Abstract
Algae include a diverse array of photosynthetic eukaryotes excluding land plants. Explaining the origin of algal plastids continues to be a major challenge in evolutionary biology. Current knowledge suggests that plastid primary endosymbiosis, in which a single-celled protist engulfs and “enslaves” a cyanobacterium, likely occurred once and resulted in the primordial alga. This eukaryote then gave rise through vertical evolution to the red, green, and glaucophyte algae. However, some modern algal lineages have a more complicated evolutionary history involving a secondary endosymbiotic event, in which a protist engulfed an existing eukaryotic alga (rather than a cyanobacterium), which was then reduced to a secondary plastid. Secondary endosymbiosis explains the majority of algal biodiversity, yet the number and timing of these events is unresolved. Here we analyzed a five-gene plastid data set to show that a taxonomically diverse group of chlorophyll c2-containing protists comprising cryptophyte, haptophyte, and stramenopiles algae (Chromista) share a common plastid that most likely arose from a single, ancient (≈1,260 million years ago) secondary endosymbiosis involving a red alga. This finding is consistent with Chromista monophyly and implicates secondary endosymbiosis as an important force in generating eukaryotic biodiversity.
Keywords: cryptophyte, haptophyte, plastid evolution, secondary endosymbiosis, stramenopiles
A plastid is the site where the energy of photons is captured and used to power the synthesis of sugars. Many photosynthetic proteins, and the rRNAs and tRNAs are encoded on the circular plastid genome, whereas many genes have been transferred to the nucleus (1–3). All plastids are believed to trace their origins to a primary endosymbiotic event in which a previously nonphotosynthetic single-celled protist engulfed a cyanobacterium that eventually became a photosynthetic organelle (4–6). The primordial alga resulting from this primary endosymbiosis then putatively gave rise through vertical evolution to the Chlorophyta, Rhodophyta, and the Glaucophyta (6).
A large body of molecular, phylogenetic, and ultrastructural data suggests that members of the Chlorophyta and Rhodophyta then gave rise to most other algal plastids through secondary endosymbiosis (7, 8). In secondary endosymbiosis, a previously nonphotosynthetic single-celled protist engulfs an existing alga that is then reduced to a secondary plastid (7–9). The red algae are particularly noteworthy in this respect because they are believed to have contributed plastids to at least five evolutionarily distantly related lineages [cryptophytes, haptophytes, stramenopiles (6, 10, 11), apicomplexa (12, 13), and dinoflagellates (14)]. Our laboratory has recently confirmed the red algal origin of the ancestral dinoflagellate plastid by using phylogenetic analyses, but surprisingly, it was found to have arisen through a tertiary endosymbiosis (uptake of a plastid of secondary endosymbiotic origin) of a haptophyte alga (15). In this case, the haptophyte plastid replaced the original dinoflagellate plastid, both of which trace their roots to the red algae. These data underline the importance of plastid endosymbiosis in the evolutionary history of eukaryotes and lead to the present study in which we address a long-standing issue in algal biology, the Chromista hypothesis (10).
Cavalier-Smith (10) proposed that the cryptophytes, haptophytes, and stramenopiles share a common ancestor and together form the kingdom Chromista. These taxa were united primarily on the basis of plastid characters, most importantly the presence of chlorophyll c2 in a four-membrane bound plastid that was located in the lumen of the endoplasmic reticulum. The cryptophytes were posited as the early divergence in this group with the retention of the remnant endosymbiont nucleus (the nucleomorph) being an ancestral character (10). However, there is presently no convincing phylogenetic evidence in support of the monophyly of the Chromista “host” cells to the exclusion of other eukaryotes (e.g., ref. 6). Therefore, it is not known whether all of the chromist plastids have arisen from multiple, independent endosymbioses (e.g., refs. 3 and 6), or whether some of them (and the host cells that contain them) trace their origins to a single endosymbiotic event followed by separation of the nuclear lineages over evolutionary time (e.g., ref. 13). Assessing chromist monophyly is important not only for algal taxonomy but more generally for understanding the frequency of secondary plastid establishment in protists. Each secondary endosymbiosis entails the stable inheritance of a foreign cell, the large-scale movement of genes from the endosymbiont to the host nucleus, and the reimport of the gene products required for photosynthesis into the organelle to facilitate plastid function (1, 12, 13). Previous phylogenetic analyses of single plastid genes, although clearly supportive of a red algal origin of the secondary plastids of the Chromista, are ambiguous about the number of events that gave rise to them (e.g., refs. 16 and 17). A more recent analysis of 41 proteins from 15 complete plastid genomes, however, with limited taxon sampling of chromists and red algae has suggested that the plastid in cryptophytes and stramenopiles has independent origins (3). And finally, the organization of the plastid genomes in each of these algae is sufficiently different to preclude a clear understanding of their interrelationships (18, 19).
A direct approach to resolving this central problem in organelle evolution is to sample a broad range of plastid genes from red algae and Chromista members to test the hypothesis of independent secondary plastid origins in cryptophytes, haptophytes, and stramenopiles. Such analyses could have three possible outcomes. (i) Demonstration of a well-supported sister-group relationship of individual Chromista plastids with different red algae in plastid gene trees, which would suggest that chromist secondary endosymbioses are independent events. (ii) Alternatively, if the chromist plastid sequences form a strongly supported monophyletic group after extensive taxon sampling, then we would accept the hypothesis that a single secondary endosymbiosis likely explains plastid origin in these algae. Another less parsimonious interpretation of this result is that the same or closely related bangiophytes gave rise independently to the plastids. (iii) A third possibility is that two of the chromist lineages may share a single secondary endosymbiosis and the third gained its plastid although an independent event. To test the Chromista hypothesis, we sequenced 13 small subunit (SSU) rRNA, 29 tufA (plastid elongation factor Tu), and 1 rbcL (ribulose-1,5-bisphosphate carboxylase/oxygenase) plastid-encoded coding region from various red and chromist algae. These sequences were added to a published data set of 36 taxa for which we also had the sequences of psaA (photosystem I P700 chlorophyll a apoprotein A1), psbA (photosystem II reaction center protein D1), and “Form I” rbcL (15), and used to infer a DNA-based phylogeny of red and chromist plastids in a context of broad taxon sampling.
Materials and Methods
Algal Cultures and Sequencing.
Red algae were chosen for the plastid phylogeny that includes representatives of all of the orders and different phylogenetic lineages of Bangiophycidae, including the Cyanidiales (17, 20). Several Cyanidiales used in our trees were collected in nature and are maintained at the Dipartimento di Biologia Vegetale culture collection at the University of Naples, Italy. Although many unicellular forms (e.g., other species of Porphyridium and Rhodosorus) are not present in our trees, a much larger data set of nuclear-encoded SSU rDNA shows that all of these taxa are robustly placed in one of the clades resolved with the present analysis (17). We have not, therefore, excluded members of any known unicellular bangiophyte lineage (i.e., a putative plastid donor) when estimating the position of the chromist plastids. In this study, a total of 43 SSU rRNA, tufA, and rbcL coding regions were isolated and sequenced from 33 red and chromist algae. For strain identifications and GenBank numbers, see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. The algal cultures were frozen in liquid nitrogen, and ground with glass beads by using a glass rod and/or MiniBeadBeater (Biospec Products, Bartlesville, OK). Total genomic DNA was extracted by using the DNeasy Plant Mini kit (Qiagen, Valencia, CA). PCRs were done with specific primers for each of the plastid genes (see Table 2, which is published as supporting information on the PNAS web site). Because introns were found in the tufA gene of some red algae, the RT-PCR method was used to isolate cDNA. For the RT-PCR, total RNA was extracted by using the RNeasy Mini kit (Qiagen). To synthesize cDNA from total RNA, M-MLV Reverse Transcriptase (GIBCO/BRL, Gaithersburg, MD) was used following the manufacturer's protocol. PCR products were purified by using the QIAquick PCR Purification kit (Qiagen), and were used for direct sequencing using the BigDye Terminator Cycle Sequencing kit (Applied Biosystems), and an ABI-3100 at the Center for Comparative Genomics at the University of Iowa. Some PCR products were cloned into pGEM-T vector (Promega) before sequencing.
Phylogenetic Analyses.
Sequences were manually aligned by using seqpup (21). The data sets used in the phylogenies are available from D.B. In the first analysis, we prepared a collection of concatenated partial SSU rRNA (1,285 nt), psaA (1,395 nt), psbA (957 nt), rbcL (1,215 nt), and tufA (975 nt) coding regions (total of 5,827 nt) from 20 red algae, 5 cryptophytes, 4 haptophytes, 4 stramenopiles, 2 chlorophytes, and the glaucophyte Cyanophora paradoxa as the outgroup. In subsequent analyses, we removed one or more of the plastid genes from the data set to test their effects on the inferred phylogenies. Because the rbcL gene of the green and glaucophyte algae are of a cyanobacterial origin, whereas those in the red algae and red algal-derived plastids are of proteobacterial origin (e.g., ref. 22), the evolutionarily distantly related green and glaucophyte rbcL sequences were coded as missing data in the phylogenetic analyses.
Trees were inferred with the minimum evolution (ME) method using general time reversible (GTR) + I + Γ and LogDet (ME-gtr, ME-ld) distances (23) and the PAUP*4.0b8 (24) computer program. The parameter estimates for the GTR + Γ + I model were estimated by using paup and a starting ME tree built with HKY-85 distances. Ten heuristic searches with random-addition-sequence starting trees and tree bisection-reconnection (TBR) branch rearrangements were done to find the optimal ME tree. Best scoring trees were held at each step. To test the stability of monophyletic groups in the ME trees, 10,000 bootstrap replicates were analyzed (25) with the DNA data set. The maximum likelihood (ML) method was also used to infer a tree using the five-gene data set. The ML tree was built with stepwise addition and rearranged with the TBR branch-swapping algorithm (24). In addition, we did Bayesian analysis of the DNA data (mrbayes version 2.0, ref. 26) using the GTR + I + Γ model. Bayesian posterior probabilities are roughly equivalent to maximum likelihood bootstrap analysis (27, 28). Metropolis-coupled Markov chain Monte Carlo (MCMCMC) from a random starting tree was initiated in the Bayesian inference and run for 1,000,000 generations. A consensus tree was made with the MCMCMC trees after convergence. We also analyzed a four-gene data set of protein-coding regions (psaA, psbA, rbcL, tufA) by using Bayesian inference of the DNA data and the GTR model with site-specific γ parameters for each codon site.
The combinability of the five plastid genes was tested with the incongruence length-difference (ILD) test (29, 30) using paup (the ILD test is called the partition homogeneity test in this program package). One thousand replicates were run with each pair of plastid data sets to infer the significance of the observed ILDs. Taxa for which we had no sequence data available (i.e., psaA for Chilomonas paramecium), or only partial data (i.e., psaA and SSU rRNA for Rhodomonas abbreviata), or for which the rbcL data were coded as “missing” (i.e., two green and one glaucophyte algae) were excluded from the ILD tests. This resulted in a total of 31 taxa being compared for each pair of data partitions.
We used the tree length distribution nonparametric bootstrap (TLDB) test (31) to examine the significance of alterations in the inferred tree topologies. In this method, we compared the ME scores of rearranged trees to the distribution of the suboptimal ME-gtr bootstrap tree scores. Rearranged trees were deemed to be significantly different from the best tree when their ME scores fell outside of the 95th quantile of the bootstrap tree distribution. And finally, the Shimodaira–Hasegawa (SH) nonparametric bootstrap test was used to compare alternative phylogenetic hypotheses regarding the positions of chromist plastids (32). The SH-test was performed by using paup, with RELL (resampling estimated log-likelihood) optimization and 100,000 bootstrap replicates.
Molecular Clock Calculations.
We used the ML method to infer the divergence times of chromist plastids. Three different constraints were used in this analysis. The first constraint was a date of 1,174–1,222 million years ago (Ma), which is widely regarded as the first appearance of taxonomically resolved eukaryotic fossils of multicellular red algae that reproduce sexually and closely resemble (both in morphology and ontogeny) present-day Bangia (33). The second constraint was a fossil date of 550–590 Ma to mark the radiation of the morphologically complex Florideophycidae red algae (i.e., Chondrus, Thorea, Palmaria, ref. 34). The final constraint was a date of 1,576 Ma as the maximum age of taxa in our tree because a recent molecular clock study dates the plant-animal-fungal divergence at this time (ref. 35, but see ref. 36). To date divergences in the ML tree of the five-gene data set, we used the Langley–Fitch method with a “local molecular clock” and the Powell search algorithm (37). Local rates were calculated for 10 different clades (e.g., for each of the chromist plastid lineages and for the Cyanidiales). We chose a method that relaxes the assumption of a constant molecular clock across the tree because the likelihood ratio test showed significant departure, in our data set, from clock-like behavior (P < 0.005). Ninety-five percent confidence intervals on divergence dates were calculated by using a drop of two (s = 2) in the log likelihood units around the estimates (38).
Results
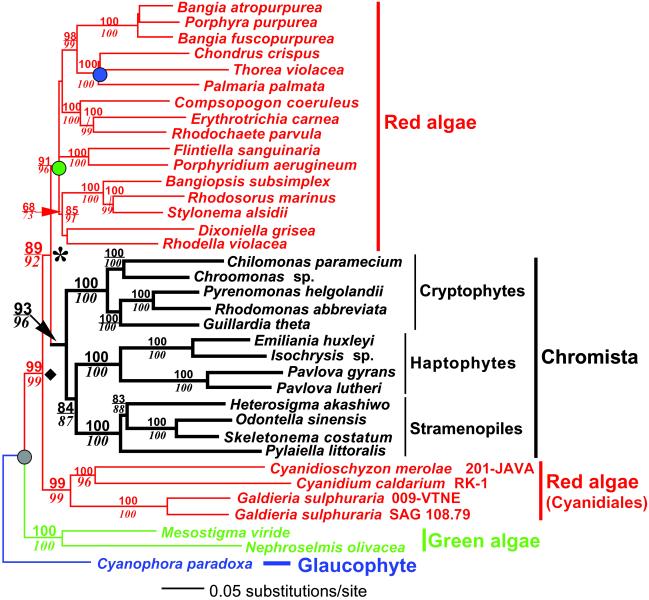
To test the monophyly of chromist plastids, we analyzed the plastid SSU rRNA, psaA, psbA, rbcL, and tufA sequences from 33 red algae and chromists, 2 green algae, and the glaucophyte, Cyanophora paradoxa, as the outgroup. The phylogenetic analyses (Fig. 1) show strong support for the monophyly of 3 different plastid clades. The first [Bayesian posterior probability with the five-gene data set (Pp5) = 1.0 and four-gene data (Pp4 = 1.0), bootstrap support for ME-gtr = 99%, ME-ld = 99%] comprises the Cyanidiales [members of the 3 recognized genera in this order; Cyanidium, Cyanidioschyzon, and Galdieria (39) were included in the analysis], the second (Pp5 = 1.0, Pp4 = 1.0, ME-gtr = 93%, ME-ld = 96%) is of the chromist plastids, and the third (Pp5 = 1.0, Pp4 = 1.0, ME-gtr = 91%, ME-ld = 96%) comprises the remainder of the red algae. The consensus of 9,800 trees resulting from the Bayesian analysis of the five- and four-gene data sets are shown in Fig. 3, which is published as supporting information on the PNAS web site. The ME, ML, and Bayesian MCMCMC consensus trees differed solely with respect to the interrelationships of the basal lineages of the non-Cyanidiales red algae (i.e., where the bootstrap values are missing in Fig. 1) and the switching of the positions of Bangia atropurpurea and Porphyra purpurea.
Fig 1.
Phylogenetic relationships of red algal (shown in red typeface) and chromist (shown in black typeface) plastids inferred from a minimum evolution analysis of the combined DNA sequences of 16S rRNA, psaA, psbA, rbcL, and tufA. Results of a ME-gtr bootstrap analysis are shown above the branches, and bootstrap values from a ME-ld analysis are shown below the branches in italics. This tree is rooted on the branch leading to the Cyanophora paradoxa sequence. The branch lengths are proportional to the number of substitutions per site (see scale in figure). The filled circles indicate nodes that were constrained for molecular clock analyses (gray = 1,576 Ma; green = 1,174–1,222 Ma; blue = 550–590 Ma). The estimated nodes are indicated by the diamond (1,342 ± 22 Ma) and the asterisk (1,261 ± 28 Ma).
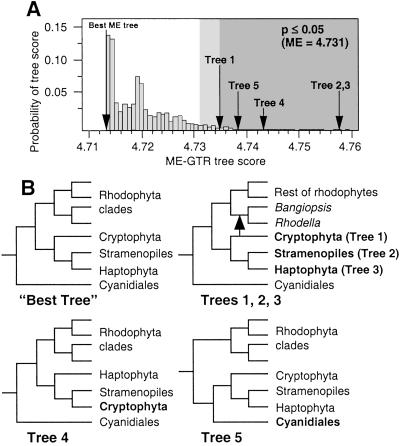
We tested the position of the chromist plastids by rearranging the best ME-gtr tree that was inferred from the original data (ME score = 4.713). The scores of the rearranged trees were compared with the distribution of the suboptimal ME-gtr bootstrap tree scores using the TLDB test (31). Three alternative hypotheses about plastid evolution were tested: (i) the different chromists branch within the derived red algae (i.e., test a later origin of chromist plastids), (ii) cryptophytes branch within the haptophytes-stramenopiles (i.e., test whether cryptophyte plastid characters are ancestral to the Chromista), and (iii) Cyanidiales and chromists are monophyletic (i.e., test the possibility of a putative thermoacidophilic algal endosymbiont). Our analysis showed that positioning either the cryptophyte (ME = 4.735), haptophyte (ME = 4.758), or stramenopiles (ME = 4.758) plastids within the derived red algae (as sister to the clade containing Bangiopsis and Rhodella) resulted in trees (i.e., trees 1, 2, and 3 in Fig. 2) that were significantly worse than the best tree (tree 1, P < 0.05; trees 2 and 3, P < 0.01). Breaking haptophyte-stramenopiles monophyly by uniting the stramenopiles and cryptophytes (tree 4 in Fig. 2, ME = 4.743) or forcing Cyanidiales and chromist monophyly (tree 5 in Fig. 2, ME = 4.738) was also not supported by this analysis (P < 0.01). We also tested the hypothesis of a paraphyletic Chromista by forcing the 3 plastid groups to diverge as independent lineages between the Cyanidiales and the non-Cyanidiales red algae. Regardless of the order of divergence, the TLDB test showed these rearranged trees to be significantly worse than the best tree shown in Fig. 1 (at P < 0.05 or 0.01). The SH-test corroborated these results by showing that placement of the cryptophyte, haptophyte, or stramenopiles plastids within the derived red algae (trees 1, 2, and 3 in Fig. 2) resulted in trees that were significantly worse than that shown in Fig. 1 (P = 0.021, P < 0.000, P < 0.000, respectively).
Fig 2.
Analysis of rearranged plastid trees using the TLDB test. (A) The distribution of suboptimal ME-gtr bootstrap trees. The area shown in light gray falls outside the 95th quantile (ME score0.05 = 4.73), whereas the darker area defines the region outside the 99th quantile (P ≤ 0.01). The score for the best ME-gtr tree is shown, as are the scores for the rearranged trees. (B) Schematic drawings of tree rearrangements used to test Chromist plastid monophyly. The “Best” ME-gtr is shown, as are trees 1, 2, and 3, in which the cryptophytes, haptophytes, and stramenopiles, respectively, are placed within the derived red algae (see arrow). Trees 4 and 5 test cryptophyte–stramenopiles and Chromista–Cyanidiales monophyly, respectively.
Combinability of the Plastid Genes.
The ILD test indicated that combinations of most of the data partitions did not result in significantly incongruent trees (see Table 3, which is published as supporting information on the PNAS web site). We used a significance threshold of P ≤ 0.01 to identify incongruence, because the ILD is believed to be a conservative test of data combinability (for details, see refs. 40 and 41). According to this criterion, one data partition, tufA, showed P < 0.01 in three of the four tests and merited closer inspection. To determine whether tufA was misleading our phylogenies, we inferred ME-gtr and ME-ld trees using the other four genes. These analyses (see Fig. 4, which is published as supporting information on the PNAS web site) resulted in nearly identical trees to that shown in Fig. 1 with comparable bootstrap support values for the critical nodes in the phylogeny (e.g., monophyly of Chromista plastids, ME-gtr = 97%, ME-ld = 96%; monophyly of haptophytes and stramenopiles, ME-gtr = 87%, ME-ld = 88%; monophyly of non-Cyanidiales red algae, ME-gtr = 83%, ME-ld = 85%). These findings suggest that the tufA sequences, although the least congruent with the other data partitions, do not significantly alter the bootstrap support for groups resolved in the five-gene tree and should, therefore, be retained in the analysis. We also tested for the presence of, and level of support for Chromista plastid monophyly in ME-gtr and ME-ld bootstrap tests using all possible two-gene data sets. In support of the tree shown in Fig. 1, this analysis shows that all but the 16S-tufA and 16S-psbA combinations resolve Chromista monophyly with moderate to strong bootstrap support (see Table 3).
Discussion
Congruence of the Five-Gene Plastid Data Set.
The finding of tree incongruence in phylogenetic analyses may be explained by two phenomena: (i) different evolutionary histories of the genes under consideration, and/or (ii) violation of the assumptions of the phylogenetic methods that are used to infer the trees (40). With regard to the first explanation, the observation that the genes we have studied trace their origin to the haploid, nonrecombining plastid genome of the ancestral red alga suggests that these linked loci share the same evolutionary history. The level of support for nodes in the tree shown in Fig. 1 varies, therefore, from gene to gene (see Table 3), but this most certainly reflects varying signal-to-noise ratios among these coding regions (barring lateral transfer) and not alternate evolutionary histories. Second, the plastid genes used here have seen broad application in phylogenetics (see ref. 42) and are not expected, therefore, to grossly violate the assumptions of the tree-building methods. Therefore, the second explanation for tree incongruence is also likely not germane to our analysis. These observations lead us to postulate that the phylogenetic hypothesis shown in Fig. 1 is a reasonable platform for studying the evolution of red and chromist plastids.
The Phylogeny of Red Algal and Chromist Plastids.
Given the robust phylogenetic hypothesis shown in Fig. 1, the position of the Cyanidiales within the tree confirms speculation that these thermoacidophiles are evolutionarily distantly related to other red algae (43). Exclusion of the chromist sequences from the Bayesian–MCMCMC and ME analyses shows that the Cyanidiales remain a monophyletic lineage (Pp5 = 1.0, Pp4 = 1.0, ME-gtr = 100%, ME-ld = 100%) that is sister to the remaining red algae. The Cyanidiales are asexual, unicellular organisms that live in high-salt environments with temperatures ranging from 37 to 45°C and pH between 1 and 2 (39). In comparison, the first taxonomically resolved eukaryotic fossils are of multicellular red algae that reproduce sexually and closely resemble (both in morphology and ontogeny) present-day Bangia (33). These taxa existed in the Mesoproterozoic and were likely present at the node marked with the green circle in Fig. 1. We used a fossil date of 1,174–1,222 Ma (33) to constrain this node and a fossil date of 550–590 Ma (34) to constrain the node (blue circle in Fig. 1) marking the radiation of the morphologically complex Florideophycidae red algae (i.e., Chondrus, Thorea, Palmaria), to calculate different divergence dates in our tree (37). In these analyses, the basal split of the green and red algae (gray circle in Fig. 1) was constrained at a maximum age of 1,576 Ma because a recent molecular clock study dates the plant-animal-fungal divergence at this time (ref. 35, but see ref. 36). Red algae and plants (including green algae) are believed to be sister groups (44, 45). With these constraints in place, we found the deepest split within the red algae, that between the Cyanidiales and all other red algal or red algal-derived plastids (marked with a diamond in Fig. 1) occurred 1,342 ± 22 Ma.
With regard to chromist plastids, our analysis confirms solidly their secondary endosymbiotic origin from red algae. Most importantly, their sister group relationship is consistent with a monophyletic origin of the host cells containing these organelles, and therefore a single secondary endosymbiosis in their common ancestor. The basal position of the cryptophytes in the Chromista (Pp5 = 1.0, Pp4 = 1.0, ME-gtr = 84%, ME-ld = 87%) suggests that retention of the remnant red algal nucleus (nucleomorph) in the periplastid space (former cytoplasm of the symbiont), the presence of phycobilin pigments, and the storage of photosynthates as starch are ancestral endosymbiont characters that were likely lost after the divergence of the cryptophytes within the Chromista (46). In addition, the cryptophytes differ with respect to one very important host character shared by the haptophytes and stramenopiles; the cryptophytes have mitochondria with flattened cristae, whereas the other chromists (and alveolates) have tubular cristae (47). If our tree is correct, then flattened mitochondrial cristae may also have been an ancestral chromist character. Consistent with these ideas, placement of the cryptophyte plastids in a derived position as sister to the stramenopiles within the Chromista resulted in a significantly worse tree (see Fig. 2). We calculated the split of the cryptophytes and haptophytes-stramenopiles to have occurred 1,136 ± 35 Ma. The haptophyte-stramenopiles split was 988 ± 50 Ma. The earliest origin of the chromist plastid (marked with an asterisk in Fig. 1) is estimated at 1,261 ± 28 Ma. These dates, although considerably older than a previous proposal of ≈600 Ma for the origin of chromist plastids (46), agree well with the fossil record, which shows the appearance of a diversity of algae and protists near the Mesoproterozoic/Neoproterozoic boundary ≈1,000 Ma (48, 49).
Recently, there have been realistic concerns that molecular clock methods overestimate evolutionary time (36). We have taken several steps to strengthen our analysis and test its robustness. The tree was calibrated on two nodes with fossil data and we have set a maximum age near the base of the tree, thereby constraining nodes both older and younger those that were estimated. Furthermore, we have conducted this analysis with a large data set (5,827 nt) with broad taxon sampling to break long branches. Rodriguez-Trelles et al. (36) also showed that dating accuracy increases with the size of the data set. Our data set is, therefore, considerably larger than the largest data set tested in that study (500 aa).
However, to address the possibility that one of our constraints is having an inordinately strong effect on the estimation of the date of chromist plastid origin, we released the constraints on each of our calibration points individually to assess the affect on the dating estimates. When the constraints on the node marking the radiation of the red algae (1,174–1,222 Ma, green circle in Fig. 1) are removed, we estimate the earliest origin of the complex plastids to be 1,037 ± 96 Ma. When the constraints on the node marking the radiation of the Florideophycidae (550–590 Ma, blue circle in Fig. 1) are removed, the origin of the complex plastids is estimated to be 1,261 ± 19 Ma. Lastly, when the constraint on the split of the red and green algae (<1,576 Ma, gray circle in Fig. 1) is released, we estimate this data to be 1,294 ± 31 Ma. These consistent results indicate that our best estimate for the origin of the complex plastids (1,261 ± 28 Ma) does not rely strongly on any one of the three calibration dates.
Despite the robustness of the tree presented in Fig. 1, comparison of this phylogeny with the plastid trees recently published by our lab (15) shows an important conflict in the position of the cryptophytes. Analysis of a broadly diverse set of chromist, dinoflagellate, and red algal psaA and psbA sequences in the latter study recovered most of the relationships shown in Fig. 1; however, the cryptophytes diverged at the base of the non-Cyanidiales red algae with moderate bootstrap support [e.g., figures 1 and 2 of ref. 15). This result is not consistent with the current analyses that included three additional coding regions (16S, rbcL, and tufA). As explanation for this discrepancy, we suggest that the length of the sequence data has a significant impact on the placement of the ancient cryptophyte clade within the red algal plastid tree. In support of this hypothesis, addition of the plastid-encoded psaB sequence (1,269 nt, H.S.Y. and D.B., unpublished data) for 22 of the 36 taxa shown in Fig. 1 (primarily the chromists and Cyanidiales, plus 5 non-Cyanidiales red algae) and recalculation of the bootstrap and Bayesian posterior probabilities (as described above) with this 7,096 nt data set shows increased support for Chromist monophyly. The ME-gtr and ME-ld bootstrap support for the Chromista plastid lineage increases from 93% and 96% (see Fig. 1) to 99% and 99%, respectively, whereas the posterior probability remains at P = 1.0 (results not shown). Similarly, bootstrap support for the monophyly of the haptophytes and stramenopiles increases from 84% and 87% to 94% and 93%, respectively. These data clearly show increasing support for chromist monophyly as the plastid data are augmented and suggest that the alternative placement of the cryptophytes in Yoon et al. (15) likely reflects insufficient phylogenetic signal to resolve this ancient relationship.
Our results, therefore, are consistent with the monophyly of chromist plastids and provide the first robust phylogenetic support for the Chromista hypothesis (10). There is presently weaker support from the analysis of SSU rRNA (50) and a combined data set of EF-1α, actin, α-tubulin, and β-tubulin amino acid sequences (that did not include haptophytes, ref. 51) for a sister-group relationship between stramenopiles and alveolates. Although preliminary, this finding is intriguing because it suggests the monophyly of chromists and alveolates (chromalveolates, ref. 46). The chromalveolate hypothesis, although still speculative, is also supported by analyses of GAPDH that show dinoflagellates, apicomplexans, cryptophytes, and stramenopiles to share a homologous plastid-targeted copy of this gene, duplicated in a common ancestor that contained a red algal-derived secondary plastid (13, 52). If these results hold up, then they imply a fundamental event in eukaryotic evolution: a single, ancient red algal secondary endosymbiosis that gave rise to the photosynthetic common ancestor of a protist superassemblage defined by the chromalveolates. The red algae are central to this idea, having given rise to the plastids in the Chromista through secondary endosymbiosis and those in the dinoflagellates through tertiary endosymbiosis.
Major Insights into Plastid Evolution.
Corroboration of the Chromista (10) leads to three major insights into plastid evolution. First, loss of plastids may have been common after secondary endosymbiosis [e.g., in aplastidial stramenopiles such as oomycetes, ciliates (13), in some dinoflagellates (53), and Cryptosporidium parvum (54)]. Supporting this idea is a recent analysis of the 6-phosphogluconate dehydrogenase (gnd) gene of cyanobacterial (i.e., plastid) origin in different protists. These data suggest that the parasitic stramenopile, Phytophtora infestans, was likely once photosynthetic because its gnd gene is closely related to the homologue in photosynthetic members of this lineage (55). Second, the Chromista must share a homologous plastid protein import system that evolved in their common ancestor. This system is necessary to import the products of the hundreds of photosynthetic genes that were presumably transferred from the nucleus of the algal symbiont to that of the host. Furthermore, an additional N-terminal domain (ER-type signal sequence) must evolve in these coding regions to target the proteins to the three- or four-membraned secondary plastid (56). These seemingly unlikely evolutionary events may be a significant bottleneck in establishing secondary endosymbioses. A direct prediction of this hypothesis is that chromist plastids trace their ancestry to a single endosymbiotic event (46). Our findings support this hypothesis. And third, chlorophyll c2 appears to have evolved only once in the common ancestor of the Chromista. Phylogenetic analysis of chlorophyll a/b and chlorophyll a/c light harvesting complex proteins supports this idea, showing that the chlorophyll a/c–binding proteins in chromists (and dinoflagellates) form a monophyletic group that traces its origin to a red algal-like ancestor (57).
In summary, our results provide an important piece of evidence for consolidating a substantial portion of the eukaryotic host tree (Chromista) through an ancient plastid secondary endosymbiosis in their common ancestor. This hypothesis awaits confirmation with the analysis of multigene data sets of nuclear or mitochondrial genes from a broad diversity of protists to test the monophyly of the chromist host cells. Regarding the recent analysis of 15 complete plastid genomes (3) that suggested separate origins of the red algal-derived plastids in a cryptophyte (Guillardia theta) and in a stramenopile (Odontella sinensis), we suggest that this result is likely explained by poor sampling of the genetically divergent and clearly ancient red algal and chromist lineages. Our expectation is that the accuracy of plastid trees using concatenated genome data will increase substantially with increased taxon sampling, an idea that has strong support in recent analyses of the causes of phylogenetic tree error (e.g., ref. 58).
Supplementary Material
Acknowledgments
We thank E. H. Bae, H.-G. Choi, K. Müller, and D. W. Freshwater for providing red algal thalli or genomic DNAs, the Sammlung von Algenkulturen Göttingen for the gift of algal cultures, and P. Cennamo for organizing the shipment of the Cyanidiales cultures and DNA. This work was supported by National Science Foundation Grants DEB 01-07754 and MCB 01-10252 (to D.B.) and a postdoctoral fellowship from the Korean Science and Engineering Foundation (to H.S.Y.).
Abbreviations
psaA, photosystem I P700 chlorophyll a apoprotein A1
psbA, photosystem II reaction center protein D1
rbcL, ribulose-1,5-bisphosphate carboxylase/oxygenase
tufA, plastid elongation factor Tu
ME, minimum evolution
GTR, general time reversible
ML, maximum likelihood
MCMCMC, Metropolis-coupled Markov chain Monte Carlo
Ma, million years ago
SH, Shimodaira–Hasegawa
TLDB, tree length distribution nonparametric bootstrap
ILD, incongruence length-difference
References
- 1.Martin W., Stoebe, B., Goremykin, V., Hansmann, S., Hasegawa, M. & Kowallik, K. (1998) Nature 393 162-165. [DOI] [PubMed] [Google Scholar]
- 2.De Las Rivas J., Lozano, J. J. & Ortiz, A. R. (2002) Genome Res. 12 567-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin W., Rujan, T., Richly, E., Hansen, A., Cornelson, S., Lins, T., Leister, D., Stoebe, B., Hasegawa, M. & Penny, D. (2002) Proc. Natl. Acad. Sci. USA 99 12246-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavalier-Smith T. (1982) Biol. J. Linn. Soc. 17 289-306. [Google Scholar]
- 5.Gray M. W. (1992) Int. Rev. Cytol. 141 233-357. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharya D. & Medlin, L. (1995) J. Phycol. 31 489-498. [Google Scholar]
- 7.Gibbs S. P. (1993) in Origins of Plastids, ed. Lewin, R. A. (Chapman and Hall, New York), pp. 107–121.
- 8.Gilson P. R. & McFadden, G. I. (1997) BioEssays 19 167-173. [DOI] [PubMed] [Google Scholar]
- 9.Ludwig M. & Gibbs, S. P. (1987) Ann. N.Y. Acad. Sci. 501 198-211. [Google Scholar]
- 10.Cavalier-Smith T. (1986) in Progress in Phycological Research, eds. Round, F. E. & Chapman, D. J. (Biopress, Bristol, U.K.), Vol. 4, pp. 309–347. [Google Scholar]
- 11.Douglas S. E., Murphy, C. A., Spencer, D. F. & Gray, M. W. (1991) Nature 350 148-151. [DOI] [PubMed] [Google Scholar]
- 12.McFadden G. I. & Waller, R. F. (1997) BioEssays 19 1033-1040. [DOI] [PubMed] [Google Scholar]
- 13.Fast N. M., Kissinger, J. C., Roos, D. S. & Keeling, P. J. (2001) Mol. Biol. Evol. 18 418-426. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z., Green, B. R. & Cavalier-Smith, T. (1999) Nature 400 155-159. [DOI] [PubMed] [Google Scholar]
- 15.Yoon H. S., Hackett, J. & Bhattacharya, D. (2002) Proc. Natl. Acad. Sci. USA 99 11724-11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helmchen T. A., Bhattacharya, D. & Melkonian, M. (1995) J. Mol. Evol. 41 203-210. [DOI] [PubMed] [Google Scholar]
- 17.Müller K. M., Oliveira, M. C., Sheath, R. G. & Bhattacharya, D. (2001) Am. J. Bot. 88 1390-1400. [PubMed] [Google Scholar]
- 18.Kowallik K. V. (1997) in Eukaryotism and Symbiosis, eds. Schenk, H. E. A., Herrmann, K. W., Jeon, K. W., Muller, N. E. & Schwemmler, W. (Springer, Berlin), pp. 3–23.
- 19.Douglas S. E. & Penny, S. L. (1999) J. Mol. Evol. 48 236-244. [DOI] [PubMed] [Google Scholar]
- 20.Garbary D. J. & Gabrielson, P. W. (1990) in Biology of the Red Algae, eds. Cole, K. M. & Sheath, R. G. (Cambridge Univ. Press, Cambridge, U.K.), pp. 477–498.
- 21.Gilbert D. G., (1995) SEQPUP: A Biological Sequence Editor and Analysis Program for Macintosh Computer (Indiana University, Bloomington).
- 22.Valentin K. & Zetsche, K. (1990) Plant Mol. Biol. 15 575-584. [DOI] [PubMed] [Google Scholar]
- 23.Lockhart P. J., Steel, M. A., Hendy, H. D. & Penny, D. (1994) Mol. Biol. Evol. 11 605-612. [DOI] [PubMed] [Google Scholar]
- 24.Swofford D. L., (2002) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.0b8.
- 25.Felsenstein J. (1985) Evolution (Lawrence, Kans.) 39 783-791. [DOI] [PubMed] [Google Scholar]
- 26.Huelsenbeck J. P. & Ronquist, F. (2001) Bioinformatics 17 754-755. [DOI] [PubMed] [Google Scholar]
- 27.Huelsenbeck J. P., Ronquist, F., Nielsen, R. & Bollback, J. P. (2001) Science 294 2310-2314. [DOI] [PubMed] [Google Scholar]
- 28.Larget B. & Simon, D. L. (1999) Mol. Biol. Evol. 16 750-759. [Google Scholar]
- 29.Mickevich M. & Farris, J. S. (1981) Syst. Zool. 30 351-370. [Google Scholar]
- 30.Farris J. S., Kallersjo, M., Kluge, A. G. & Bult, C. (1994) Cladistics 10 315-319. [DOI] [PubMed] [Google Scholar]
- 31.Miadlikowska J. & Lutzoni, F. (2000) Int. J. Plant Sci. 161 925-958. [Google Scholar]
- 32.Shimodaira H. & Hasegawa, M. (1999) Mol. Biol. Evol. 16 1114-1116. [Google Scholar]
- 33.Butterfield N. J. (2000) Paleobiology 26 386-404. [Google Scholar]
- 34.Xiao S., Zhang, Y. & Knoll, A. H. (1998) Nature 391 553-558. [Google Scholar]
- 35.Wang D. Y., Kumar, S. & Hedges, S. B. (1999) Proc. R. Soc. London B 266 163-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-Trelles F., Tarrio, R. & Ayala, F. J. (2002) Proc. Natl. Acad. Sci. USA 99 8112-8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanderson M. J., (2002) r8s (University of California, Davis), Version 1.06 (beta).
- 38.Cutler D. J. (2000) Mol. Biol. Evol. 17 1647-1660. [DOI] [PubMed] [Google Scholar]
- 39.Albertano P., Ciniglia, C., Pinto, G. & Pollio, A. (2000) Hydrobiologia 433 137-143. [Google Scholar]
- 40.Cunningham C. W. (1997) Mol. Biol. Evol. 14 733-740. [DOI] [PubMed] [Google Scholar]
- 41.Barker F. K. & Lutzoni, F. M. (2002) Syst. Biol. 51 625-637. [DOI] [PubMed] [Google Scholar]
- 42.Bhattacharya D. (1997) in Origins of Algae and Their Plastids, ed. Bhattacharya, D. (Springer, New York), pp. 1–11.
- 43.Seckbach J. (1987) in Endocytobiology III, eds. Lee, J. J. & Frederick, J. F. (Ann. N.Y. Acad. Sci., New York), pp. 424–437.
- 44.Burger G., Saint-Louis, D., Gray, M. W. & Lang, B. F. (1999) Plant Cell 11 1675-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreira D., Le Guyader, H. & Phillippe, H. (2000) Nature 405 69-72. [DOI] [PubMed] [Google Scholar]
- 46.Cavalier-Smith T. (2000) Trends Plant Sci. 5 174-182. [DOI] [PubMed] [Google Scholar]
- 47.Gray M. W., Lang, B. F., Cedergren, R., Golding, G. B., Lemieux, C., Sankoff, D., Turmel, M., Brossard, N., Delage, E., Littlejohn, T. G., et al. (1998) Nucleic Acids Res. 26 865-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knoll A. H. (1992) Science 256 622-627. [DOI] [PubMed] [Google Scholar]
- 49.Xiao S., Knoll, A. H. & Yuan, X. (1998) J. Paleontol. 72 1072-1086. [Google Scholar]
- 50.Van de Peer Y. & De Wachter, R. (1997) J. Mol. Evol. 45 619-630. [DOI] [PubMed] [Google Scholar]
- 51.Baldauf S. L., Roger, A. J., Wenk-Siefert, I. & Doolittle, W. F. (2000) Science 290 972-977. [DOI] [PubMed] [Google Scholar]
- 52.Liaud M. F., Lichtle, C., Apt, K., Martin, W. & Cerff, R. (2000) Mol. Biol. Evol. 17 213-223. [DOI] [PubMed] [Google Scholar]
- 53.Saldarriaga J. F., Taylor, F. J., Keeling, P. J. & Cavalier-Smith, T. (2001) J. Mol. Evol. 53 204-213. [DOI] [PubMed] [Google Scholar]
- 54.Zhu G., Keithly, J. S. & Philippe, H. (2000) Int. J. Syst. Evol. Microbiol. 50 1673-1681. [DOI] [PubMed] [Google Scholar]
- 55.Andersson J. O. & Roger, A. J. (2002) Curr. Biol. 12 115-119. [DOI] [PubMed] [Google Scholar]
- 56.McFadden G. I. (1999) J. Eukaryotic Microbiol. 46 339-346. [DOI] [PubMed] [Google Scholar]
- 57.Durnford D. G., Deane, J. A., Tan, S., McFadden, G. I., Gantt, E. & Green, B. R. (1999) J. Mol. Evol. 48 59-68. [DOI] [PubMed] [Google Scholar]
- 58.Zwickl D. J. & Hillis, D. M. (2002) Syst. Biol. 51 588-598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.