Abstract
RING domains act in a variety of unrelated biochemical reactions, with many of these domains forming key parts of supramolecular assemblies in cells. Here, we observe that purified RINGs from a variety of functionally unrelated proteins, including promyelocytic leukemia protein, KAP-1/TIF1β, Z, Mel18, breast cancer susceptibility gene product 1 (BRCA1), and BRCA1-associated RING domain (BARD1), self-assemble into supramolecular structures in vitro that resemble those they form in cells. RING bodies form polyvalent binding surfaces and scaffold multiple partner proteins. Separation of RING bodies from monomers reveals that self-assembly controls and amplifies their specific activities in two unrelated biochemistries: reduction of 5′ mRNA cap affinity of eIF4E by promyelocytic leukemia protein and Z, and E3 ubiquitin conjugation activity of BARD1:BRCA1. Functional significance of self-assembly is underscored by partial restoration of assembly and E3 activity of cancer predisposing BRCA1 mutant by forced oligomerization. RING self-assembly creates bodies that act structurally as polyvalent scaffolds, thermodynamically by amplifying activities of partner proteins, and catalytically by spatiotemporal coupling of enzymatic reactions. These studies reveal a general paradigm of how supramolecular structures may function in cells.
Keywords: protein association, supramolecular scaffold, catalytic surface
In addition to well-defined cellular organelles, eukaryotic cells also contain a number of supramolecular structures variably termed dots, speckles, and bodies. It is unknown whether these structures are supramolecular assemblies involved in organization of discrete sets of biochemical reactions or are merely nonfunctional aggregates. The significance of this question is readily apparent in the case of nuclear structures formed by promyelocytic leukemia protein (PML) and breast cancer susceptibility gene product 1 (BRCA1), where mutations in these proteins disrupt their respective nuclear structures and likely play a role in human disease, e.g., in acute promyelocytic leukemia (fusion of PML with RARα) and in breast and ovarian cancers (BRCA1 RING mutations; refs. 1 and 2). In cells, the structural integrity of the RING domains of PML and BRCA1 is required both for the integrity of their nuclear structures and for the cellular functions of the respective proteins (1, 2).
Over 70 proteins observed in intracellular supramolecular assemblies contain RING domains (http://atlas.physbio.mssm.edu/∼kbgroup/rings). RINGs are small zinc-binding domains that coordinate two zinc ions using a cross-brace topology (3) and constitute the third most abundant class in common in eukaryotic genomes annotated to date (IRP001814, Saccharomyces cerevisiae, Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens in the InterPro classification of SWISS-PROT/TrEMBL database; www.ebi.ac.uk/proteome). Determining a general function for these domains is complicated by the extraordinary functional diversity of the proteins that contain them (3, 4). Only three biochemical activities have been ascribed to RINGs: transcriptional repression by KAP-1/TIF1β (5), translational repression by PML and Z (6), and ubiquitin (Ubq) conjugation by a variety of RINGs, including BRCA1 and BRCA1-associated RING domain (BARD1) (7). These activities have no apparent common features. The frequent occurrence of RING-containing proteins in supramolecular assemblies in cells led us to investigate whether RING domains play a general role in supramolecular assembly. Furthermore, the correlation between the structural integrity of supramolecular bodies and their cellular functions led us to examine whether these assemblies are biochemically functional entities.
Methods
Proteins were bacterially expressed as GST and (His)6 fusions and purified to homogeneity (6, 8). The human PML construct contains the RING, B-boxes, and a portion of the coiled-coil (residues 49–257; ref. 9). Human KAP-1 contains the RING, B-boxes, and coiled-coil (residues 22–418; ref. 5). The lymphocytic choriomeningitis virus Z (Armstrong strain) construct contains full-length protein (residues 1–90; ref. 6). The human BRCA1 construct contains the RING and flanking helical bundle (residues 1–101) or a longer fragment comprising residues 1–639, and the human BARD1 construct contains residues 26–119 or full-length protein (10). The UbcH5C construct contains residues 1–147 (11). Mouse eIF4E is full length and differs from human eIF4E by four amino acids (6). m7GpppG was obtained from New England Biolabs. Rabbit E1 is 96% identical to human E1 and was purchased from Affiniti (Mamhead, U.K.). Bovine Ubq (Sigma) was purified by reverse-phase HPLC. Mouse anti-Ubq antibody (Zymed) was used as characterized (12). Nanogold, Nano-W, and NanoVan were purchased from Nanoprobes (Yaphank, NY). Colloidal gold (gold) was obtained from Electron Microscopy Sciences (Fort Washington, PA). All other reagents were of American Chemical Society grade and were purchased from Sigma. Circular dichroism and fluorescence spectroscopies were performed as described (6). Size exclusion chromatography and electron microscopy were performed as described (8). Details of experimental methods and analyses are published as supporting information on the PNAS web site, www.pnas.org.
Results and Discussion
Initially, we set out to examine whether purified fragments containing RING domains (RINGF) of a variety of unrelated proteins form supramolecular assemblies in vitro similar to those formed by their full-length counterparts in cells. In cells, eIF4E-interacting growth regulatory protein PML and arenaviral protein Z, transcriptional repressor KAP-1/TIF1β, polycomb group protein Mel18, and tumor suppressors BRCA1 and BARD1 form PML nuclear bodies (NBs; ref. 13), centromeric foci (14), polycomb repressive complex (15), and BRCA1 nuclear dots (NDs; 16), respectively. Despite their differences, all of these structures are roughly spherical and 0.1–1 μm in diameter, as observed using confocal microscopy. Thus, we purified RINGFs and full-length proteins and examined their ability to form bodies in vitro using negative staining electron microscopy (EM; Fig. 1A). In all four cases, purified RINGFs, both independently in the case of Mel18 and in the context of larger RBCC/TRIM motifs of PML and KAP-1/TIF1β, self-assemble into ≈50-nm spherical structures in vitro (Fig. 1A) that approximate structures formed by their respective proteins in cells (13–16). These structures are similar in size and morphology to those formed by the full-length arenaviral RING protein Z (8). In cells, structural integrity of the RING domain in the context of the full-length proteins is required for the formation of supramolecular structures and their functional activity (3). Thus, we investigated whether structural integrity of the RING was required for its assembly in vitro. Zinc-coordinating Cys residues in the first and second zinc-binding sites were mutated to Ala, and abilities of mutants to self-assemble in vitro and for full-length counterparts to form bodies and function biologically were assessed. Site I RINGF mutants are only partially structured as ascertained by circular dichroism spectroscopy, whereas site II mutants possess mean secondary structure content indistinguishable from that of wild-type proteins (Fig. 5, which is published as supporting information on the PNAS web site). Site I mutants of PML, Z, and KAP-1/TIF1β fail to self-assemble in vitro as observed by EM (Fig. 1); do not form PML or Z NBs and KAP-1/KRAB complexes in cells as full-length proteins; and fail to function in vivo in growth suppression, translational repression, and transcriptional repression, respectively (5, 13). In contrast, site II mutants disrupt neither the structure of RINGF nor its ability to self-assemble in vitro (Fig. 1A) or form supramolecular structures and function in vivo as full-length proteins (5, 8, 17). These data provide a strong correlation between the structural integrity of the RING, supramolecular RING assembly, and cellular function.
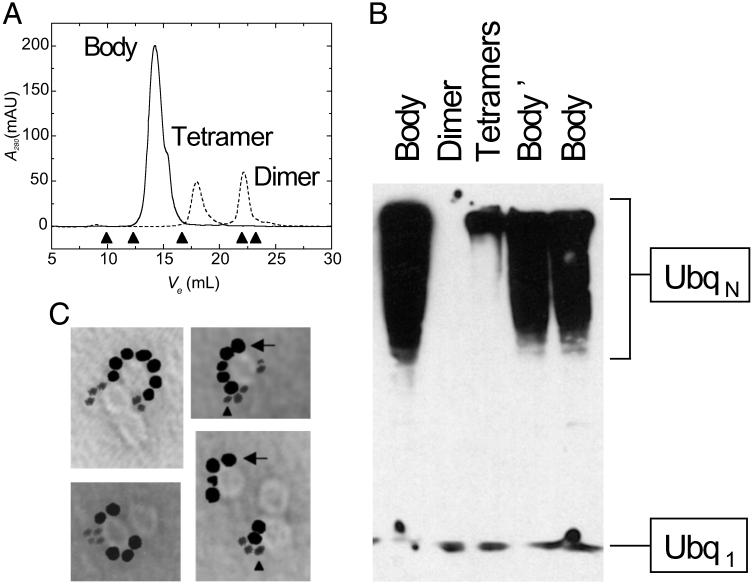
Fig 1.
A variety of unrelated RING domains self-assemble into spherical and ring-shaped bodies, and scaffold multiple RING partner proteins on their surface. (A) Single-particle EM micrographs of RING bodies at a nominal magnification of ×80,000, using uranyl acetate counterstain. (Bottom Right) Micrograph is of RING of Mel18, whereas the rest are of PML, Z, and KAP-1/TIF1β and their site I and site II mutants. (B) Single-particle EM micrographs of BARD1 homomeric bodies, BRCA1 homomeric bodies, BARD1:BRCA1 heteromeric bodies, and BARD1:BRCA1 C64G at a nominal magnification of ×80,000, using methylamine tungstenate counterstain. (C and D) EM micrographs of Z and gold-eIF4E (C) and PML and gold-eIF4E (D), counterstained with uranyl acetate. Multiple gold-eIF4E molecules are scaffolded on the surface of both Z bodies and PML bodies, in agreement with gel filtration measurements (Fig. 7A). Note that heterogeneity in the number of molecules scaffolded by RING bodies is likely due to low (femtomolar) protein concentrations required for single-particle EM measurements that are several orders of magnitude below Kd for the respective associations, as well as stochastic heterogeneity on the microscopic level. The median diameter of gold particles is 6 nm.
This correlation is particularly striking in the case of BRCA1 RINGF, which self-assembles into 13-nm ring-shaped structures in vitro (Fig. 1B), forms NDs, and functions as a tumor suppressor in the context of the full-length protein in cells, being one of the principal susceptibility genes for breast and ovarian cancers (16). Longer constructs of BRCA1 exhibit similar behavior (Fig. 6, which is published as supporting information on the PNAS web site). The cancer-predisposing C64G mutation of BRCA1 RING abolishes self-assembly in vitro of both RINGF as well as longer constructs (Fig. 1B), and formation of NDs, and tumor suppression in vivo by full-length proteins (16). In all, purified RINGFs from proteins with diverse biochemical and cellular functions self-assemble in vitro into high-order oligomers that resemble supramolecular structures formed by the corresponding full-length RING-containing proteins in cells.
To characterize the functional significance of high-order RING assembly, we examined whether RING bodies function as architectural scaffolds by arranging partner proteins on their surface. In vitro, both PML RING and full-length Z repress translation of a selected subset of mRNAs by directly binding translation initiation factor eIF4E (6). To monitor the ability of PML and Z bodies to scaffold eIF4E, we labeled eIF4E with gold (gold-eIF4E, 6 nm median diameter) and purified PML: and Z:gold-eIF4E complexes by gel filtration (Fig. 7A, which is published as supporting information on the PNAS web site). We verified that eIF4E was singly conjugated with gold by using gel filtration, UV/Vis spectroscopy, and EM (Fig. 7B). Multiple gold-eIF4E molecules are bound by RING bodies formed by both PML and Z, as observed by EM (Fig. 1 C and D), with the degree of scaffolding consistent with the subunit stoichiometry of RING bodies. In cells, such architectural assemblies may be used to sequester into and titrate partner proteins out of nonfunctional storage sites, as has been proposed for PML NBs and other nuclear organelles such as splicing speckles and Cajal bodies (18). Alternatively, RING bodies may be directly functional by virtue of forming polyvalent binding surfaces, in addition to the indirect and purely architectural functions described above.
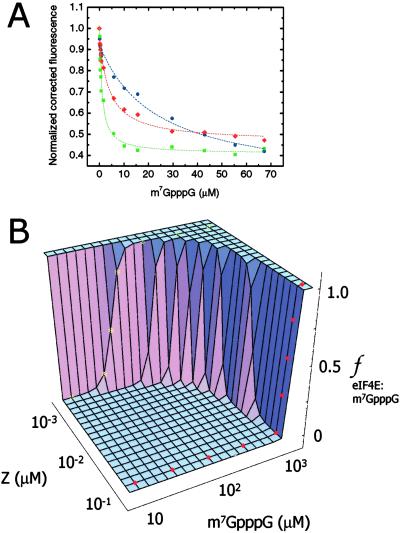
Thus, we examined the possibility that polyvalent binding by RING supramolecular assemblies amplifies the specific activity of RING partner proteins. Both PML RINGF and Z reduce the affinity of eIF4E for its ligand, 7-methyl guanosine 5′ cap of mRNA [m7GpppG (6)]. This antagonism of eIF4E activity is in part responsible for translational repression and host cell shutoff by arenaviral Z (6) and for inhibition of nucleocytoplasmic mRNA transport and growth suppression by PML (19). We fractionated Z bodies from monomers by gel filtration (Fig. 7A) and examined the affinity of Z bodies and monomers for eIF4E using difference circular dichroism spectroscopy (Fig. 8D, which is published as supporting information on the PNAS web site). The affinity of Z monomer- and Z body-bound eIF4E for m7GpppG was evaluated by using fluorescence spectroscopy (Fig. 2A). Binding of eIF4E to monomeric Z reduces its affinity for m7GpppG by 20-fold, and this effect is greatly potentiated when Z is assembled into bodies, reducing cap affinity by nearly 250-fold (Table 1, which is published as supporting information on the PNAS web site). To understand this phenomenon in depth, we constructed a thermodynamic linkage model, which relates RING assembly, RING partner binding, and RING partner activity. At low concentrations where Z is largely monomeric, eIF4E binds m7GpppG with relatively high affinity and relatively low cooperativity, signified by left-shifted sigmoidal binding curves (yellow * in Fig. 2B). However, at high concentrations where Z is largely assembled into bodies, eIF4E activity is both greatly inhibited and rendered extremely cooperative, as indicated by right-shifted step function curves that bind m7GpppG in all-or-none fashion (red • in Fig. 2B). Indeed, Z self-assembly also amplifies the effect of mutating the m7GpppG-binding site in eIF4E (Fig. 8, Table 1). Mutation of W56A in eIF4E reduces its affinity for m7GpppG by 50-fold, as compared with wild-type eIF4E (Fig. 8, Table 1). Cap affinity is reduced 150-fold by binding of Z monomers, whereas binding of Z bodies reduces cap affinity by >1,500-fold (Fig. 8, Table 1). In this manner, RING assembly into bodies amplifies the effect of individual RING monomers, increasing their specific activity and, in the case of Z, greatly potentiating and regulating its ability to antagonize eIF4E. In addition to spatially and energetically organizing RING partner proteins on the polyvalent RING bodies, scaffolds formed by RING domains may also temporally couple time-dependent enzymatic reactions on their surfaces.
Fig 2.
Z bodies amplify specific activity of individual Z monomers as a result of thermodynamic linkage between self-assembly and RING partner binding and convert Z from a low-affinity low-cooperativity eIF4E inhibitor into a high-affinity switch-like antagonist. (A) Normalized corrected Trp fluorescence emission quenching curves of eIF4E and their fits, in free form (green ▪) and when bound to kinetically captured Z monomers (red ♦) and Z bodies (blue •) in the presence of increasing concentrations of m7GpppG. All proteins were used at 2 μM. (B) Thermodynamic linkage model describing the relationship among RING body assembly, RING partner binding, and RING partner activity for Z, eIF4E, and eIF4E ligand m7GpppG. Fraction of eIF4E (ƒ) bound to m7GpppG (y axis) and m7GpppG binding curves (x axis) are plotted as a function of increasing concentrations of Z (z axis) that lead to self-assembly of Z monomers into bodies, which scaffold multiple molecules of eIF4E.
To elucidate whether RING supramolecular assemblies have catalytic effects, we examined the ability of RING bodies and monomers to support Ubq conjugation and polymerization. A number of RING proteins act as E3 Ubq ligating enzymes, whereby they cooperate directly with E2 conjugating and indirectly with E1 activating enzymes to carry out conjugation and polymerization of activated Ubq thioesters into poly-Ubq isopeptide chains (20). To test the above hypothesis, we used the RINGFs of BRCA1 and BARD1, because together they constitute an E3 enzyme (7), autoubiquitinate (11), and undergo self-assembly in vitro (Fig. 1B). Although many RINGs act as E3 enzymes (20), their precise enzymatic role in the ubiquitination reactions is poorly understood, despite availability of atomic level structural data (21). It has been proposed that RINGs function as E3s by allosterically activating E2s (22). This appears unlikely, at least with respect to large-scale allosteric effects, because the structure of the UbcH7 E2 is virtually identical in free and Cbl RING-bound states (21, 23). Alternatively, it is possible that RING E3s polymerize Ubq by coupling multiple Ubq thioester-bound E2 enzymes to each other, as a result of polyvalent binding by RING bodies and increased specific activities of RING monomers and E2 enzymes.
Thus, we characterized self-assembly of BRCA1 and BARD1 RINGFs using gel filtration to fractionate bodies from low-order oligomers and monomers. Consistent with previous studies of Z bodies (8), BRCA1 and BARD1 RINGFs rapidly form dimers and tetramers and assemble into 12-meric 150 kDa bodies on the timescale of hours (Fig. 3A), which appear as ≈13 nm ring-shaped structures by EM (Fig. 1B). Similarly, full-length BRCA1 and BARD1 also form bodies (Fig. 6). Because heterodimerization of BRCA1 and BARD1 RINGFs is absolutely required for efficient E3 activity (7), we verified heteromerization by analytical gel filtration, electrospray ionization mass spectrometry of BRCA1:BARD1 heterodimers and bodies (Figs. 9 and 10, which are published as supporting information on the PNAS web site) and atomic absorption spectrometry of heterodimers and bodies using differential labeling of BRCA1 and BARD1 with Zn2+ and Co2+, respectively (Table 2, which is published as supporting information on the PNAS web site), confirming that BRCA1 and BARD1 RINGFs are present in both dimers and bodies in ≈1:1 ratio. We fractionated BRCA1:BARD1 heterodimers from bodies by gel filtration and examined their E3 activities relative to each other in an ubiquitination assay in vitro, resolving the reaction products by SDS/PAGE and blotting them using an antibody that recognizes both monomeric and polymerized forms of Ubq (Fig. 3B). BRCA1:BARD1 bodies exhibit much higher activity than tetramers or BRCA1:BARD1 heterodimers (Fig. 3B). BRCA1:BARD1 bodies constitute genuine and specific E3 enzymes as determined from their dependence on ATP, E1, and E2 enzymes (Fig. 10, which is published as supporting information on the PNAS web site). Thus, supramolecular assemblies formed by BRCA1:BARD1 RINGFs are more efficient than their unassembled heterodimers, presumably because they couple multiple E2 Ubq thioesters as a result of polyvalent binding of E2s by RING bodies.
Fig 3.
BARD1:BRCA1 support Ubq polymerization by self-assembling into bodies that spatiotemporally couple multiple molecules of UbcH5C and form a catalytic polyvalent binding surface. (A) Size exclusion chromatography of BARD1:BRCA1 as a function of elution volume (Ve) immediately on initiation of assembly to kinetically capture dimers and tetramers (dashed), and, after incubation for 24 h, obtain bodies (solid). Bodies elute with globular molecular mass of 150 kDa, corresponding to 12 mers, as determined by using molecular weight standards represented by arrowheads (left to right: thyroglobulin, 667 kDa; catalase, 232 kDa; albumin, 67 kDa; chymotrypsinogen A, 25 kDa; RNase A, 14 kDa). (B) Relative activation of UbcH5C and poly-Ubq formation by BARD1:BRCA1 dimers, tetramers, and bodies as purified by gel filtration. Body′ represents freshly purified BARD1:BRCA1 12 mers, whereas Body represents 12 mers stored at −80°C, demonstrating that storage conditions do not affect activity. (C) Single-particle EM micrographs of products of ubiquitination assay of gel filtration purified BARD1:BRCA1:gold-UbcH5C complex using nanogold-Ubq, counterstained with methylamine tungstenate and methylamine vanadate, allowing visualization of both colloidal and nanogold particles. Multiple molecules of gold-UbcH5C (arrow, median diameter of 6 nm) are scaffolded by BARD1:BRCA1 ring-shaped bodies ≈13 nm in diameter, with multiple polynanogold-Ubq chains in the vicinity (arrowhead, 1.4-nm diameter). Heterogeneity in the number of molecules scaffolded by RING bodies is likely due to low (femtomolar) protein concentrations required for single-particle EM measurements that are several orders of magnitude below Kd for the respective associations, as well as stochastic heterogeneity on the microscopic level. Note that the difference in contrast between gold and nanogold is due to the difference in electron scattering power of the gold and nanogold particles.
To directly examine whether BRCA1:BARD1 bodies scaffold multiple E2 molecules on their surface, we labeled their cognate E2, UbcH5C, with gold (gold-UbcH5C, 6 nm median diameter) and Ubq with nanocrystalline gold (nanogold–Ubq, 1.4 nm diameter) and used them in the ubiquitination assay, visualizing the products by EM. We verified that both UbcH5C and Ubq were singly conjugated by analytical gel filtration, UV/Vis spectroscopy, and EM (Fig. 6 C and D). Both gold-UbcH5C and nanogold-Ubq are active components of the ubiquitination reaction in an E3-dependent manner (Fig. 12, which is published as supporting information on the PNAS web site). We observe scaffolding of multiple gold-UbcH5C molecules on the RING body surface, with several chains of polymerized nanogold-Ubq nearby (Fig. 3C). Thus, BRCA1:BARD1 RINGFs cooperate to form an E3 Ubq ligase as a result of polyvalent binding of E2 on the surface of their catalytic bodies. Because the cancer predisposing C64G mutation in BRCA1 abolishes self-assembly of RINGF and full-length proteins in vitro (Fig. 1B) and in vivo (16), and impairs E3 activity (7), we investigated whether E3 activity of BRCA1 C64G can be restored by forced oligomerization.
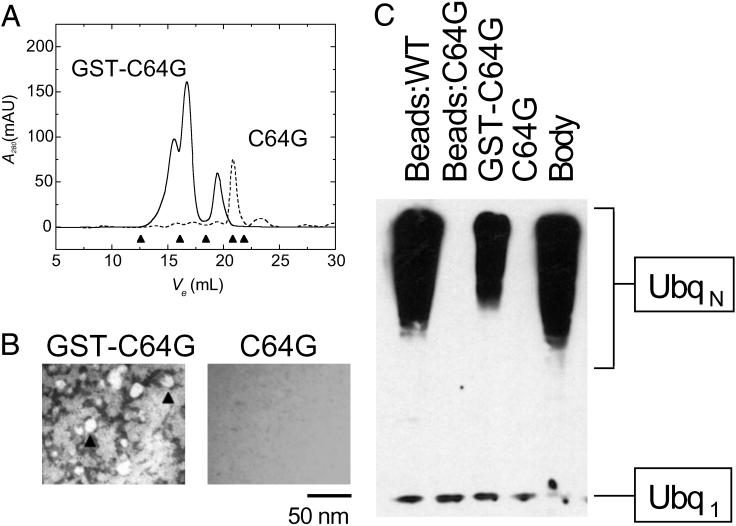
To this end, we fused BRCA1 C64G RINGF to GST (GST-BRCA1 C64G), because GST forms high-affinity dimers in solution (24) and may therefore provide an oligomerization interface to replace the one putatively disrupted by cancer predisposing C64G mutation. BRCA1 C64G RINGF retains wild-type secondary structure content (Fig. 5B) and its ability to heterodimerize with BARD1 (25). Examination of BARD1:BRCA1 C64G by gel filtration fails to detect any high-order oligomers, with most of the protein eluting as heterodimers (Fig. 4A), which are not visible by EM (Fig. 4B), in agreement with previous studies of this mutant (25). On the other hand, BARD1:GST-BRCA1 C64G elutes as a mixture of heterodimers and high-order oligomers of heterogeneous globular molecular weights of 170–500 kDa (Fig. 4A), which appear as a mixture of amorphous aggregates and spherical bodies roughly 15 nm in diameter by EM (Fig. 4B), similar to those formed by wild-type BARD1:BRCA1 (Fig. 1B). Remarkably, BARD1:GST-BRCA1 C64G exhibits partial E3 activity as compared with wild-type (Fig. 4C). It is noteworthy that BARD1:GST-BRCA1 C64G immobilized on glutathione-sepharose beads exhibits no detectable activity relative to wild-type (Fig. 4C), in agreement with previous studies (7). Lack of activity in the immobilized state likely reflects steric effects of Sepharose bead binding on RING assembly and suggests that spatial organization, in addition to spatial restriction, contributes to the catalytic enhancement due to RING self-assembly. The enhanced E3 activity of supramolecular assemblies formed by BRCA1:BARD1 RINGs (Fig. 3B), near wild-type secondary structure content of BRCA1 C64G (Fig. 5B), its virtually identical affinity for BARD1 (25), and the ability to partially restore its E3 activity through forced oligomerization (Fig. 4C), collectively support the notion that the loss of E3 activity by BRCA1 C64G mutation is due to its disruption of self-assembly of BRCA1 into an active polyvalent catalytic surface in cooperation with BARD1.
Fig 4.
Self-assembly of carcinogenic BRCA1 C64G mutant can be restored by forced oligomerization, leading to restoration of its ubiquitination activity. (A) Size exclusion chromatography profiles as a function of elution volume (Ve) of BARD1:BRCA1 C64G (dashed line) and BARD1:GST-BRCA1 C64G (solid line). Fusion of BRCA1 C64G to GST leads to formation of 170- to 500-kDa molecular mass species. (B) EM micrographs of BARD1:GST-BRCA1 C64G (GST-C64G) and BARD1:BRCA1 C64G (C64G), both at a nominal magnification of ×15,000, counterstained with uranyl acetate. Fusion of BRCA1 C64G to GST leads to formation of amorphous aggregates and round bodies ≈15 nm in diameter (arrowheads) that resemble those formed by wild-type BARD1:BRCA1 (Fig. 1B), whereas BARD1:BRCA1 C64G does not assemble. (C) Activation of UbcH5C and Ubq polymerization by BARD1/BRCA1 C64G (C64G), BARD1:GST-BRCA1 C64G (GST-C64G), as visualized by SDS/PAGE/Western blotting. Fusion of BRCA1 C64G to GST restores partial ubiquitination activity, as compared with wild-type soluble BARD1:BRCA1 (Body), and GST-BARD1:GST-BRCA1 immobilized on glutathione-Sepharose (Beads:WT). Interestingly, Sepharose-immobilized BARD1:GST-BRCA1 C64G (Beads:C64G) does not support Ubq polymerization, in agreement with other reports (7).
In all, this work demonstrates that a variety of functionally unrelated RING domains form high-order assemblies that polyvalently scaffold partner proteins on their surface and amplify and control both their thermodynamic and catalytic specific activities. To our knowledge, this is the first example of a small domain increasing its specific activity and the activities of its partners through high-order supramolecular self-assembly. Furthermore, to our knowledge this is the first exposition of the functional linkage between assembly and partner activity and control of biological activity by supramolecular assembly, a phenomenon originally predicted by Wyman and colleagues over 30 years ago (26). Formation of supramolecular structures by RINGs enhances specific activities of their partner proteins in two unrelated biochemical processes: (i) reduction of 5′ mRNA cap affinity of eIF4E by PML and Z, and (ii) E3 Ubq conjugation activity of BRCA1:BARD1. Although the contribution of BARD1:BRCA1 E3 activity to tumor suppression and maintenance of genome integrity is currently poorly understood, it is reassuring that BRCA1 functions in cells require its assembly into supramolecular foci at sites of DNA damage, and that two recently identified substrates of BARD1:BRCA1 E3 ligase in vivo, H2AX and FANCD2 in addition to itself, are both involved in chromatin remodeling at sites of DNA damage during BRCA1-dependent DNA repair (27). Notwithstanding, functional importance of self-assembly is highlighted by the observation that forced oligomerization of the cancer predisposing BRCA1 C64G mutant partially restores its E3 activity, suggesting a potential means of restoring the activity of carcinogenic BRCA1 mutants in vivo. Supramolecular structures formed by different RING domains have distinct features, having different orders of assembly (24 in the case of Z, and 12 in the case of BRCA1:BARD1, for example), binding specific partners (eIF4E in the case of PML and Z and UbcH5C in the case of BRCA1:BARD1), and differing in responses to sites I and II RING mutations. Only future studies will reveal whether other RINGs self-assemble in a manner similar to those examined in this study, whether self-assembly is a general property of RING domains, and what fraction of the rich physical repertoire that RINGs exhibit in vitro occurs in vivo.
High-order RING assembly into functional structures has widespread implications for how supramolecular structures may function in cells. It is hotly debated whether supramolecular assemblies such as PML NBs and BRCA1 NDs are directly active nuclear organelles or merely storage sites. The latter notion arises, in part, from the traditional view that proteins are functionally active in monomeric or low-order states. Our current studies suggest that PML NBs and BRCA1 NDs form polyvalent surfaces that act directly in diverse biochemical reactions, critical for their cellular functions. Indeed, formation of supramolecular structures through high-order self-assembly may serve as a molecular mechanism of compartmentalization of biochemical reactions for subcellular organelles that are not membrane bound, such as those found in the nucleus. Moreover, insofar as PML NBs are disrupted in the majority of cases of acute promyelocytic leukemia and cancer predisposing mutations in BRCA1 result in disruption of BRCA1 NDs (1, 2), RING self-assembly may be essential for normal cellular function, where it may serve to organize, control, and integrate networks of biochemical reactions that collectively underlie biological phenomena. Taken together, these results provide a fundamentally new conceptual framework for understanding how supramolecular assemblies may function in cells, how these functions may be regulated, and how they can be artificially restored when disrupted by disease.
Supplementary Material
Acknowledgments
We are indebted to Roman Osman, Neville Kallenbach, Maria Salvato, Allan Capili, and Alexander Varshavsky for invaluable discussions; Zhen-Qiang Pan and Angus Chen (Mt. Sinai School of Medicine) for advice about ubiquitination and the gift of UbcH5C; Frank Rauscher (Wistar Institute) for KAP-1; Jonathan Licht (Mt. Sinai School of Medicine) for BRCA1 and its mutants; Rachel Klevit (University of Washington) for BARD1; Sandy Ross, Bill Laws (University of Montana), and Dixie Goss (Hunter College) for assistance with fluorescence experiments; Cliff Soll (Hunter College) for help with mass spectrometry; and Joseph Samet (Mt. Sinai School of Medicine) for EM technical support. We are grateful to Alexander Varshavsky, Ravi Iyengar, Harel Weinstein, Aneel Aggarwal, Rachel Klevit, and Ze'ev Ronai for critical reading of the manuscript. A.K. is supported by the National Institutes of Health Medical Scientist Training Program. K.L.B.B. is a scholar of the Leukemia and Lymphoma Society. This work was funded by National Institutes of Health Grants CA 80728 and CA 88991.
Abbreviations
Ubq, ubiquitin
NB, nuclear body
ND, nuclear dot
gold, colloidal gold
nanogold, nanocrystalline gold
EM, electron microscopy
PML, promyelocytic leukemia protein
BRCA1, breast cancer susceptibility gene product 1
BARD1, BRCA1-associated RING domain
References
- 1.Melnick A. & Licht, J. D. (1999) Blood 93 3167-3215. [PubMed] [Google Scholar]
- 2.Chen Y., Chen, C. F., Riley, D. J., Allred, D. C., Chen, P. L., Von Hoff, D., Osborne, C. K. & Lee, W. H. (1995) Science 270 789-791. [DOI] [PubMed] [Google Scholar]
- 3.Kentsis A. & Borden, K. L. B. (2000) Curr. Protein Pept. Sci. 1 49-73. [DOI] [PubMed] [Google Scholar]
- 4.Saurin A. J., Borden, K. L., Boddy, M. N. & Freemont, P. S. (1996) Trends Biochem. Sci. 21 208-214. [PubMed] [Google Scholar]
- 5.Peng H., Begg, G. E., Schultz, D. C., Friedman, J. R., Jensen, D. E., Speicher, D. W. & Rauscher, F. J., III (2000) J. Mol. Biol. 295 1139-1162. [DOI] [PubMed] [Google Scholar]
- 6.Kentsis A., Dwyer, E. C., Perez, J. M., Sharma, M., Chen, A., Pan, Z. Q. & Borden, K. L. (2001) J. Mol. Biol. 312 609-623. [DOI] [PubMed] [Google Scholar]
- 7.Hashizume R., Fukuda, M., Maeda, I., Nishikawa, H., Oyake, D., Yabuki, Y., Ogata, H. & Ohta, T. (2001) J. Biol. Chem. 276 14537-14540. [DOI] [PubMed] [Google Scholar]
- 8.Kentsis A., Gordon, R. E. & Borden, K. L. (2002) Proc. Natl. Acad. Sci. USA 99 667-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borden K. L., Campbelldwyer, E. J., Carlile, G. W., Djavani, M. & Salvato, M. S. (1998) J. Virol. 72 3819-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brzovic P. S., Rajagopal, P., Hoyt, D. W., King, M. C. & Klevit, R. E. (2001) Nat. Struct. Biol. 8 833-837. [DOI] [PubMed] [Google Scholar]
- 11.Chen A., Kleiman, F. E., Manley, J. L., Ouchi, T. & Pan, Z. Q. (2002) J. Biol. Chem. 277 22085-22092. [DOI] [PubMed] [Google Scholar]
- 12.Sternsdorf T., Puccetti, E., Jensen, K., Hoelzer, D., Will, H., Ottmann, O. G. & Ruthardt, M. (1999) Mol. Cell. Biol. 19 5170-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borden K. L., Boddy, M. N., Lally, J., O'Reilly, N. J., Martin, S., Howe, K., Solomon, E. & Freemont, P. S. (1995) EMBO J. 14 1532-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuda E., Agata, Y., Sugai, M., Katakai, T., Gonda, H. & Shimizu, A. (2001) J. Biol. Chem. 276 14222-14229. [DOI] [PubMed] [Google Scholar]
- 15.Kanno M., Hasegawa, M., Ishida, A., Isono, K. & Taniguchi, M. (1995) EMBO J. 14 5672-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Y., Xu, X. L., Yang, M. C., Wei, F., Ayi, T. C., Bowcock, A. M. & Baer, R. (1997) Proc. Natl. Acad. Sci. USA 94 12075-12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell Dwyer E. J., Lai, H., MacDonald, R. C., Salvato, M. S. & Borden, K. L. (2000) J. Virol. 74 3293-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maul G. G., Negorev, D., Bell, P. & Ishov, A. M. (2000) J. Struct. Biol. 129 278-287. [DOI] [PubMed] [Google Scholar]
- 19.Cohen N., Sharma, M., Kentsis, A., Perez, J. M., Strudwick, S. & Borden, K. L. (2001) EMBO J. 20 4547-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weissman A. M. (2001) Nat. Rev. Mol. Cell. Biol. 2 169-178. [DOI] [PubMed] [Google Scholar]
- 21.Zheng N., Wang, P., Jeffrey, P. D. & Pavletich, N. P. (2000) Cell 102 533-539. [DOI] [PubMed] [Google Scholar]
- 22.Pickart C. M. (2001) Annu. Rev. Biochem. 70 503-533. [DOI] [PubMed] [Google Scholar]
- 23.Huang L., Kinnucan, E., Wang, G., Beaudenon, S., Howley, P. M., Huibregtse, J. M. & Pavletich, N. P. (1999) Science 286 1321-1326. [DOI] [PubMed] [Google Scholar]
- 24.Nishida M., Harada, S., Noguchi, S., Satow, Y., Inoue, H. & Takahashi, K. (1998) J. Mol. Biol. 281 135-147. [DOI] [PubMed] [Google Scholar]
- 25.Brzovic P. S., Meza, J. E., King, M. C. & Klevit, R. E. (2001) J. Biol. Chem. 276 41399-41406. [DOI] [PubMed] [Google Scholar]
- 26.Wyman J. & Gill, S. J., (1990) Binding and Linkage: Functional Chemistry of Biological Macromolecules (University Science Books, Mill Valley, CA).
- 27.Baer R. & Ludwig, T. (2002) Curr. Opin. Genet. Dev. 12 86-91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.