Abstract
Several low-fidelity DNA polymerases have recently been discovered that are able to bypass DNA lesions during DNA synthesis in vitro. The efficiency and accuracy of lesion bypass is, however, both polymerase and lesion specific. For example, in vitro studies revealed that human DNA polymerase κ (Polκ) is unable to insert a base opposite a cis-syn thymine-thymine dimer or cisplatin adduct, yet can bypass some DNA lesions such as abasic site and acetylaminofluorene-adducted guanine in an error-prone manner. More importantly, Polκ is able to bypass benzo[a]pyrene (B[a]P)-adducted guanine accurately and efficiently. To investigate the biological function of Polκ, we have generated mouse embryonic stem (ES) cells deficient in the Polk gene encoding the enzyme. Polk-deficient ES cells grow normally and their sensitivities to UV and x-ray radiation are only slightly affected. In contrast, the mutant cells are highly sensitive to both killing and mutagenesis induced by B[a]P. Furthermore, the spectrum of mutations recovered in the Polk-deficient cells is different from that in the wild-type cells. Thus, our results indicate that Polκ plays an important role in suppressing mutations at DNA lesions generated by B[a]P.
Chromosomal DNA in living organisms is continually exposed to a vast variety of damaging agents from exogenous and endogenous sources. Most DNA lesions are repaired by one of multiple DNA repair pathways that are found in all prokaryotic and eukaryotic cells (1). However, repair is often slow; for example, between the two major DNA lesions produced by UV-irradiation, (6–4) photoproducts that generate substantial distortions in the DNA double-helix structure are rapidly detected and repaired, but cyclobutane pyrimidine dimers (CPDs) that distort the DNA to a lesser extent are repaired much more slowly (2). When the replication complex encounters a persisting lesion, it often stalls at the site because replicative DNA polymerases have a high selectivity for the correct normal base and cannot insert any base opposite most lesions. To avoid an aberrant cessation of the cell cycle caused by such a blockage of DNA replication, the stalled DNA polymerase needs to be transiently replaced by another DNA polymerase that is capable of bypassing the lesion (3). Because such lesion-bypass enzymes do not have a proofreading function, translesion synthesis (TLS) is inevitably accompanied by mutations at a frequency that depends on the type of DNA damage and the particular polymerase(s) involved.
Recently, a number of previously unrecognized DNA polymerases, which are likely to have evolved specially to carry out TLS, have been identified in both prokaryotes and eukaryotes (3–7). These enzymes are collectively designated the Y-family DNA polymerases (8), because they share multiple common motifs in their primary sequences that are distinct from those of the previously known A-, B-, C-, and X-families of DNA polymerases (9). Nevertheless, they retain tertiary structures conserved in most DNA polymerases, right-hand architecture with fingers, palm, and thumb subdomains (ref. 10 and references therein). Human cells have multiple Y-family enzymes, among which Polη bypasses thymine-thymine (T-T) CPDs efficiently and accurately in vitro, in most cases inserting two As opposite the T-T dimers (11, 12). Polη cannot, however, bypass past (6–4) photoproducts, which distort the DNA structure more than CPDs. Patients with the variant form of xeroderma pigmentosum (XP-V) lack Polη (13, 14). As a consequence they are highly mutable by UV light and predisposed to sunlight-induced skin cancer (15). These facts clearly demonstrate a crucial role for Polη in the response to UV damage. Another human Y-family enzyme, Polκ, exhibits a different specificity for lesion bypass in vitro (16–21). Polκ bypasses abasic sites and acetylaminofluorene adducts in an error-prone manner, but is not able to bypass either T-T CPDs or (6–4) photoproducts or cisplatin adducts (17).
Benzo[a]pyrene (B[a]P) and other polycyclic aromatic hydrocarbons (PAHs) are ubiquitous environmental carcinogens that are present in cigarette smoke and air pollutants (1, 22). B[a]P is thought to be responsible for the p53 mutations detected in the lung tumors of smokers (23). B[a]P and other PAHs are converted to active mutagens and carcinogens when metabolized by intracellular processes mediated by the arylhydrocarbon receptor (AhR; ref. 24). Activated B[a]P introduces bulky adducts into cellular DNA, predominantly at the 2-amino position of guanine and less frequently at the 6-amino position of adenine, and these lesions can, like UV-induced DNA damage, be repaired by nucleotide excision repair (NER). Under in vitro experimental conditions, Polη bypasses B[a]P-adducted guanines with low efficiency and fidelity (25, 26), but Polκ efficiently bypasses these lesions in a mostly error-free manner by inserting C opposite the bulky lesion (18, 27). We have recently shown that expression of the mouse Polk gene is, at least partially, under the control of AhR, being induced by 3-methylcholanthrene, a PAH (28). These findings have suggested that Polκ might participate in the bypass of DNA damage produced by B[a]P and other PAHs. Furthermore, Polκ expression is elevated in human lung cancer tissues compared with their matched normal tissue counterparts, whereas no apparent correlation was found between Polκ expression level and smoking history of the 29 patients examined (29).
To determine any biological significance from these findings related to Polκ and B[a]P, we constructed mutants from a mouse embryonic stem (ES) cell line with both copies of the Polk gene disrupted, and examined such mutants for sensitivity to various DNA-damaging agents, including B[a]P. We show here that Polk-deficient mutant cells are hypersensitive to B[a]P and accumulate more B[a]P-induced mutations than the parental cells. Thus, our results provide strong evidence that Polκ does play an important role in accurately bypassing DNA lesions induced by B[a]P in mammalian cells.
Materials and Methods
Construction of the Mouse Polk Gene Targeting Vector.
A λ phage genomic DNA library from a TT2 mouse ES cell line (an F1 embryo from a cross between a C57BL/6 female and a CBA male; ref. 30) was screened with a Polk-cDNA fragment. Phage clones that hybridized to the probe were subcloned into the vector pZERO2 (Invitrogen). Genomic structure of the mouse Polk gene was compared with the human POLK sequence (see AB040752 to AB040765 in the GenBank/DDBJ/EMBL DNA database). To construct pTKLS1 for targeting the mouse Polk gene, the 5-kb SpeI–XhoI fragment from intron 3 to intron 4 and the 3-kb SmaI–BamHI fragment in intron 6 were inserted into the SalI and XhoI site of the pLNTK vector (31), respectively (see Fig. 1b).
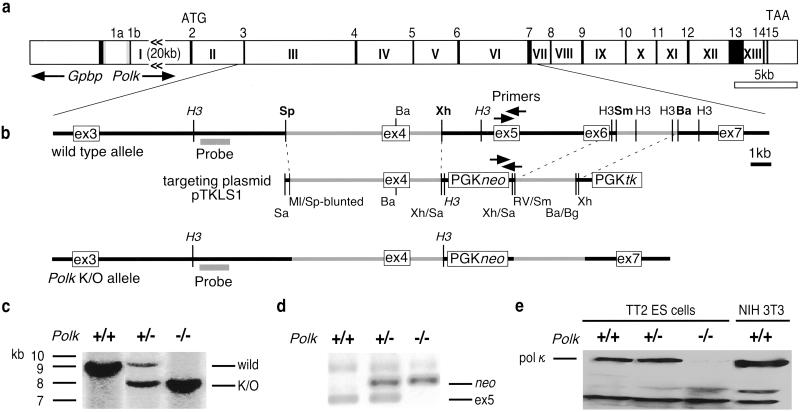
Fig 1.
Genomic structure of the mouse Polk gene and construction of Polk−/− ES cells. (a) Mouse Polk genomic structure. Arabic and roman numbers indicate exons and introns, respectively. The Gpbp gene, which encodes Goodpasture antigen binding protein, lies immediately upstream of the mouse Polk gene, as in the human chromosome (see AB036934 in the GenBank/DDBJ/EMBL DNA database). (b) Construction of Polk-knockout targeting plasmid pTKLS1. Restriction sites of the endogenous and targeting DNA are indicated: H3, HindIII; Sp, SpeI; Ba, BamHI; Xh, XhoI; Sm, SmaI; Sa, SalI; Ml, MluI; RV, EcoRV; Bg, BglII. The sites used for the construction are indicated in boldface. Phosphoglycerate kinase (PGK)-promoter-driven G418-resistance neo gene and ganciclovir-sensitive thymidine kinase (tk) gene are indicated as PGKneo and PGKtk, respectively. Arrows indicate PCR primers used to confirm the structure of the Polk gene. A 1-kb probe (denoted by gray line) was used to detect the 11- or 9.5-kb fragment from the intact or disrupted allele, respectively, after HindIII digestion (sites are indicated in italics) of ES cell genomic DNA. (c) DNA blot of Polk+/+ (TT2), Pol+/− (no. 48), and Polk−/− (A71) ES cells. Genomic DNA extracted from the cells was digested with HindIII and probed as indicated with radiolabeled DNA. Bands corresponding to the endogenous or the targeted allele are indicated. (d) PCR analysis of TT2, no. 48, and A71 ES cells. Disruption of Polk exon 5 was confirmed by PCR using exon 5-specific primers and PGKneo-amplifying primers (indicated by arrows in b). (e) Protein immunoblot analysis of TT2, no. 48, and A71 ES cells. Whole cell extracts (50 μg) prepared from ES cells and NIH 3T3 cells were separated by SDS/8% PAGE, transferred to a poly(vinylidene difluoride) (PVDF) membrane, and probed with anti-Polκ primary antibody and horseradish peroxidase (HRP)-conjugated donkey anti-rabbit secondary antibody. Polκ protein was detected by the ECL detection system (Amersham Pharmacia).
Generation of Polk-Targeted ES Cells.
To generate Polk-deficient ES cells, 20 μg of pTKLS1 DNA linearized by SalI cleavage was transfected into 2 × 107 TT2 ES cells by electroporation. The transfected cells were cultured in ES medium [DMEM, 15% FBS, and 1,000 units/ml murine leukemia inhibitory factor (ESGRO), both from GIBCO] containing G418 (300 μg/ml) and ganciclovir (1.5 μM) for 8 days. Resistant clones were identified and amplified. Polk+/− clones were identified by DNA blot analysis using a radiolabeled 1-kb genomic DNA fragment upstream of the 5′ SpeI site as a probe (see Fig. 1b). Of the 160 G418/ganciclovir-resistant clones, three clones (nos. 48, 131, and 151) had a disrupted Polk locus. To isolate Polk−/− ES cells, Polk+/− ES clones were cultured in medium containing 1.8 mg/ml G418 for 8 days, and resistant clones were analyzed by DNA blotting. Of the 43 clones thus obtained, four clones (A71 and A72, derivative of no. 48, and F21 and F22, derivative of no. 151) contained only the targeted Polk allele. Absence of the wild-type Polk allele was confirmed by PCR using the primer set of mmPolk5–5 (5′-GCTACTTCGAATTACCATGCAAGG-3′) and mmPolk5–3 (5′-CTCCTGTACTCACAGCTCTATATTTGTCAAA-3′) that amplify endogenous Polk exon5, or of G3 (5′-GGGCCAGCTCATTCCTCCACTCA-3′) and mmPolkSm (5′-GCAGGGTTGGAAACAGCCACAC-3′) that amplify the neomycin-resistance gene. The absence of Polκ protein was also confirmed by immunoblotting using anti-Polκ antibodies (28).
Establishment of Xpa−/− ES Cells.
The Xpa−/− ES cells were established by double targeting of the Xpa gene in F1/1 cells (32). The secondary targeting vector was constructed by replacing the neomycin-resistance gene of the primary targeting vector, pNeoXP5.7-TK (33) with the hygromycin-resistance gene. The secondary targeting vector was electroporated into the Xpa+/− ES cells, and G418 (175 μg/ml)- and hygromycin (110 μg/ml)-resistant ES clones were selected. Of 185 clones resistant to both G418 and hygromycin, one was targeted in both alleles and is Xpa−/− (data not shown). This cell line is 176-20.
Construction of Polk−/− ES Cells Expressing Wild-Type Polk cDNA.
To generate G418-sensitive Polk−/− ES cells, the PGKneo gene flanked by the loxP recombination sites in the targeted allele was removed by infection with adenovirus expressing the site-specific Cre recombinase protein (34). The isolated clone was designated A71-2. The wild-type Polk-expressing plasmid pc821 (35) was transfected into A71-2 ES cells. Transfected ES cells were selected with G418 (300 μg/ml). Of the 24 G418-resistant clones, six Polk-expressing clones (821-13, -16, -19, -22, and -23) were identified by RT-PCR. These clones were tested for sensitivity to 40 μM B[a]P. All clones were less sensitive than parental A71-2 cells, and one clone (821-23) was examined for B[a]P sensitivity in detail.
Cell Survival Assay.
The sensitivity of Polk-targeted ES cells to ionizing radiation, UV, and B[a]P was determined by measuring the colony-forming efficiency of cells treated over a range of doses. Cells were grown on feeder plates supplied with ES medium and maintained by passaging every third day to keep logarithmic-phase growth. Trypsinized cells were suspended in ES medium or PBS and exposed to ionizing or UV irradiation. Control and irradiated cells were plated on feeder plates. Before B[a]P treatments, cells were allowed to adhere to feeder plates for 6 h and then treated with the drug for 20 h. B[a]P (Nacalai Tesque, Kyoto) was prepared in DMSO and activated by the S-9 fraction of rat liver homogenates (Japan Immunoresearch Laboratories, Gunma, Japan). Control cells were also treated with S-9 (0.1% DMSO/0.1% S-9). After treatment, cells were grown for 10–14 days on feeder plates in ES medium containing 1 mM caffeine, fixed, stained, and counted. All experiments were carried out in triplicate; data points are indicated as mean with standard error.
B[a]P-Induced Mutation Frequency and Mutation Spectrum at the Hprt Locus.
B[a]P-treated cells were cultured for 8 days to express the Hprt-defective phenotype. After this expression period, 5 × 105 cells were plated on a gelatinized 10-cm dish (without feeder cells) in the presence of ES medium containing 40 μM 6-thioguanine (6-TG, Nacalai Tesque). Several dilution cultures were also plated without 6-TG medium to calculate the colony-forming efficiency. After selection for 2 weeks, 6-TG-resistant (6-TGR) colonies were counted and cloned. 6-TGR cells were amplified and total RNA was isolated. RT-PCR and direct sequencing were performed as described (35).
Results
Construction of Polk-Deficient Cells.
The mouse Polk and human POLK genes have a similar structure that includes 15 exons (Fig. 1a and our unpublished data). Exons 5 and 6 include the conserved motif sequences II and III, respectively, which are essential for enzymatic activity (16). A targeting plasmid, pTKLS1, was constructed by replacing a 10-kb genomic fragment, including exons 5 and 6 of the Polk gene, with the PGKneo gene (see Materials and Methods). The plasmid also contains the HSV-tk gene for counterselection against nonspecific integrants by ganciclovir. pTKLS1 was transfected into TT2 ES cells by electroporation and cells resistant to both G418 (300 μg/ml) and ganciclovir (1 mM) were selected. One hundred and sixty clones were selected and characterized by Southern blotting and PCR analysis (Fig. 1 c and d). The Polk+/−clones thus identified were cultured in ES medium containing 1.8 mg/ml G418 to select for clones with higher resistance to the drug. Forty-three clones survived the higher G418 dose and four of these clones (A71 and A72 from no. 48, and F21 and F22 from no. 151) had a disruption in both copies of the Polk gene (Fig. 1 c and d). Polκ protein was undetectable in the Polk−/− cells by Western blot analysis (Fig. 1e).
Properties of Polk-Deficient Cells.
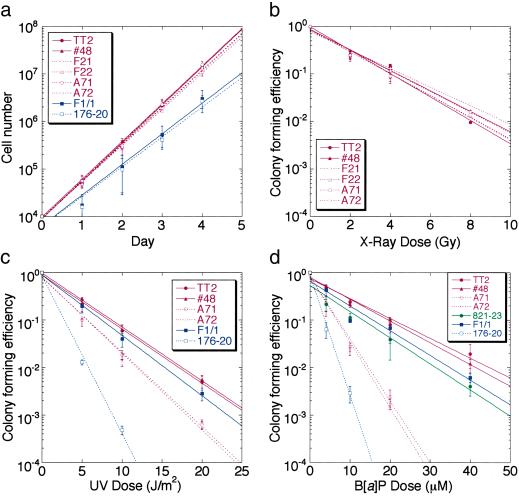
The Polk-deficient cells and the parental Polk-proficient cells proliferated at similar rates on feeder plates (Fig. 2a). Another wild-type mouse ES cell line, F1/1 (32), and an Xpa−/− derivative of this cell line, 176-20, both grew at a slightly slower rate than TT2 cells or cells derived from TT2 (Fig. 2a). Xpa−/− cells are defective in NER and consequently very sensitive to various DNA-damaging agents (33). The data shown in Fig. 2a demonstrate unequivocally that neither Polκ nor Xpa is required for cell proliferation. Polk+/+, Polk+/−, and Polk−/− had similar levels of sensitivity to x-rays; in contrast, Polk−/− cells were slightly but significantly sensitive to UV. Polk−/− cells were much less sensitive to UV than were NER-deficient Xpa−/− cells. The UV sensitivity associated with the Polk−/− phenotype is a somewhat unexpected result, because previous in vitro results (16–19) have indicated that Polκ by itself does not bypass UV-induced DNA damage. Our results implicate at least a minor role for Polκ in the in vivo bypass of UV damage.
Fig 2.
Growth of Polk−/− ES cells and sensitivity to DNA-damaging treatments. (a) Growth curve. Approximately 5 × 104 wild-type (TT2 and F1/1), Polk+/− (no. 48), Polk−/− (F21, F22, A71, and A72), and Xpa−/− (176-20 derivative of F1/1) ES cells were plated on feeder cells and the number of viable cells was counted at indicated times after plating. (b) X-ray sensitivity. Trypsinized cells were x-irradiated and allowed to form colonies on feeder cells. The relative colony-forming efficiency is plotted against the x-ray dose. (c) UV sensitivity. Trypsinized cells were resuspended in PBS before UV irradiation. The relative colony-forming efficiency is plotted against the UV dose. (d) B[a]P sensitivity. Cells were allowed to adhere to feeder plates 6 h before drug treatment. After 20 h in culture in B[a]P/S-9-containing ES medium, cells were washed three times with PBS and then cultured in ES medium. Rescue of B[a]P sensitivity of the Polk−/− (A71) cells by a Polk-expressing cDNA was also examined. Construction of the 821-23 ES cell line expressing the wild-type Polk cDNA is described in Materials and Methods. All experiments were carried out in triplicate and each data point is the mean of three independent measurements. Error bars indicate standard error.
Polk−/− cells were also examined for sensitivity to B[a]P and compared with Xpa−/− cells. Remarkably Polk−/− cells were three times more sensitive to B[a]P treatment than Polk+/+ and Polk+/− cells (Fig. 2d). This sensitivity approached the 5-fold hypersensitivity of Xpa−/− cells, which are completely unable to remove B[a]P adducts. In most cases, the B[a]P sensitivity of Polk−/− cells was restored to normal when cells were transfected with a clone expressing wild-type Polκ. A typical result with one such case (821-23) is shown in Fig. 2d. The results shown in Fig. 2d provide very strong evidence for a major role for Polκ in the response of mammalian cells to B[a]P damage.
Frequencies of B[a]P-Induced Mutations in Polk-Proficient and -Deficient Cells.
In view of the striking hypersensitivity of Polk−/− cells to killing by B[a]P, we next investigated their response to its mutagenic effects. B[a]P-induced mutagenesis was studied in Polk-proficient and -deficient cells by measuring the incidence of mutations at the Hprt locus that confer 6-TG resistance (Fig. 3a). The frequency of spontaneous 6-TGR mutations was very low (<10−6) in the wild-type and Polk−/− cells, and much higher in Xpa−/− cells (7 × 10−6). As expected, Xpa−/− cells were highly prone to B[a]P-induced mutagenesis. When exposed to 0.5 μM B[a]P, Xpa−/− cells had a mutation frequency of 1.3 × 10−4, which did not increase significantly with higher doses. In Polk−/− cells, the mutation frequency was ≈1 × 10−6 in cells exposed to 0.5 μM B[a]P and it increased to ≈1 × 10−4 in cells exposed to 10 μM B[a]P (>100-fold higher than the spontaneous mutation frequency). At saturation, the mutation frequency was similar in Xpa−/− and Polk−/− cells. Wild-type and Polk+/− cells were ≈10-fold less sensitive to B[a]P-induced mutagenesis than Polk-deficient cells (mutation frequency of 1 × 10−5 at 10 μM B[a]P; Fig. 3a and also see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). These data show that Polκ protects cells not only from the lethal effects of B[a]P, but also from its mutagenic effects.
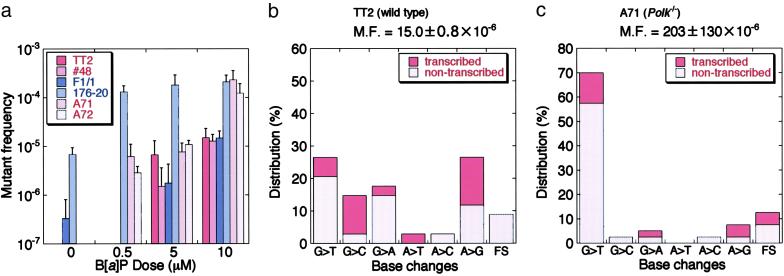
Fig 3.
Frequency and pattern of 6-TGR mutations induced by B[a]P. (a) Induction of 6-TGR mutants after treatment with different doses of B[a]P. (b) 6-TGR mutations in the TT2 ES cells induced by 10 μM B[a]P treatment. Thirty-seven independent clones were analyzed, of which 31 base substitutions, 3 frameshifts (FSs), and 5 unidentified splicing mutations were identified. Ratio of base substitution, FS mutations, and strand with the purine presumably adducted by B[a]P are indicated. MF, 6-TGR mutant frequency. (c) 6-TGR mutation pattern in Polk−/− A71 ES cells induced by 10 μM B[a]P treatment. A total of 38 independent clones were analyzed, and 35 base substitutions, 5 FSs, and 2 unidentified splicing mutations were observed. One mutant had no mutation in the sequence region analyzed.
B[a]P-Induced Mutations Generated in Polk-Proficient and -Deficient Cells.
To gain further insight into the mutations induced by B[a]P in the presence and absence of Polκ, we analyzed the mutation spectrum in the wild-type and Polk−/− cells treated with B[a]P. About 40 clones each resistant to 6-TG were isolated from the wild-type and Polk−/− cells treated with 10 μM B[a]P, and Hprt mRNAs were amplified by RT-PCR for direct DNA sequencing. With one exceptional case, single or multiple alterations were identified in the Hprt-cDNA sequence analyzed, and the results are summarized in Fig. 3 b and c (also see Table 2, which is published as supporting information on the PNAS web site). Base-substitution mutations predominated over frameshift (FS) mutations in both cell types; the ratio of base-substitution to FS mutations was 31:3 and 35:5 in wild-type and Polk−/− cells, respectively (see Table 2). Such base substitutions occurred at G about twice more frequently than at A in wild-type cells (20 vs. 11), but they occurred predominantly at G in Polk−/− cells (31 vs. 4). Furthermore, whereas G-to-T transversions and A-to-G transitions were observed almost equally in wild-type cells (9 and 8, respectively, of the total 31 mutations), G-to-T transversions were overwhelmingly predominant in Polk−/− cells (28 of 35). Another feature of the mutation spectra is that the majority of mutations can be attributed to adduct in the nontranscribed strand. The ratio of mutations from the nontranscribed strand vs. the transcribed strand is 19:12 in the wild-type cells and 27:8 in Polk−/− cells. This result implies that the bulky adducts on the transcribed strand are repaired more rapidly than those on the nontranscribed strand by the transcription-coupled repair mechanism (36–38). From these results, we conclude that Polκ plays a crucial role in correctly bypassing dG-N2-B[a]P-7,8-diol-9,10-epoxide (BPDE) adducts, the major products generated by B[a]P, and that in Polk-deficient cells another TLS enzyme(s) is involved in the bypass in an error-prone manner, generating mostly G-to-T transversions.
Discussion
The results presented here show that Polk−/− cells are hypersensitive to both the lethal and the mutagenic effects of B[a]P. This phenotype is reminiscent of the response of Polη-deficient XP-V cells to UV light, although Polk−/− cells are more sensitive to the killing effects of B[a]P than are XP-V cells to UV light. The increased mutability of Polk−/− cells in response to B[a]P provides a further example of the apparently paradoxical situation of an error-prone DNA polymerase actually protecting cells from inducing mutations. This finding can be explained by the specificities of the enzymes in vitro when presented with different damaged templates. Polκ is able to carry out TLS past B[a]P-adducted guanines efficiently in vitro, inserting predominantly C opposite each of the four different stereoisomers of dG-N2-BPDE adducts that result from reaction of DNA with anti-BPDE (27). For example, the insertion efficiency (Fins as measured by Vmax/Km in steady-state kinetic experiments) of correct C opposite (+)-trans-dG-N2-BPDE (the predominant product by anti-BPDE, ref. 35) was two to three orders of magnitude higher than that of A, G, or T misinsertion opposite the lesion. Moreover, the extension efficiency (Fext) from C/(+)-trans-dG-N2-BPDE pair is also higher that that of A, G, or T paired with the same lesion by 230-, 1,100- or 27-fold, respectively. Consequently, the overall bypass efficiency (Fins × Fext) for C/(+)-trans-dG-N2-BPDE is at least three orders of magnitude higher than that of the other pairs. From these in vivo and in vitro results, we conclude that Polκ is crucial for accurate bypass of dG-N2-BPDE adducts, and its absence leads to less efficient and more error-prone bypass of the major BPDE adducts by other TLS enzyme(s).
It seems likely that Polη is involved in the mutagenic bypass of dG-N2-BPDE adducts, especially when Polκ is absent. An in vitro experiment (26) showed that Polη inserted A, G, or T more frequently (by 22-, 5.5-, or 1.5-fold, respectively) than C opposite (+)-trans-dG-N2-BPDE (= Trans S stereoisomer in ref. 26), although in a sequence context (5′-CG*A-3′, in which G* is the modified base) different from that used for the above experiment with Polκ (5′-CG*C-3′). The extension efficiency (Fext) from A, G, or T paired with the lesion was a few- to severalfold higher than that of the correct C pair. Our finding that the big majority of the mutations (28 of 35) induced by B[a]P in Polk-deficient ES cells were G-to-T transversions is consistent with the idea that Polη carries out TLS past dG-N2-BPDE adducts in the absence of Polκ. The overall bypass efficiency (Fins × Fext) of (+)-trans-dG-N2-BPDE by Polη was highest when A was incorporated opposite the lesion, but the value was less than 10−5 of that for C incorporation opposite a nondamaged template by Polη (26). This result is in sharp contrast to the situation with T-T CPD; Polη synthesizes DNA opposite a CPD T-T dimer with the same accuracy and efficiency as opposite the nondamaged DNA template in vitro (11, 12). Polη appears to have a structure adapted for bypassing CPD and it can also bypass various other lesions correctly at reduced efficiencies (20, 39, 40), but dG-N2-BPDE adducts seem to be very poor substrates for the enzyme to bypass correctly. More recently, Rechkoblit et al. (41) compared the bypass of (+)- and (−)-trans-dG-N2-BPDE in the same sequence context by human Polκ, Polη, Polι, and yeast Polζ, although not quantitatively by steady-state kinetic experiments. The overall bypass of either of the two stereoisomers was higher with Polκ than with Polη; however, Fins appeared to be to the contrary. Polη inserted a base indiscriminately, especially A in preference to others, opposite either one of the above two dG-N2-BPDE adducts, whereas Polκ inserted the correct C predominantly at lower efficiency. Neither Polι nor yeast Polζ, either alone or in combination, was effective in TLS past the dG-N2-BPDE adducts.
The human REV1 protein, another Y-family enzyme, was shown to insert C opposite (+)-trans-dG-N2-BPDE in a sequence context (5′-CG*C-3′), whereas not extending any further from the inserted site (42). The Fins was 0.15 of that for C insertion opposite the nondamaged template. This value is much higher than the corresponding value of 2.8 × 10−3 for Polκ (27). Although no experimental evidence is currently available to support a role for REV1 in the bypass of BPDE-adducts in vivo, it is possible that the correct bypass of dG-N2-BPDE adducts might be carried out in vivo not only by Polκ alone as discussed above, but also by the two-polymerase two-step mechanism, i.e., the insertion of C opposite the lesions by REV1 and subsequent extension by Polκ (42, 43). Mutagenic bypass of dG-N2-BPDE adducts in wild-type cells may be because of sequential action of Polη and Polκ (43). Polη inserts A, G, or T more frequently than C opposite dG-N2-BPDE adducts, but extending further less effectively (41), and subsequent extension of the misincorporated bases by Polκ may lead to generating base-substitution mutations. The increased proportions of G-to-T transversions in the mutations among the survivors of B[a]P-treated Polk-deficient ES cells (Fig. 3c) may reflect in vivo bypass of dG-N2-BPDE adducts that depends mainly on Polη. It is not known whether human Polζ can insert a base opposite the dG-N2-BPDE adducts or extend from an inserted base by other enzyme(s) because the human Polζ enzyme has not yet been purified.
Until recently, it has not been known which enzyme(s) is involved in the bypass of dA-N6-BPDE adducts, the minor products generated by B[a]P. A very recent paper by Rechkoblit et al. (41) reported that Polη bypassed (+)-trans-dA-N6-BPDE adduct but not (−)-trans-dA-N6-BPDE adduct in vitro, whereas Polκ was completely blocked by both of the lesions. They also showed that neither human Polι nor yeast Polζ, either alone or in combination, was effective in TLS past the dA-N6-BPDE adducts, although Polι was able to insert T opposite (−)-trans-dA-N6-BPDE but not opposite (+)-trans-dA-N6-BPDE adduct. Another study by a different group (E. Frank, J. Sayer, D. Jerina, and R. Woodgate, personal communication) indicated that Polι inserted T efficiently opposite (+)- and (−)-trans-dA-N6-BPDE adducts in a different sequence context, with a misincorporation frequency ranging between 2 × 10−3 and 6 × 10−4, although further extension was relatively poor. Again, it seems possible that Polκ might be involved in catalyzing extension from a base inserted opposite dA-N6-BPDE adducts by Polι.
Our finding that Polk-deficient ES cells show moderate sensitivity to UV radiation (Fig. 2c) was unexpected, because all of the in vitro data (16–19) have indicated that Polκ by itself cannot bypass T-T CPD or (6–4) photoproducts. Our in vitro experiment (17) indicated that, at a high enzyme concentration, Polκ inefficiently inserted one base opposite the 3′T of a T-T (6–4) photoproduct, but could not subsequently insert a base opposite the 5′T of the lesion. At the same enzyme concentration, synthesis by Polκ was completely blocked at one base before 3′T of a T-T CPD. However, a recent paper (44) has shown that Polκ can efficiently catalyze extension from G or A (but not from C or T) inserted opposite the 3′T of a T-T CPD, whereas it does not extend from any base inserted opposite the 3′T of a T-T (6–4) photoproduct. Our in vivo results shown in Fig. 2c, taken together with these in vitro data, suggests that Polκ plays a role in bypass of UV-induced damage, albeit a minor one compared with that of Polη, and its role might become more important when Polη is absent, as in XP-V cells.
In conclusion, our results provide strong evidence that Polκ plays a major role in protecting cells from the genotoxic effects of B[a]P, and, by implication, of other PAHs. This finding is also consistent with our earlier observations that the promoter region of the mouse Polk gene contains two copies of the arylhydrocarbon receptor binding site and that Polκ is induced on exposure of mice to a polycyclic hydrocarbon (28).
Supplementary Material
Acknowledgments
We thank M. Fukuda of the Institute for Virus Research for her excellent technical assistance, and E. Ohashi, M. Moriya, R. Woodgate, and A. R. Lehmann for valuable comments that improved the manuscript. This work was supported in part by Grants-In-Aid for Scientific Research of Priority Area from the Ministry of Education, Culture, Sports, and Science of Japan.
Abbreviations
B[a]P, benzo[a]pyrene
BPDE, B[a]P-7,8-diol-9,10-epoxide
CPD, cyclobutane pyrimidine dimer
ES, embryonic stem
NER, nucleotide excision repair
PAH, polycyclic aromatic hydrocarbon
Polκ, DNA polymerase κ
6-TG, 6-thioguanine
6-TGR, 6-TG resistance
TLS, translesion synthesis
XP-V, xeroderma pigmentosum variant
Fins, insertion efficiency
Fext, extension efficiency
References
- 1.Friedberg E. C., Walker, G. C. & Siede, W., (1995) DNA Repair and Mutagenesis (Am. Soc. Microbiol., Washington, DC).
- 2.Katsumi S., Kobayashi, N., Imoto, K., Nakagawa, A., Yamashina, Y., Muramatsu, T., Shirai, T., Miyagawa, S., Sugiura, S., Hanaoka, F., et al. (2001) J. Invest. Dermatol. 117 1156-1161. [DOI] [PubMed] [Google Scholar]
- 3.Woodgate R. (1999) Genes Dev. 13 2191-2195. [DOI] [PubMed] [Google Scholar]
- 4.Johnson R. E., Washington, M. T., Prakash, S. & Prakash, L. (1999) Proc. Natl. Acad. Sci. USA 96 12224-12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodman M. F. & Tippin, B. (2000) Nat. Rev. Mol. Cell Biol. 1 101-109. [DOI] [PubMed] [Google Scholar]
- 6.Friedberg E. C., Feaver, W. J. & Gerlach, V. L. (2000) Proc. Natl. Acad. Sci. USA 97 5681-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgers P. M., Koonin, E. V., Bruford, E., Blanco, L., Burtis, K. C., Christman, M. F., Copeland, W. C., Friedberg, E. C., Hanaoka, F., Hinkle, D. C., et al. (2001) J. Biol. Chem. 276 43487-43490. [DOI] [PubMed] [Google Scholar]
- 8.Ohmori H., Friedberg, E. C., Fuchs, R. P., Goodman, M. F., Hanaoka, F., Hinkle, D., Kunkel, T. A., Lawrence, C. W., Livneh, Z., Nohmi, T., et al. (2001) Mol. Cell 8 7-8. [DOI] [PubMed] [Google Scholar]
- 9.Ito J. & Braithwaite, D. K. (1991) Nucleic Acids Res. 19 4045-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beard W. A. & Wilson, S. H. (2001) Structure (London) 9 759-764. [DOI] [PubMed] [Google Scholar]
- 11.Masutani C., Araki, M., Yamada, A., Kusumoto, R., Nogimori, T., Maekawa, T., Iwai, S. & Hanaoka, F. (1999) EMBO J. 18 3491-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson R. E., Washington, M. T., Prakash, S. & Prakash, L. (2000) J. Biol. Chem. 275 7447-7450. [DOI] [PubMed] [Google Scholar]
- 13.Masutani C., Kusumoto, R., Yamada, A., Dohmae, N., Yokoi, M., Yuasa, M., Araki, N., Iwai, S., Takio, K. & Hanaoka, F. (1999) Nature 399 700-704. [DOI] [PubMed] [Google Scholar]
- 14.Johnson R. E., Kondratick, C. M., Prakash, S. & Prakash, L. (1999) Science 285 263-265. [DOI] [PubMed] [Google Scholar]
- 15.Maher V., Ouellette, L. M., Curren, R. D. & McCormick, J. J. (1976) Nature 261 593-595. [DOI] [PubMed] [Google Scholar]
- 16.Johnson R. E., Prakash, S. & Prakash, L. (1999) Proc. Natl. Acad. Sci. USA 97 3838-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohashi E., Ogi, T., Kusumoto, R., Iwai, S., Masutani, C., Hanaoka, F. & Ohmori, H. (2000) Genes Dev. 14 1589-1594. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Yuan, F., Wu, X., Wang, M., Rechkoblit, O., Taylor, J.-S., Geacintov, N. E. & Wang, Z. (2000) Nucleic Acids Res. 28 4138-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlach V. L., Feaver, W. J., Fischhaber, P. L. & Friedberg, E. C. (2001) J. Biol. Chem. 276 92-98. [DOI] [PubMed] [Google Scholar]
- 20.Levine R. L., Miller, H., Grollman, A., Ohashi, E., Ohmori, H., Masutani, C., Hanaoka, F. & Moriya, M. (2001) J. Biol. Chem. 276 18717-18721. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki N., Ohashi, E., Hayashi, K., Ohmori, H., Grollman, A. P. & Shibutani, S. (2001) Biochemistry 40 15176-15183. [DOI] [PubMed] [Google Scholar]
- 22.Phillips D. H. (1983) Nature 303 468-472. [DOI] [PubMed] [Google Scholar]
- 23.Denissenko M. F., Pao, A., Tang, M. & Pfeifer, G. P. (1996) Science 274 430-432. [DOI] [PubMed] [Google Scholar]
- 24.Hankinson O. (1995) Annu. Rev. Pharmacol. Toxicol. 35 307-340. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Yuan, F., Wu, X., Rechkoblit, O., Taylor, J.-S., Geacintov, N. E. & Wang, Z. (2000) Nucleic Acids Res. 28 4717-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiapperino D., Kroth, H., Kramarczuk, I. H., Sayer, J. M., Masutani, C., Hanaoka, F., Jerina, D. M. & Cheh, A. M. (2002) J. Biol. Chem. 277 11765-11771. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki N., Ohashi, E., Kolbanovskiy, A., Geacintov, N. E., Grollman, A. P., Ohmori, H. & Shibutani, S. (2002) Biochemistry 41 6100-6106. [DOI] [PubMed] [Google Scholar]
- 28.Ogi T., Mimura, J., Hikida, M., Fujimoto, H., Fujii-Kuriyama, Y. & Ohmori, H. (2001) Genes Cells 6 943-953. [DOI] [PubMed] [Google Scholar]
- 29.O-Wang J., Kawamura, K., Tada, Y., Ohmori, H., Kimura, H., Sakiyama, S. & Tagawa, M. (2001) Cancer Res. 61 5366-5369. [PubMed] [Google Scholar]
- 30.Yagi T., Tokunaga, T., Furuta, Y., Nada, S., Yoshida, M., Tsukada, T., Saga, Y., Takeda, N., Ikawa, Y. & Aizawa, S. (1993) Anal. Biochem. 214 70-76. [DOI] [PubMed] [Google Scholar]
- 31.Gorman J. R., van der Stoep, N., Monroe, R., Cogne, M., Davidson, L. & Alt, F. W. (1996) Immunity 5 241-252. [DOI] [PubMed] [Google Scholar]
- 32.Tokunaga T. & Tsunoda, Y. (1992) Dev. Growth Differ. 34 561-566. [DOI] [PubMed] [Google Scholar]
- 33.Nakane H., Takeuchi, S., Yuba, S., Saijo, M., Nakatsu, Y., Murai, H., Nakatsuru, Y., Ishikawa, T., Hirota, S., Kitamura, Y., et al. (1995) Nature 377 165-168. [DOI] [PubMed] [Google Scholar]
- 34.Kanegae Y., Takamori, K., Sato, Y., Lee, G., Nakai, M. & Saito, I. (1996) Gene 181 207-212. [DOI] [PubMed] [Google Scholar]
- 35.Ogi T., Kato, T., Jr., Kato, T. & Ohmori, H. (1999) Genes Cells 4 607-618. [DOI] [PubMed] [Google Scholar]
- 36.Cheng S. C., Hilton, B. D., Roman, J. M. & Dipple, A. (1989) Chem. Res. Toxicol. 2 334-340. [DOI] [PubMed] [Google Scholar]
- 37.Denissenko M., Pao, A., Pfeifer, G. P. & Tang, M.-S. (1998) Oncogene 16 1241-1247. [DOI] [PubMed] [Google Scholar]
- 38.Hanawalt P. C. (1994) Science 266 1957-1958. [DOI] [PubMed] [Google Scholar]
- 39.Masutani C., Kusumoto, R., Iwai, S. & Hanaoka, F. (2000) EMBO J. 19 3100-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kusumoto R., Masutani, C., Iwai, S. & Hanaoka, F. (2002) Biochemistry 41 6090-6099. [DOI] [PubMed] [Google Scholar]
- 41.Rechkoblit O., Zhang, Y., Guo, D., Wang, Z., Amin, S., Kreminsky, J., Louneva, N. & Geacintov, N. E. (2002) J. Biol. Chem. 277 30488-30494. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y., Wu, X., Rechkoblit, O., Geacintov, N. E., Taylor, J.-S. & Wang, Z. (2002) Nucleic Acids Res. 30 1630-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Wu, X., Guo, D., Rechkoblit, O. & Wang, Z. (2002) DNA Repair 1 559-569. [DOI] [PubMed] [Google Scholar]
- 44.Washington M. T., Johnson, R. E., Prakash, L. & Prakash, S. (2002) Proc. Natl. Acad. Sci. USA 99 1910-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.