Abstract
The pineal gland, sharing morphological and biochemical similarities with the retina, plays a unique and central role in the photoneuroendocrine system. The unique development of the pineal gland is directed by a specific combination of the expressed genes, but little is known about the regulatory mechanism underlying the pineal-specific gene expression. We isolated a 1.1-kbp fragment upstream of the zebrafish exo-rhodopsin (exorh) gene, which is expressed specifically in the pineal gland. Transgenic analysis using an enhanced green fluorescent protein reporter gene demonstrated that the proximal 147-bp region of the exorh promoter is sufficient to direct pineal-specific expression. This region contains three copies of a putative cone rod homeobox (Crx)/Otx-binding site, which is known to be required for expression of both retina- and pineal-specific genes. Deletion and mutational analyses of the exorh promoter revealed that a previously uncharacterized sequence TGACCCCAATCT termed pineal expression-promoting element (PIPE) is required for pineal-specific promoter activity in addition to the Crx/Otx-binding sites. By using the zebrafish rhodopsin (rh) promoter that drives retina-specific expression, we created a reporter construct having ectopic PIPE in the rh promoter at a position equivalent to that in the exorh promoter by introducing five nucleotide changes. Such a slight modification in the rh promoter induced ectopic enhanced green fluorescent protein expression in the pineal gland without affecting its retinal expression. These results identify PIPE as a critical cis-element contributing to the pineal-specific gene expression, in combination with the Crx/Otx-binding site(s).
Living organisms use environmental light signals for multiple physiological functions such as vision, photoentrainment of circadian rhythms, regulation of body color, and detection of seasonal changes in photoperiod. These diverse functions are mediated not only by the retina but also by extra-ocular photoreceptive organs, such as the pineal gland. The retina and pineal gland probably arose via divergence from a common ancestral photoreceptive organ, and consistently, the pineal gland acts as a photosensory organ in the lowest vertebrate (1–3). In the course of vertebrate evolution, the physiological role of the pineal gland has been changed from a photosensory organ to a photoendocrinal organ in the lower vertebrates and eventually to a neuroendocrinal organ in mammals (4, 5). Generally, the retina receives visual images and transmits them to the brain, whereas the primary role of the pineal gland is the rhythmic production of circulating melatonin, which regulates numerous physiological activities (6). Despite such a dynamic change in the physiological function, the pineal gland displays many similarities to the retina in tissue/cellular morphology and biochemical properties (7, 8). In fish, amphibians, and lacertilian reptiles, their pineal photoreceptor cells possess well developed and lamellar outer segments, which are homologous to the outer segments of retinal photoreceptor cells. These photoreceptor cells form an ordered layer structure in both tissues. Biochemically, the retinal phototransduction proteins such as opsin, α-subunit of transducin, arrestin, and recoverin also have been localized in the pineal photoreceptor cells (9–13). These observations illustrate remarkable parallels in the pineal and retinal phototransduction pathways, but very little is known about the molecular basis that accounts for the characteristic development of the pineal gland. The regulatory mechanism responsible for tissue-specific gene expression and its evolutionary background are, therefore, important issues providing clues to the developmental control specifying the pineal or retinal identity.
In recent years, a number of studies have been reported about transcriptional regulation of the retinal genes (reviewed in ref. 14), among which the rhodopsin (rh) promoter has been studied extensively. Biochemical approaches and in vitro transcription assays have identified several cis-acting DNA elements, such as Ret-1 (15), BAT-1 (16), and Ret-4 (17). These studies suggest a cooperative interplay of multiple cis-acting elements and transcription factors for the retinal photoreceptor cell-specific gene expression. Consistent with this idea, several transcription factors have been identified and shown to regulate the rh gene expression. Subtraction cDNA cloning resulted in identification of neural retina leucine zipper (Nrl), a basic leucine zipper (bZIP) transcription factor that is expressed in all cell layers of adult mammalian retina (18, 19). Nrl binds to a well conserved cis-acting element Nrl response element (NRE) and transactivates the rh promoter (20, 21). On the other hand, cone rod homeobox (Crx) is an Otx-related homeodomain protein expressed exclusively in the retinal photoreceptor cells and pinealocytes (22–24). Crx transactivates the rh promoter by binding to BAT-1 and Ret-4 (22) and is implicated in the regulation of photoreceptor cell-specific gene expression (25). In addition to these transcription factors, Rx and Erx seem to participate in the regulation of the rh gene expression (26–28).
In contrast to these advances in the studies on the retina-specific gene expression, transcriptional regulation of the pineal gene remains poorly understood. The only cis-acting element identified so far is a pineal regulatory element (PIRE), which is recognized by Crx (29, 30). PIRE with a consensus sequence of TAATC/T is present in 5′-flanking regions of several pineal genes such as rat arylalkylamine-N-acetyltransferase and human hydroxyindole-O-methyltransferase genes (29). Recently, it has been reported that circadian gene expression in the zebrafish pineal complex requires Otx5, which is closely related to Crx (31). These studies suggest that a member of the Crx/Otx family and its binding site(s) play a common role in the transcription of both pineal and retinal genes. It should be stressed, however, that the transcriptional regulation operated by Crx/Otx is inadequate to explain the mechanism segregating the pineal- and retina-specific gene expression. As yet unidentified transcription factor(s) and cis-acting element(s) should strictly determine pineal-specific gene expression, most probably in combination with the Crx/Otx-dependent regulation.
We previously demonstrated that the zebrafish has two distinct rhodopsin genes that are highly similar in coding sequence to each other (74% identical) but show unique tissue distributions (32). One is the canonical rh gene expressed only in the retina and the other is exo-rhodopsin (exorh) gene expressed specifically in the pineal gland. A phylogenetic analysis indicated that rh and exorh genes were produced by gene duplication that occurred early in the ray-finned fish lineage (32). Such a close kinship between the two genes, together with their specificities in tissue distribution, prompted us to investigate the evolutionary scenario of tissue-specific promoters. Because the proximal promoter sequences of rh genes are highly conserved among vertebrates including the zebrafish (33), we expected that the zebrafish exorh promoter sequence should provide an invaluable information about the mechanism segregating the pineal- and retina-specific gene expression. The zebrafish is an excellent animal model suitable for an in vivo promoter analysis with an (E)GFP reporter gene, because of its feasibility of transgenesis and transparency of embryos and larvae (34, 35). Taking into account these advantages, the present study undertook the isolation and in vivo analyses of the zebrafish exorh promoter, and we identified a previously uncharacterized cis-acting element mediating the pineal-specific gene expression. We named it pineal expression-promoting element (PIPE).
Materials and Methods
Isolation of 5′-Flanking Regions of the Zebrafish exorh, Zebrafish rh, and European Eel exorh Genes.
A 1,076-bp region upstream from the ATG initiation codon of the zebrafish exorh gene (GenBank accession no. AB079551) was obtained by three rounds of PCR-based genome walking with the LA PCR in vitro Cloning Kit (Takara Shuzo, Kyoto). Subsequent PCR with zebrafish genome and a pair of primers (5′-GCTCA GCTGG CAGTA CTACC-3′ and 5′-GCAGC TTCTT GTGCT GCACC-3′) amplified a genomic fragment containing a 1,055-bp upstream region together with a short coding sequence of the zebrafish exorh gene. The amplified product was then subcloned into pCR2.1-TOPO vector (Invitrogen). Six clones were obtained from three independent amplification reactions and sequenced to confirm the sequence with no PCR error. In a similar manner, a 238-bp region upstream from the ATG initiation codon of the European eel exorh gene (GenBank accession no. AB079552) was obtained from the eel genomic DNA by using the LA PCR in vitro Cloning Kit and sequenced. On the other hand, screening of λ-Fix II zebrafish genomic library resulted in isolation of a 10.5-kbp fragment containing a 4,558-bp sequence upstream of the rh coding region.
Microinjection and Generation of Germ-Line Transgenic Zebrafish.
Multiple DNA constructs were generated for microinjection (see Results and Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org). They were prepared with the Plasmid Midi Kit (Qiagen, Chatsworth, CA). Rh(−1084), Rh(−1084)/PIPE, and PIPE-Rh(−1084) were treated with SacI, and the other constructs were treated with EcoRI for linearization of each plasmid (cut at a unique site located upstream of the promoter). The linearized DNA was purified by phenol-chloroform extraction and subsequently by chloroform extraction and precipitated with ethanol. The purified DNA was dissolved at a final concentration of 25 ng/μl either in distilled water containing 0.05% phenol red (for transient expression assay) or in 0.1 M KCl/0.05% phenol red solution (for production of transgenic fish). Each DNA construct was microinjected into one-cell-stage embryos of WT zebrafish by using Transjector 5246 and Micromanipulator 5171 (Eppendorf). F0 founder fish were identified by PCR analysis of the genomic DNA pool of 2- to 3-day-old F1 embryos with primers specific for the enhanced GFP (EGFP) coding sequence. We established three, four, three, one, and three independent transgenic lines of the zebrafish with DNA constructs Ex(−1055), Ex(−301), Ex(−147), Rh(−1084), and Rh(−1084)/PIPE, respectively.
Results
Characterization of the 5′-Flanking Region of the Zebrafish exorh Gene.
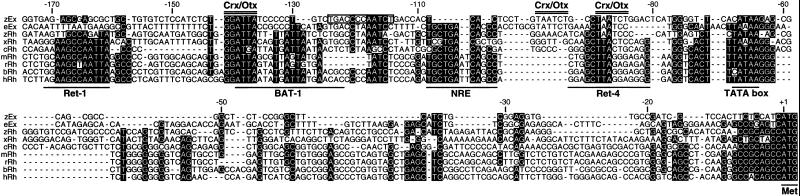
We isolated a 1.1-kbp genomic DNA fragment upstream of the coding region of the zebrafish exorh gene (Fig. 5, which is published as supporting information on the PNAS web site). The putative promoter sequence contained five copies of TAATCC/T sequence, a potential binding site of Crx/Otx (22, 24) that contributes to the pineal gene expression (25, 29–31). When the sequence in the proximal promoter region (−175 to −1 having three potential sites for Crx/Otx-binding) was aligned with those of the rh promoters from various vertebrates (Fig. 1), we found a single conserved site for Crx/Otx-binding at position −142 to −137 (inverted form). Another site (−90 to −85) was conserved in the fish rh and exorh genes, and the third site (−99 to −94) was found only in the zebrafish exorh gene (Fig. 1). A TATA-like sequence in the zebrafish exorh promoter was aligned with TATA box conserved among rh genes. On the other hand, Nrl response element (NRE) that is present in many rh genes (Fig. 1) and is important for retinal gene expression was not found in the zebrafish exorh promoter (Fig. 1 and Fig. 5).
Fig 1.
The proximal promoter sequences of exorh and rh genes of vertebrates. The upstream sequence of the zebrafish exorh (zEx) was aligned with those of rh genes of the zebrafish (zRh), Xenopus (xRh), chicken (cRh), mouse (mRh), rat (rRh), bovine (bRh), and human (hRh). The upstream sequence of the European eel exorh (eEx) was determined in the present study and included in the alignment. Nucleotides conserved among at least six sequences are shown with white characters on black backgrounds. Horizontal lines indicate TATA box, ATG initiation codon, and conserved cis-elements identified previously in the rh promoters. Potential Crx/Otx-binding sites found in the zebrafish exorh promoter are double-lined. The PIPE sequence in the zebrafish exorh gene is boxed. The nucleotide numbers are relative to the translation initiation site of the zebrafish exorh gene. Accession numbers of the sequences obtained from GenBank are U23808 (xRh), M98497 (cRh), M55171 (mRh), U22180 (rRh), and U49742 (hRh). The bRh sequence was obtained from the original paper (17).
A 1,055-bp Fragment Upstream of the exorh-Coding Region Directs Pineal-Specific Gene Expression.
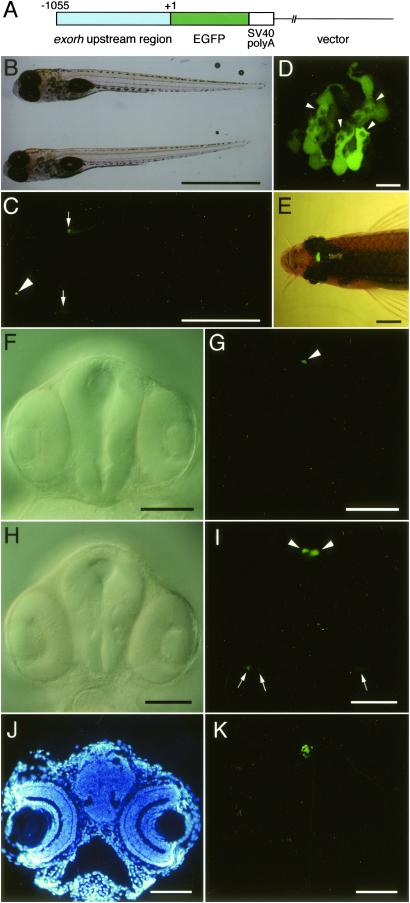
To investigate whether the 1.1-kbp upstream region of the zebrafish exorh promoter is sufficient to direct pineal-specific gene expression, we generated a reporter construct Ex(−1055) by ligating EGFP gene to the 1,055-bp fragment upstream from the exorh translation initiation site (Fig. 2A) and established three lines of the transgenic zebrafish having this construct. The transgenic larvae from every line at 7 days postfertilization (dpf) showed EGFP fluorescence signals in the pineal gland with no detectable signal in the other tissues (Fig. 2 B and C). Each of the EGFP-positive pineal cells displayed highly differentiated morphology with an outer segment-like extrusion (Fig. 2D), a structure characteristic of the (pineal) photoreceptor cell (36). No EGFP signal was detectable within another type of the pineal neurons that project their axons outside the pineal gland (37), and this pineal projection neuron never emitted EGFP signals throughout maturation (data not shown). On the other hand, strong EGFP signals were sustained in the photoreceptor-like pineal cells of matured transgenic fish (6 months old; Fig. 2E), indicating that the 1,055-bp fragment upstream from the exorh translation initiation site is sufficient to maintain pineal-specific gene expression in the zebrafish.
Fig 2.
Pineal-specific EGFP expression in Ex(−1055) transgenic zebrafish. (A) Schematic representation of Ex(−1055) construct in a linearized form used for microinjection. (B) Dorsolateral views (bright field image) of a WT larva (upper) and Ex(−1055) transgenic larva (lower) at 7 dpf. (C) Fluorescent image of B. The transgene-dependent fluorescence signal was observed specifically in the pineal gland of Ex(−1055) transgenic larva (arrowhead), and autofluorescence signals observed in transgenic and WT fish are marked by arrows. (D) High-magnification confocal image (dorsal view) of EGFP-positive pineal cells of 7-dpf-larva, with anterior to the left. The outer segment-like extrusion is indicated by each arrowhead. (E) Dorsal view of Ex(−1055) transgenic adult fish illuminated with both tungsten lamp and blue light. EGFP fluorescence signals were observed only in the pineal gland throughout its life. (F–I) Frontal views (dorsal up) of living transgenic embryos observed at 28 hpf (F and G) or 43 hpf (H and I) by using Nomarski optics (F and H) or fluorescence microscopy (G and I). Arrowheads in G and I indicate EGFP-positive cells in the pineal gland, and arrows in I point to EGFP-positive cells in the retina. The embryos in G and I were photographed under the same exposure conditions. (J) 4′,6-diamidino-2-phenylindole staining of a 10-μm-thick cross-section of the head of Ex(−1055) transgenic larva at 7 dpf. (K) EGFP fluorescent image of J. [Bars = 1 mm (B and C), 10 μm (D), 2 mm (E), and 100 μm (F–K).] For a clear demonstration, pigmentation of the embryos and larvae was reduced by treatment with 0.003% 1-phenyl-2-thiourea (Nacalai Tesque, Kyoto).
Developmental change in EGFP expression pattern was examined in Ex(−1055) transgenic embryos at earlier stages. The EGFP-positive cell in the pineal gland was first detected at around 26 hours postfertilization (hpf; Fig. 2 F and G), which is consistent with the previous study showing the differentiation of the pineal photoreceptor cells at a similar timing (38). Intensity of the pineal EGFP fluorescence signals became stronger at 43 hpf (Fig. 2 H and I). Although weak fluorescence signals were observed in the ventral retina at 43 hpf (Fig. 2 H and I), the retinal signals disappeared by 7 dpf (Fig. 2 J and K) and were not observed at any later stages. These results indicated that the 1,055-bp region of the exorh promoter should be a good target for investigating the regulatory mechanism of pineal-specific gene expression.
The 147-bp Proximal Region of the Zebrafish exorh Promoter Is Sufficient for Pineal-Specific Gene Expression.
To localize cis-acting DNA element(s) required for the pineal-specific gene expression, we generated two reporter constructs, Ex(−301) and Ex(−147), by ligating EGFP gene to 301-bp and 147-bp fragments of the exorh promoter, respectively (see Fig. 6A, which is published as supporting information on the PNAS web site). Then, the transgenic zebrafish were established for Ex(−301) and Ex(−147), respectively. Both of the constructs drove the pineal-specific EGFP expression in the transgenic animals (Fig. 6 D–G), and the EGFP-positive cells in these Ex(−301) and Ex(−147) transgenic larvae showed morphological features of the pineal photoreceptor cell (data not shown). The number of the EGFP-positive cells within each pineal gland of these larvae was also indistinguishable from that of Ex(−1055) transgenic larvae. These results demonstrate that the short promoter region of exorh gene (−147 to −1) retains the DNA element(s) essential for the pineal cell-specific gene expression. Among these larvae, the intensity of the pineal fluorescence signal tended to decrease as the promoter region of the transgene became shorter (Fig. 6 C, E, and G), although the signal intensity varied slightly among individuals from different lines that were produced with the same construct. The length-dependent change in signal intensity suggests that the region between −1055 and −148 contributes to enhancement of exorh gene expression because of the two putative Crx/Otx-binding sites (Fig. 5) and/or unknown elements in this region.
Internal Deletion and Mutational Analyses Revealed a Positive Regulatory Element in the exorh Promoter.
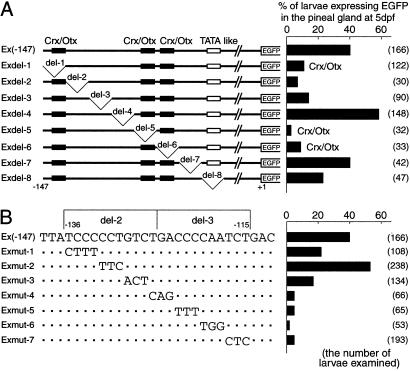
We focused on identification of the pineal-specific cis-acting element(s) presumably present in the short upstream region (−147 to −1) of exorh gene. A series of systematic deletion constructs (Exdel-1 to Exdel-8) was generated by introducing consecutive 11-bp internal deletions (del-1 to del-8) into the promoter region of Ex(−147) construct (Fig. 3A). These deletions cover the 88-bp region (−147 to −60) of the exorh proximal promoter without any overlap nor gap between the neighbors. Embryos microinjected with each construct were raised to 5 dpf, and we scored the percentage of the larvae expressing EGFP fluorescence signals in the pineal gland. In this transient expression assay, the parental construct Ex(−147) induced the pineal EGFP expression in 40% of 166 larvae examined (Fig. 3A), and six of the eight deletion constructs showed lower percentages (3–23%). Among these, Exdel-8 (23%) lacks the TATA-like sequence (Fig. 3A), and Exdel-1 (11%), Exdel-5 (3%) and Exdel-6 (9%) carry a deletion of a potential Crx/Otx-binding site (Fig. 3A), an element that is important for the gene expression in the pineal gland as well as in the retina (25, 29–31). Notably, the promoter activities were significantly reduced in Exdel-2 (7%) and Exdel-3 (14%), of which the deleted sequences in tandem (see Fig. 3B) showed no homology with any known regulatory element, and it is most probable that the 22-bp region (from −136 to −115) contains a positive regulatory element(s) indispensable for pineal-specific gene expression.
Fig 3.
Effects of internal deletions (A) and site-directed mutations (B) in the exorh promoter sequence. (Right) Bar graphs indicate the percentage of larvae that expressed EGFP fluorescence signals in the pineal gland at 5 dpf, and the number of larvae examined at 5 dpf is shown in parentheses. (A Left) The parental construct Ex(−147) and its deletion mutants (Exdel-1 to Exdel-8) are schematically depicted. Black and white boxes indicate the positions of three putative Crx/Otx-binding sites and the TATA-like sequence, respectively. (B Left) The partial nucleotide sequences of the parental construct Ex(−147) and its seven mutant constructs, Exmut-1 to Exmut-7, with dots representing unaltered nucleotides. Numbers at the top indicate the positions of nucleotides relative to the translation initiation site.
To define the element(s), the parental construct Ex(−147) was mutated in the 22-bp region to generate another series of constructs (Exmut-1 to Exmut-7), in each of which consecutive 3–4 nucleotides were systematically mutated (Fig. 3B). Every construct was microinjected into the zebrafish embryos for the transient expression assay at 5 dpf, and we observed strikingly reduced promoter activities in the four constructs, Exmut-4 to Exmut-7 (Fig. 3B) that have mutations at the 12-bp cluster (−126 to −115) covering del-3 region together with an end of del-2 sequence. On the other hand, mutations at positions covering most of del-2 region (−136 to −127) had less effect on the promoter function (Fig. 3B, Exmut-1, -2, and -3). These results of the transient expression assay altogether demonstrate that the pineal(-specific) expression of exorh gene highly depends on the 12-bp DNA sequence TGACCCCAATCT (−126 to −115), which we termed PIPE in this study.
PIPE Can Direct Gene Expression in Pineal Photoreceptor Cells.
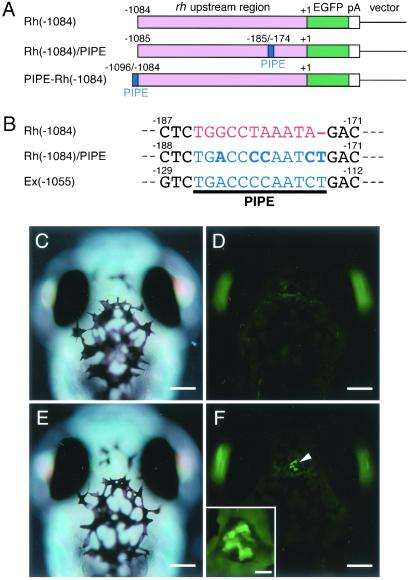
PIPE may govern directly the pineal-specific gene expression in cooperation with Crx/Otx-binding sites or, alternatively, enhance the promoter activity without contributing to the tissue-specific regulation. Considering these possibilities, we examined the effect of ectopic placement of PIPE on expression of a gene regulated by the rh promoter that structurally resembles the exorh promoter (Fig. 1) but probably drives retina-specific gene expression in the zebrafish (32). As a control parental construct, we produced a reporter Rh(−1084), in which 1,084 bps upstream of the zebrafish rh coding region were fused to the EGFP gene (Fig. 4A), and it was used to generate transgenic fish. Rh(−1084) transgenic larvae displayed strong EGFP fluorescence signals exclusively in the eyes, and no detectable signal was observed in the pineal gland (Fig. 4 C and D). A detailed examination of tissue sections prepared from the transgenic larvae revealed localization of EGFP expression in the rod photoreceptor cells (data not shown), consistent with the previous study that used a 1.2-kbp fragment of the zebrafish rh promoter (33). Then, the parental construct Rh(−1084) was modified to carry an ectopic PIPE sequence by introducing four nucleotide substitutions and one nucleotide insertion (Fig. 4B) to a region that is aligned with PIPE in the exorh promoter (Fig. 1). Three independent lines of transgenic zebrafish were established with this chimeric construct Rh(−1084)/PIPE, and all of the transgenic larvae (at 7 dpf) exhibited EGFP fluorescence signals in the pineal gland in addition to those in the eyes (Fig. 4 E and F), with no detectable signal in the other tissues. The expression pattern of EGFP signals in the pineal gland was indistinguishable from those observed in Ex(−1055), Ex(−301), and Ex(−147) transgenic larvae. The rh promoter acquired the ability to drive ectopic gene expression in the pineal gland by virtue of the mutational change of only several nucleotides, supporting the idea that newly created PIPE is functional as a cis-acting DNA element that induces pineal-specific gene expression. No significant difference was observed in retinal fluorescence signals between Rh(−1084) and Rh(−1084)/PIPE transgenic animals, and this eliminates a negative regulatory role of PIPE for retinal gene expression.
Fig 4.
Ectopic gene expression in the pineal gland driven by the zebrafish rh chimeric promoter carrying the PIPE sequence. (A) Schematic representation of the constructs, Rh(−1084), Rh(−1084)/PIPE, and PIPE-Rh(−1084). (B) Comparison of PIPE and nearby sequence in Ex(−1055) with those in the corresponding region of Rh(−1084) or Rh(−1084)/PIPE. Bold characters in Rh(−1084)/PIPE represent five nucleotides (four substitutions and a single insertion) modified from Rh(−1084) to create an ectopic PIPE sequence. (C–F) Dorsal views (anterior up) of Rh(−1084) (C and D) and Rh(−1084)/PIPE (E and F) transgenic larvae at 7 dpf. Nomarski (C and E) and fluorescent (D and F) images were taken at the same focal plane without moving the larvae. EGFP fluorescence signals in the pineal gland of Rh(−1084)/PIPE transgenic larva are marked by an arrowhead (F). (F Inset) High-magnification image of the EGFP-positive pineal structure. The larvae in C–F were not treated with 1-phenyl-2-thiourea, so that strong EGFP fluorescence signals in the pigmented eyes (D and F) are only visible through the pupil. [Bars = 100 μm (C–F), 20 μm (F Inset).]
Finally, we asked whether the above mutations introduced into Rh(−1084) might have disrupted an unknown DNA element(s) responsible for pineal-specific repression of the rh gene expression. To demonstrate the positive regulatory role of PIPE more directly, we generated an additional chimeric construct PIPE-Rh(−1084), which has one copy of PIPE upstream of Rh(−1084) construct (Fig. 4A). In transient expression assay, ≈60% of larvae injected with PIPE-Rh(−1084) expressed EGFP fluorescence signals both in the pineal gland and in the retina at 5 dpf. Control injection of the parental construct Rh(−1084) induced no detectable expression of EGFP signals in the pineal gland at 5 dpf. Together, these results demonstrate that PIPE can direct gene expression in the pineal gland. We concluded that PIPE is the major contributor to the pineal-specific gene expression mediated by the zebrafish exorh promoter.
Discussion
In the present study, we accomplished detailed analyses of the exorh promoter without assistance of in vitro assay, by taking advantage of zebrafish transgenesis technology. This may represent an excellent example of zebrafish transgenesis to delineate a cis-acting DNA element in vivo, and the identification of PIPE corroborates the prominent aspect of the strategy (39). We used the rapidity of the transient expression assay for scanning the exorh promoter region (Fig. 3) while we established multiple transgenic lines to prove eventually the crucial role of PIPE in the pineal-specific gene expression (Fig. 4).
Our systematic deletion analysis revealed that, just like regulation of the rh promoter, the pineal-specific activity of the exorh promoter required the putative Crx/Otx-binding sites to be present in the proximal region (Fig. 3A). This finding is consistent with previous studies on several genes expressed in the pineal gland of the rat, mouse, chicken, and zebrafish (25, 29–31). In addition to the Crx/Otx-binding sites, our analysis identified PIPE as a major contributor to the pineal-specific gene expression, suggesting strongly that the pineal-specific expression of the zebrafish exorh gene is mediated by a combination of those cis-acting elements and presumably by multiple transcription factors. Such a mode of transcriptional regulation operated by multiple cis-elements and transcription factors is generally seen in a variety of tissue-specific promoters including the rh promoter, which contains a number of evolutionarily conserved cis-acting elements such as BAT-1, Ret-4, and NRE (16, 17, 20, 21). Indeed, rh gene is transactivated cooperatively by Crx and Nrl proteins through these sites (22). We speculate that the Crx/Otx-binding site would provide a basis for photoreceptor cell (cell type)-specific expression in both the retina and pineal gland, whereas NRE and PIPE would serve for determination of tissue-specificity, i.e., expression in the retina and pineal gland, respectively.
It is possible that PIPE also plays a role in transcriptional regulation of other genes expressed in the zebrafish pineal gland. For example, we found a 12-bp DNA sequence (TGACCCCTCTCT) similar to PIPE in the proximal promoter region of the zebrafish floating head, an important gene that is expressed from early stages in the pineal gland and is required for differentiation of most pineal neurons (37). On the other hand, PIPE or PIPE-related sequence is not found in a 2.1-kbp fragment upstream of the chicken pineal-specific gene, pinopsin (12, 40), and PIPE may be present at a more distal region or downstream of the translation initiation site. Further sequence analysis of pineal-specific genes in other vertebrates as well as in the zebrafish should help to evaluate the general role of PIPE in pineal-specific gene expression.
PIPE is most likely recognized by the transcription factor(s) expressed specifically in the pineal gland, but none of the pineal-specific transcription factors has been identified yet. One noticeable feature of PIPE is a CCAAT motif (Fig. 5), a binding site for many transcription factors such as CBF/NF-Y, CTF/NF1, and C/EBP (41). Thus, it is possible that PIPE serves as a functional CCAAT box for the pineal-specific expression. This idea is, however, at odds with our sequence analysis of the exorh promoter of the European eel, Anguilla anguilla, in which a CCAAT motif is not found at the equivalent site (Fig. 1). The consensus sequence between the zebrafish PIPE and the PIPE-like element in the eel exorh is TGACCNNAATCN. The conserved nine nucleotides rather than the CCAAT motif might be essential for binding of a common transcription factor(s), although it remains to be determined whether the PIPE-like element in the eel exorh promoter serves for pineal-specific expression.
We previously suggested that teleost exorh and rh genes were produced from an ancestral gene by gene duplication that occurred in the ray-finned fish lineage before the teleost radiation (32). It is conceivable that the ancestral gene was expressed in both the retina and pineal gland, and that the two descendent genes, rh and exorh genes, became segregated from one another with respect to the localization and function. This possibility is consistent with the duplication-degeneration-complementation (DDC) model (42), which predicts that complementary nonfunctionalization of the cis-acting elements can produce the spatio-temporal partitioning of the ancestral functions between the descendents. The present study showed complementary absence of the cis-elements, PIPE and NRE, in the proximal promoters of the zebrafish rh and exorh genes, respectively, as predicted by the DDC model. This model leads to the speculation that the promoter region of the common ancestor of rh and exorh genes formerly contained both PIPE and NRE. Subsequent loss of a functional PIPE in the rh promoter could restrict the rh gene expression to the retina, whereas loss of NRE might lead to the pineal-specific expression of exorh gene. Consistently, a sequence comparison revealed significant homology in nucleotide sequence between PIPE and the equivalent site in the zebrafish rh promoter (Fig. 1), and experimentally we showed that only several nucleotide changes within the site of the rh promoter induced the pineal expression (Fig. 4). Interestingly, the eel exorh promoter has a sequence highly similar to NRE at a site corresponding to that of rh NRE (Fig. 1), although no NRE-like sequence is found in the zebrafish exorh promoter. These observations seem to support the idea that a functional NRE was originally present in the ancestral exorh promoter and then lost in the Euteleost lineage after the divergence between the Anguilliformes and the Euteleostei. Further study of the molecular basis for similarity and difference between the pineal and retinal gene expression could provide an important insight into the evolution of the vertebrate photoreceptive organs.
Supplementary Material
Acknowledgments
We thank Dr. K. Kawakami (National Institute of Genetics) for helpful advice about maintenance of fish. We thank Dr. H. Okamoto (Brain Science Institute, RIKEN) for providing the zebrafish strain Michigan and the zebrafish genomic library. We are grateful to Dr. David R. Hyde (University of Notre Dame) for his kind gifts of antibodies to the zebrafish opsins. We also thank Dr. T. Okano for helpful comments and discussion and K. Imazato for assistance with fish maintenance. This work was supported in part by Grants-in-Aid from the Japanese Ministry of Education, Science, Sports, and Culture.
Abbreviations
rh, rhodopsin
Nrl, neural retina leucine zipper
NRE, Nrl response element
Crx, cone rod homeobox
exorh, exo-rhodopsin
PIPE, pineal expression-promoting element
EGFP, enhanced GFP
dpf, days postfertilization
hpf, hours postfertilization
References
- 1.Pu G. A. & Dowling, J. E. (1981) J. Neurophysiol. 46 1018-1038. [DOI] [PubMed] [Google Scholar]
- 2.Cole W. C. & Youson, J. H. (1982) Am. J. Anat. 165 131-163. [DOI] [PubMed] [Google Scholar]
- 3.Tamotsu S. & Morita, Y. (1986) J. Comp. Physiol. A 159 1-5. [DOI] [PubMed] [Google Scholar]
- 4.Collin J. P., Voisin, P., Falcón, J., Faure, J. P., Brisson, P. & Defaye, J. R. (1989) Arch. Histol. Cytol. 52 441-449. [DOI] [PubMed] [Google Scholar]
- 5.Shedpure M. & Pati, A. K. (1995) Indian J. Exp. Biol. 33 625-640. [PubMed] [Google Scholar]
- 6.Falcón J. (1999) Prog. Neurobiol. 58 121-162. [DOI] [PubMed] [Google Scholar]
- 7.Meissl H. (1997) Biol. Cell 89 549-554. [PubMed] [Google Scholar]
- 8.Kusmic C. & Gualtieri, P. (2000) Micron 31 183-200. [DOI] [PubMed] [Google Scholar]
- 9.Mirshahi M., Faure, J. P., Brisson, P., Falcón, J., Guerlotte, J. & Collin, J. P. (1984) Biol. Cell 52 195-198. [DOI] [PubMed] [Google Scholar]
- 10.Vigh-Teichmann I. & Vigh, B. (1990) J. Pineal Res. 8 323-333. [DOI] [PubMed] [Google Scholar]
- 11.Korf H. W., White, B. H., Schaad, N. C. & Klein, D. C. (1992) Brain Res. 595 57-66. [DOI] [PubMed] [Google Scholar]
- 12.Okano T., Yoshizawa, T. & Fukada, Y. (1994) Nature 372 94-97. [DOI] [PubMed] [Google Scholar]
- 13.Yoshikawa T., Yashiro, Y., Oishi, T., Kokame, K. & Fukada, Y. (1994) Zool. Sci. 11 675-680. [PubMed] [Google Scholar]
- 14.Livesey F. J. & Cepko, C. L. (2001) Nat. Rev. Neurosci. 2 109-118. [DOI] [PubMed] [Google Scholar]
- 15.Morabito M. A., Yu, X. & Barnstable, C. J. (1991) J. Biol. Chem. 266 9667-9672. [PubMed] [Google Scholar]
- 16.DesJardin L. E. & Hauswirth, W. W. (1996) Invest. Ophthalmol. Visual Sci. 37 154-165. [PubMed] [Google Scholar]
- 17.Chen S. & Zack, D. J. (1996) J. Biol. Chem. 271 28549-28557. [DOI] [PubMed] [Google Scholar]
- 18.Swaroop A., Xu, J., Pawar, H., Jackson, A., Skolnick, C. & Agarwal, N. (1992) Proc. Natl. Acad. Sci. USA 89 266-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Q., Ji, X., Breitman, M. L., Hitchcock, P. F. & Swaroop, A. (1996) Oncogene 12 207-211. [PubMed] [Google Scholar]
- 20.Kumar R., Chen, S., Scheurer, D., Wang, Q. L., Duh, E., Sung, C. H., Rehemtulla, A., Swaroop, A., Adler, R. & Zack, D. J. (1996) J. Biol. Chem. 271 29612-29618. [DOI] [PubMed] [Google Scholar]
- 21.Rehemtulla A., Warwar, R., Kumar, R., Ji, X., Zack, D. J. & Swaroop, A. (1996) Proc. Natl. Acad. Sci. USA 93 191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S., Wang, Q. L., Nie, Z., Sun, H., Lennon, G., Copeland, N. G., Gilbert, D. J., Jenkins, N. A. & Zack, D. J. (1997) Neuron 19 1017-1030. [DOI] [PubMed] [Google Scholar]
- 23.Freund C. L., Gregory-Evans, C. Y., Furukawa, T., Papaioannou, M., Looser, J., Ploder, L., Bellingham, J., Ng, D., Herbrick, J. S., Duncan, A., et al. (1997) Cell 91 543-553. [DOI] [PubMed] [Google Scholar]
- 24.Furukawa T., Morrow, E. M. & Cepko, C. L. (1997) Cell 91 531-541. [DOI] [PubMed] [Google Scholar]
- 25.Furukawa T., Morrow, E. M., Li, T., Davis, F. C. & Cepko, C. L. (1999) Nat. Genet. 23 466-470. [DOI] [PubMed] [Google Scholar]
- 26.Mathers P. H., Grinberg, A., Mahon, K. A. & Jamrich, M. (1997) Nature 387 603-607. [DOI] [PubMed] [Google Scholar]
- 27.Martinez J. A. & Barnstable, C. J. (1998) Biochem. Biophys. Res. Commun. 250 175-180. [DOI] [PubMed] [Google Scholar]
- 28.Kimura A., Singh, D., Wawrousek, E. F., Kikuchi, M., Nakamura, M. & Shinohara, T. (2000) J. Biol. Chem. 275 1152-1160. [DOI] [PubMed] [Google Scholar]
- 29.Li X., Chen, S., Wang, Q., Zack, D. J., Snyder, S. H. & Borjigin, J. (1998) Proc. Natl. Acad. Sci. USA 95 1876-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernard M., Dinet, V. & Voisin, P. (2001) J. Neurochem. 79 248-257. [DOI] [PubMed] [Google Scholar]
- 31.Gamse J. T., Shen, Y. C., Thisse, C., Thisse, B., Raymond, P. A., Halpern, M. E. & Liang, J. O. (2002) Nat. Genet. 30 117-121. [DOI] [PubMed] [Google Scholar]
- 32.Mano H., Kojima, D. & Fukada, Y. (1999) Mol. Brain Res. 73 110-118. [DOI] [PubMed] [Google Scholar]
- 33.Kennedy B. N., Vihtelic, T. S., Checkley, L., Vaughan, K. T. & Hyde, D. R. (2001) J. Biol. Chem. 276 14037-14043. [DOI] [PubMed] [Google Scholar]
- 34.Higashijima S., Okamoto, H., Ueno, N., Hotta, Y. & Eguchi, G. (1997) Dev. Biol. 192 289-299. [DOI] [PubMed] [Google Scholar]
- 35.Long Q., Meng, A., Wang, H., Jessen, J. R., Farrell, M. J. & Lin, S. (1997) Development (Cambridge, U.K.) 124 4105-4111. [DOI] [PubMed] [Google Scholar]
- 36.Kusmic C., Barsanti, L., Passarelli, V. & Gualtieri, P. (1993) Micron 24 279-286. [DOI] [PubMed] [Google Scholar]
- 37.Masai I., Heisenberg, C. P., Barth, K. A., Macdonald, R., Adamek, S. & Wilson, S. W. (1997) Neuron 18 43-57. [DOI] [PubMed] [Google Scholar]
- 38.Heisenberg C. P., Brand, M., Jiang, Y. J., Warga, R. M., Beuchle, D., van Eeden, F. J. M., Furutani-Seiki, M., Granato, M., Haffter, P., Hammerschmidt, M., et al. (1996) Development (Cambridge, U.K.) 123 191-203. [DOI] [PubMed] [Google Scholar]
- 39.Meng A., Tang, H., Ong, B. A., Farrell, M. J. & Lin, S. (1997) Proc. Natl. Acad. Sci. USA 94 6267-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takanaka Y., Okano, T., Yamamoto, K. & Fukada, Y. (2002) J. Neurosci. 22 4357-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maity S. N. & de Crombrugghe, B. (1998) Trends Biochem. Sci. 23 174-178. [DOI] [PubMed] [Google Scholar]
- 42.Force A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L. & Postlethwait, J. (1999) Genetics 151 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.