (♦), and 1.2 μM (▪). Each data set was fitted by using GRAPHPAD PRISM software with one phase exponential equation: Y = Ymax ⋅(1 − e−kx). There was no significant change in the rate constant k over all RecR protein concentrations.
Phage T4 UvsY Protein Also Anneals Complementary ssDNA.
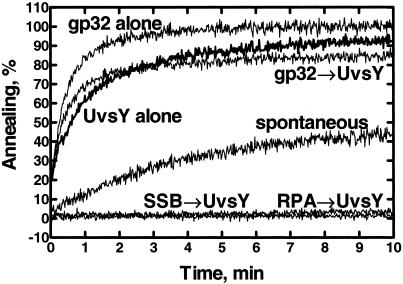
UvsY protein of bacteriophage T4 has some of the same functions of Rad52 and RecO proteins; UvsY protein helps UvsX protein to displace gp32 so that the presynaptic complex can be formed (8, 9). Because both Rad52 and RecO proteins have an annealing activity that requires its cognate ssDNA-binding protein, we tested whether UvsY protein has a similar annealing activity. Fig. 5 shows that UvsY protein alone, indeed, does possess significant annealing activity (Fig. 5, UvsY alone). As already reported (40), gp32 itself mediates ssDNA annealing (Fig. 5, gp32 alone); this behavior is in contrast to the other ssDNA-binding proteins examined in this paper, as both RPA and SSB protein inhibit spontaneous annealing. We also tested whether UvsY protein has the same species-specific requirement for an ssDNA-binding protein. UvsY protein does not anneal ssDNA complexed with either SSB protein (Fig. 5, SSB → UvsY) or RPA (Fig. 5, RPA → UvsY), demonstrating that ssDNA-binding proteins from different species cannot support UvsY protein-mediated annealing. However, whether UvsY protein can anneal gp32-coated oligonucleotides is unclear because annealing of the oligonucleotide substrates by gp32 is faster than annealing by UvsY protein.
Fig 5.
UvsY protein has ssDNA annealing activity. Annealing of oligo-25 and oligo-26 (200 nM each) was carried out with UvsY protein (UvsY alone, bold line), gp32 (gp32 alone), or gp32 and UvsY (gp32 → UvsY), or in the absence of protein (spontaneous). The above reactions were started by addition of oligo-26 to the other components that had been preincubated for 100 s in the reaction buffer. Annealing reactions using SSB-coated (SSB → UvsY) or RPA-coated (RPA → UvsY) oligonucleotides were started by the addition of UvsY protein to the oligonucleotides that were preincubated with either SSB or RPA. The final concentrations of gp32, SSB, RPA, and UvsY proteins were 133, 67, 50, and 20 nM, respectively.
RecO and Rad52 Proteins Share a Weak Sequence Similarity.
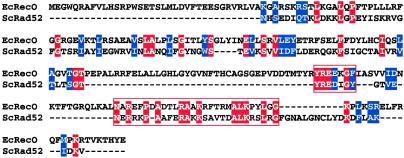
Although RecO protein and T4 UvsY protein have functional similarities to Rad52 protein, no significant similarity at the sequence level was reported. The sequence alignment of RecO protein and the most conserved region of Rad52 protein (Fig. 6) revealed regions of weak similarity; however, no similarity was found between UvsY protein and either RecO or Rad52 protein.
Fig 6.
RecO protein and the N-terminal region of Rad52 protein share a weak sequence similarity. Rad52 protein sequences from six species were aligned to determine the most conserved region of Rad52 protein. Then the sequences of residues 49–193 of ScRad52 protein and RecO protein were aligned by using the T-COFFEE program (www.ch.embnet.org/software/TCoffee.html). Identical amino acid residues are highlighted by the solid red background, and conserved residues are highlighted by the blue background. The most conserved areas are boxed: residues 135–141 and 146–172 in Rad52 protein and residues 166–172 and 192–219 in RecO protein, respectively.
Discussion
In this paper, we demonstrate that E. coli RecO protein mediates the annealing of complementary ssDNA complexed with E. coli SSB protein. Neither yeast RPA nor T4 gp32 protein can substitute for SSB protein, suggesting that a species-specific interaction between RecO and SSB proteins is important for the annealing reaction. This behavior is equivalent to the annealing reaction mediated by Rad52 protein (14, 15), which also requires its cognate ssDNA-binding protein, RPA. Therefore, we propose that RecO and Rad52 proteins are homologous with regard to two functions: (i) their ability to mediate a replacement of the cognate ssDNA-binding protein for the cognate RecA-like protein and (ii) their ability to promote annealing of ssDNA when complexed with the cognate ssDNA-binding protein. In addition to the species specificity, excess RecO protein, like Rad52 protein, slows annealing (Fig. 2). This suggests that annealing by these proteins shares a similar mechanism.
The optimum RecO concentration for annealing the 48-mer oligonucleotides depends on whether SSB protein is present (Fig. 2). In the presence of SSB protein, annealing is most rapid when SSB and RecO proteins are present at approximately equimolar amount. Even though SSB protein slows the rate of annealing by 5-fold (Fig. 2), the extent of annealing is almost the same in the presence and absence of SSB protein (Figs. 1 and 3). Together with the observation that RecO protein is incapable of annealing RPA-coated DNA (Figs. 1 and 3), we conclude that SSB protein is indeed involved in the rate-limiting step of this annealing process.
One potential difference between Rad52 and RecO proteins is that RecO protein acts in concert with RecR protein. RecR protein interacts with RecO protein (24, 28), and the RecOR complex stimulates the displacement of SSB protein from ssDNA by RecA protein better than RecO protein alone (6). Therefore, we thought that RecR protein might affect the ssDNA annealing activity of RecO protein in a similar manner. Our analyses showed that RecR protein reduces the extent of annealing by RecO protein in a RecR protein concentration-dependent manner (Fig. 4). Thus, RecR protein may be controlling the two distinct activities of RecO protein: RecR protein stimulates the mediator function of RecO protein, but it inactivates its annealing function.
We have also showed that bacteriophage T4 UvsY protein has annealing activity. Annealing occurs in the presence of T4 gp32 but not in the presence of noncognate ssDNA-binding proteins; because gp32 can anneal ssDNA, we are unable to determine which protein is acting in the presence of UvsY protein. Unlike RecO and Rad52 proteins, however, UvsY protein cannot anneal plasmid-length DNA (unpublished observations) for reasons that we do not understand. Annealing of long ssDNA by UvsY protein may not be necessary for bacteriophage T4 recombination because gp32 is capable of annealing long ssDNA. Nevertheless, UvsY protein does have an annealing activity, supporting the idea that annealing activity is conserved in all Rad52 protein counterparts tested.
A weak sequence similarity between Rad52 and RecO proteins was found (Fig. 6), but not with UvsY protein. Despite the lack of strong sequence similarity, RecO and UvsY proteins display biochemical and genetic characteristics that justify their description as counterparts of eukaryotic Rad52 protein; they all possess ssDNA- and dsDNA-binding abilities, facilitation of ssDNA-binding protein displacement by the cognate DNA strand-exchange protein, annealing of simple DNA, and annealing of complex DNA in the presence of a cognate ssDNA-binding protein.
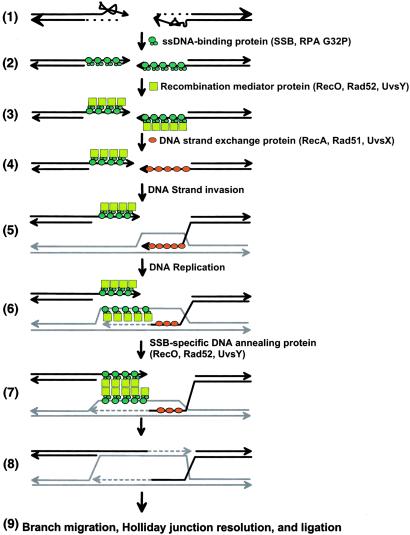
The conservation of these properties suggests that these activities comprise important biological functions. We propose that the ability to anneal DNA that is complexed with a homologous ssDNA-binding protein is necessary for two biochemical steps of double-strand DNA break (DSB) repair (41, 42): these are (i) to serve as the mediator protein that accelerates the displacement of ssDNA-binding protein by the RecA-like protein and (ii) to anneal the ssDNA within the D-loop made by strand invasion and the ssDNA of the processed dsDNA end that did not participate in DNA strand invasion (see Fig. 7). DSB repair starts by processing the DSB to produce ssDNA with 3′ overhangs (Fig. 7, step 1); this processing need not be concurrent. The resulting ssDNA has secondary structure that is removed by an ssDNA-binding protein (SSB in bacteria, RPA in eukaryotes, or gp32 in phage T4) (step 2). This ssDNA-binding protein must be replaced by the DNA strand-exchange protein, a step that is kinetically slow but that is accelerated by either RecO or RecOR protein in E. coli, Rad52 protein in eukaryotes, or UvsY protein in phage T4. (step 3). Each recombination mediator protein binds its cognate ssDNA-binding protein to recruit a DNA strand-exchange protein (RecA protein in E. coli, Rad51 protein in eukaryotes, or UvsX protein in phage T4) (10) to replace the ssDNA-binding protein on the ssDNA, thereby forming a presynaptic complex (step 4). This presynaptic complex can now invade homologous dsDNA (step 5). At the same time, the ssDNA-binding protein binds to the DNA strand displaced by the strand invasion event (step 6) (43, 44). DNA synthesis extends the 3′ end of the D-loop (step 6). The complementary complexes of ssDNA-binding protein–Rad52 homologue–ssDNA can now anneal to each other (step 7), allowing the 3′ end of the processed DSB to serve as the primer for additional DNA synthesis. In the case of phage T4, gp32, UvsY protein, or both may anneal the strands.
Fig 7.
A model for DSB repair illustrating the proposed role for annealing of SSB–ssDNA complexes by RecO, Rad52, or UvsY proteins. (1) The DSB is processed to expose ssDNA with 3′ overhangs. (2) The ssDNA is coated by an ssDNA-binding protein (SSB/RPA/gp32). (3) The recombination mediator protein (RMP: RecO/Rad52/UvsY) binds to the SSB–ssDNA complex. (4) The DNA strand-exchange protein (RecA/Rad51/UvsX) is recruited by the RMP (RecO/Rad52/UvsY) and replaces the ssDNA-binding protein at one of the processed ssDNA tails. (5) The presynaptic complex (DNA strand-exchange protein–ssDNA complex) invades homologous DNA, displacing one strand of homologous dsDNA. (6) DNA replication initiates from the invaded 3′ end within the D-loop. SSB (SSB/RPA/gp32) and RMP (RecO/Rad52p/UvsY) bind the displaced strand produced by DNA-strand invasion and DNA synthesis. (7) The complex of displaced ssDNA–SSB–RMP anneals with the ssDNA–SSB–RMP complex containing the other 3′ overhang. In the case of phage T4, gp32, UvsY protein, or both may actually anneal these strands. (8) Further DNA synthesis (9), ligation, branch migration, and resolution of double Holliday junction result in two intact homologous DNA molecules.
Thus, in our proposal, both displacement of the ssDNA-binding protein and annealing between the ssDNA produced at the second processed end of DSB and the DNA strand displaced by strand invasion are mediated by the same protein: RecO protein, Rad52 protein, or UvsY protein or their functional counterparts. This is an effective and economical way to connect two absolutely necessary steps in the DSB repair model: DNA strand invasion by ssDNA from one processed DSB and DNA annealing of the displaced strand to the second processed end of the DSB. Because all of the recombination mediator proteins possess both activities, it is likely that this model expresses a general feature of homologous recombination in many organisms. Thus, DSB repair need not occur by two DNA strand invasion events but could also occur by a single DNA strand invasion event, followed by DNA displacement and DNA annealing (42). In support of this view, Hunter and Kleckner identified a meiotic recombination intermediate called the single-end invasion product (45), which is produced by DNA-strand exchange between one processed DSB end and its homologue; formation of this intermediate precedes double-end invasion products. The DNA annealing activity of Rad52 protein and its counterparts is also likely involved in other modes of DSB repair. Recent evidence suggests that meiotic recombination in S. cerevisiae occurs by synthesis-dependent strand annealing and strand displacement-mediated crossover mechanisms (46, 47), both of which require annealing of complementary ssDNA. Rad52 protein and its homologues are well suited to these processes as well.
Acknowledgments
We thank the members of the S.C.K. laboratory, Piero Bianco, Mark Dillingham, Naofumi Handa, Katsumi Morimatsu, Jim New, and Yun Wu, for comments on the manuscript. We thank Dr. Scott Morrical (University of Vermont) for providing UvsY protein, Dr. Richard Kolodner for the RPA-overproducing strain, and Dr. Piero Bianco (University of California, Davis) for providing gp32. This work was supported by National Institutes of Health Grants AI-18987 and GM-62653.
Abbreviations
DSB, double-strand break
RPA, replication protein-A
dsDNA, double-stranded DNA
ssDNA, single-stranded DNA
[3H]SAM, S-[methyl-3H]adenosyl-l-methionine
DAPI, 4′,6-diamidino-2-phenyllindole
References
- 1.Bianco P. R., Tracy, R. B. & Kowalczykowski, S. C. (1998) Front. Biosci. 3 D570-D603. [DOI] [PubMed] [Google Scholar]
- 2.Seitz E. M., Haseltine, C. A. & Kowalczykowski, S. C. (2001) Adv. Appl. Microbiol. 50 101-169. [DOI] [PubMed] [Google Scholar]
- 3.Petes T. D., Malone, R. E. & Symington, L. S. (1991) in The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics, eds. Broach, J. R., Jones, E. & Pringle, J. (Cold Spring Harbor Lab. Press, Plainview, NY), Vol. I, pp. 407–521. [Google Scholar]
- 4.Pâques F. & Haber, J. E. (1999) Microbiol. Mol. Biol. Rev. 63 349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedberg E. C., Walker, G. C. & Siede, W., (1995) DNA Repair and Mutagenesis (Am. Soc. Microbiol., Washington, DC).
- 6.Umezu K., Chi, N. W. & Kolodner, R. D. (1993) Proc. Natl. Acad. Sci. USA 90 3875-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luisi-DeLuca C. & Kolodner, R. D. (1994) J. Mol. Biol. 236 124-138. [DOI] [PubMed] [Google Scholar]
- 8.Yonesaki T. & Minagawa, T. (1989) J. Biol. Chem. 264 7814-7820. [PubMed] [Google Scholar]
- 9.Harris L. D. & Griffith, J. D. (1989) J. Mol. Biol. 206 19-27. [DOI] [PubMed] [Google Scholar]
- 10.Sung P. (1997) J. Biol. Chem. 272 28194-28197. [DOI] [PubMed] [Google Scholar]
- 11.New J. H., Sugiyama, T., Zaitseva, E. & Kowalczykowski, S. C. (1998) Nature 391 407-410. [DOI] [PubMed] [Google Scholar]
- 12.Shinohara A. & Ogawa, T. (1998) Nature 391 404-407. [DOI] [PubMed] [Google Scholar]
- 13.Mortensen U. H., Bendixen, C., Sunjevaric, I. & Rothstein, R. (1996) Proc. Natl. Acad. Sci. USA 93 10729-10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugiyama T., New, J. H. & Kowalczykowski, S. C. (1998) Proc. Natl. Acad. Sci. USA 95 6049-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shinohara A., Shinohara, M., Ohta, T., Matsuda, S. & Ogawa, T. (1998) Genes Cells 3 145-156. [DOI] [PubMed] [Google Scholar]
- 16.Fishman-Lobell J., Rudin, N. & Haber, J. E. (1992) Mol. Cell. Biol. 12 1292-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugawara N. & Haber, J. E. (1992) Mol. Cell. Biol. 12 563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinohara A., Ogawa, H. & Ogawa, T. (1992) Cell 69 457-470. [DOI] [PubMed] [Google Scholar]
- 19.Milne G. T. & Weaver, D. T. (1993) Genes Dev. 7 1755-1765. [DOI] [PubMed] [Google Scholar]
- 20.Hays S. L., Firmenich, A. A. & Berg, P. (1995) Proc. Natl. Acad. Sci. USA 92 6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolodner R., Fishel, R. A. & Howard, M. (1985) J. Bacteriol. 163 1060-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahdi A. A. & Lloyd, R. G. (1989) Nucleic Acids Res. 17 6781-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lloyd R. G. & Buckman, C. (1991) Biochimie 73 313-320. [DOI] [PubMed] [Google Scholar]
- 24.Hegde S. P., Qin, M. H., Li, X. H., Atkinson, M. A., Clark, A. J., Rajagopalan, M. & Madiraju, M. V. (1996) Proc. Natl. Acad. Sci. USA 93 14468-14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webb B. L., Cox, M. M. & Inman, R. B. (1995) J. Biol. Chem. 270 31397-31404. [DOI] [PubMed] [Google Scholar]
- 26.Webb B. L., Cox, M. M. & Inman, R. B. (1997) Cell 91 347-356. [DOI] [PubMed] [Google Scholar]
- 27.Shan Q., Bork, J. M., Webb, B. L., Inman, R. B. & Cox, M. M. (1997) J. Mol. Biol. 265 519-540. [DOI] [PubMed] [Google Scholar]
- 28.Umezu K. & Kolodner, R. D. (1994) J. Biol. Chem. 269 30005-30013. [PubMed] [Google Scholar]
- 29.Boyle J. M. & Symonds, N. (1969) Mutat. Res. 8 431-439. [DOI] [PubMed] [Google Scholar]
- 30.Boyle J. M. (1969) Mutat. Res. 8 441-449. [DOI] [PubMed] [Google Scholar]
- 31.Formosa T., Burke, R. L. & Alberts, B. M. (1983) Proc. Natl. Acad. Sci. USA 80 2442-2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Formosa T. & Alberts, B. M. (1984) Cold Spring Harbor Symp. Quant. Biol. 49 363-370. [DOI] [PubMed] [Google Scholar]
- 33.Kanaar R. & Hoeijmakers, J. H. (1998) Nature 335 337-338. [DOI] [PubMed] [Google Scholar]
- 34.Bryant F. R. & Lehman, I. R. (1985) Proc. Natl. Acad. Sci. USA 82 297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanzetta P. A., Alvarez, L. J., Reinach, P. S. & Candia, O. A. (1979) Anal. Biochem. 100 95-97. [DOI] [PubMed] [Google Scholar]
- 36.LeBowitz J., (1985) Ph.D. thesis (Johns Hopkins University, Baltimore).
- 37.Alani E., Thresher, R., Griffith, J. D. & Kolodner, R. D. (1992) J. Mol. Biol. 227 54-71. [DOI] [PubMed] [Google Scholar]
- 38.Kowalczykowski S. C., Lonberg, N., Newport, J. W. & von Hippel, P. H. (1981) J. Mol. Biol. 145 75-104. [DOI] [PubMed] [Google Scholar]
- 39.Bittner M., Burke, R. L. & Alberts, B. M. (1979) J. Biol. Chem. 254 9565-9572. [PubMed] [Google Scholar]
- 40.Alberts B. M. & Frey, L. (1970) Nature 227 1313-1318. [DOI] [PubMed] [Google Scholar]
- 41.Szostak J. W., Orr-Weaver, T. L., Rothstein, R. J. & Stahl, F. W. (1983) Cell 33 25-35. [DOI] [PubMed] [Google Scholar]
- 42.Sun H., Treco, D. & Szostak, J. W. (1991) Cell 64 1155-1161. [DOI] [PubMed] [Google Scholar]
- 43.Chow S. A., Rao, B. J. & Radding, C. M. (1988) J. Biol. Chem. 263 200-209. [PubMed] [Google Scholar]
- 44.Lavery P. E. & Kowalczykowski, S. C. (1992) J. Biol. Chem. 267 9315-9320. [PubMed] [Google Scholar]
- 45.Hunter N. & Kleckner, N. (2001) Cell 106 59-70. [DOI] [PubMed] [Google Scholar]
- 46.Allers T. & Lichten, M. (2001) Mol. Cell 8 225-231. [DOI] [PubMed] [Google Scholar]
- 47.Allers T. & Lichten, M. (2001) Cell 106 47-57. [DOI] [PubMed] [Google Scholar]