Abstract
Among numerous established human hepatoma cell lines, none has been shown susceptible to hepatitis B virus (HBV) infection. We describe here a cell line, called HepaRG, which exhibits hepatocyte-like morphology, expresses specific hepatocyte functions, and supports HBV infection as well as primary cultures of normal human hepatocytes. Differentiation and infectability are maintained only when these cells are cultured in the presence of corticoids and dimethyl sulfoxide. The specificity of this HBV infection model was ascertained by both the neutralization capacity of HBV-envelope protein-specific antibodies and the competition with an envelope-derived peptide. HepaRG cells therefore represent a tool for deciphering the mechanism of HBV entry. Moreover, their close resemblance to normal human hepatocytes makes them suitable for many applications including drug metabolism studies.
Hepatitis B, one of the major infectious diseases worldwide, is caused by a small enveloped DNA virus, the hepatitis B virus (HBV). HBV exhibits a very narrow host range and shows a strong tropism for liver parenchymal cells. It has therefore been assumed that susceptibility to HBV infection is restricted to differentiated cells. Accordingly, it was found that only human hepatocyte primary cultures were susceptible to HBV infection (1–4). However, the use of this model is hampered by the limited availability and the inherent variability of human liver material. Even though several human hepatoma-derived cell lines support HBV replication after HBV DNA transfection (5–9), none of them are susceptible to HBV infection.
We describe here a hepatoma-derived cell line that expresses a representative panel of liver-specific genes and is susceptible to HBV infection. This goal was achieved by combining an original selection procedure applied early after the cell line establishment in culture and the use of appropriate culture conditions, allowing the commitment of these cells to an optimal differentiation status.
Methods
Isolation of the Cell Line and Culture Conditions.
Cells were isolated from a liver tumor of a female patient suffering from hepatocarcinoma and hepatitis C infection. All experimental procedures were conducted in conformity with French laws and regulations and were approved by the National Ethics Committee. The samples were minced into small pieces, washed with Hepes buffer (pH 7.7; 140 mM NaCl/2.68 mM KCl/0.2 mM Na2HPO4/10 mM Hepes), and digested with 0.025% collagenase D (Boehringer Mannheim) diluted in the same buffer supplemented with 0.075% CaCl2 under gentle stirring at 37°C. The cell suspension was washed twice in Hepes buffer and resuspended in a William's E medium supplemented with 10% FCS, 100 units/ml penicillin, 100 μg/ml streptomycin, 5 μg/ml insulin, and 5 × 10−7 M hydrocortisone hemisuccinate. Cell suspension was distributed in several dishes without any coating feeder layer. After several weeks, cell growth was sufficient to fulfill the culture dishes. Cells appeared well differentiated, with a hepatocyte-like morphology. Dishes having the most homogenous cell population were passaged by gentle trypsinization. After three passages, all cells were aliquoted and frozen in 10% DMSO and kept in liquid nitrogen vapors. After thawing, cells originating from one single dish showing a high proportion of cells with a hepatocyte morphology were further passaged in the culture medium used for their isolation.
The selection procedure relies on two main steps: (i) induction of cell differentiation with DMSO, and (ii) partial purification of the differentiated cells. Induction of differentiation was performed by treating a dish containing confluent cells with the culture medium supplemented with 2% DMSO and hydrocortisone 5 × 10−5 M for 4 weeks. The differentiated cells, showing a typical aspect of hepatocytes clustered in small colonies, were then selectively isolated from nondifferentiated cells. The procedure includes a brief trypsinization, which allows the hepatocyte-like cells to remain aggregated, whereas nondifferentiated cells were mostly present as single cells. To this end, the cell culture was washed with a phosphate saline buffer and then incubated with a commercial trypsin–EDTA solution (Invitrogen) at 37°C. When most cells were detached from the substratum, 25% FCS was added to neutralize trypsin activity. Cells were then gently collected and allowed to sediment for 10 min at 1 × g. The pellet consisting mostly of aggregated cells was resuspended, and the sedimentation procedure was repeated once. Cells selected by this procedure were plated in new dishes.
Cells were passaged every 2 weeks (1/5 dilution) by trypsinization. The phenotypic stability of the cell line was greatly favored by continuously culturing cells in the presence of a high hydrocortisone concentration (5 × 10−5 M). Therefore, the growth medium was composed of William's E medium supplemented with 10% FCS, 100 units/ml penicillin, 100 μg/ml streptomycin, 5 μg/ml insulin, and 5 × 10−5 M hydrocortisone hemisuccinate. For the routine differentiation process, a two-step procedure was used. Cells were first maintained for 2 weeks in the growth medium. Then, they were maintained in the differentiation medium (the same culture medium supplemented with 2% DMSO) for 2 more weeks. The medium was renewed every 2 or 3 days.
HBV Infection of Cell Culture.
As an infectious inoculum, a 50-fold concentrated culture supernatant of HepG2 clone 2.2.15 cells (5) was preferentially used because of an unlimited supply and a constant quality. It was prepared from freshly collected supernatants by precipitating viral particles in the presence of 6% polyethylene glycol (PEG). The pellet was resuspended in PBS containing 25% FCS. Aliquots were stored at −80°C. A concentration of 4 × 109 HBV genome equivalent per milliliter was determined by dot-blot for the inoculum; ≈106 differentiated HepaRG cells were incubated with 100 μl of this concentrated infectious source, 10-fold diluted in 1 ml of culture medium supplemented with 4% PEG 8000 (Sigma), for 20 h at 37°C. At the end of the incubation, cells were washed three times with the culture medium, maintained in the presence of 2% DMSO and 5 × 10−5 M hydrocortisone hemisuccinate, and harvested at indicated times.
Neutralization assays were performed by incubating viruses with the monoclonal hepatitis B surface antibody S 39–10 (gift of A. Budkowska, Pasteur Institute, Paris) for 1 h at room temperature before infection. Peptide competition was performed by preincubating cells with chemically synthesized myristoylated HBV preS2–78 peptide for 30 min before the addition of the inoculum. Myristoylated duck hepatitis B virus-derived peptide (AA 2–41) (10) was used as a negative control.
Enzyme Assays.
Phenacetin deethylation to paracetamol, oxidation of nifedipine to the derivative 3,5-dimethoxycarbonyl-2,6-dimethyl-4-(2-nitro-phenyl)pyridine, dextromethorphan demithylation to dextrorphan tartrate, tolbutamide hydroxylation to hydroxytolbutamide, and 4-mephenytoin hydroxylation to 4-hydroxy-mephenytoin were determined after 2–16 h of incubation with human hepatocytes by high performance liquid chromatography according to methods detailed elsewhere (11, 12). The concentrations of substrates were 2 × 10−4 mol/liter for phenacetin, 2 × 10−4 mol/liter for nifedipine, 2 × 10−4 mol/liter for dextromethorphan and mephenytoin, and 1 mmol/liter for tolbutamide. Results were expressed as picomoles of metabolites formed per hour and per microgram of DNA. GST activity was estimated using 1-chloro-2,4-dinitrobenzene (Merck) as a substrate at pH 6.5 and room temperature (13).
Assay for HBV-Specific Protein.
Hepatitis B surface antigen (HBsAg) was detected in the medium by an ELISA kit (Monolisa AgHBs plus) obtained from Bio-Rad.
Viral DNA Extraction and Analysis.
Viral replicative DNA intermediates were isolated from whole cell lysates. Cells recovered after trypsinization and one wash were lysed for 16 h at 37°C in lysis buffer (10 mM Tris⋅HCl, pH 7.4/0.5% SDS/10 mM EDTA, pH 7.4/10 mM NaCl) supplemented with proteinase K (200 μg/ml). The covalently closed circular DNA form was selectively extracted from cells recovered by trypsinization. Cells were lysed at room temperature in a lysis buffer not supplemented with proteinase K. In both cases, cellular DNA was precipitated overnight at 4°C with 1 M NaCl.
Complete viral particles were isolated from cell supernatants by immunoprecipitation with a polyclonal anti-HBs antibody (DAKO). Nucleic acids were extracted after an overnight lysis at 37°C in lysis buffer supplemented with tRNA (40 μg/ml) and proteinase K (200 μg/ml).
For all of the above procedures, DNA was extracted by phenol chloroform and precipitated by isopropanol. Nucleic acids were analyzed by Southern blot procedure on a 1.5% agarose gel. Molecular weight markers were HBV DNA restriction fragments (3,182 and 1,504 bp). Hybridization was performed with an HBV-specific probe covering the whole genome. HBV DNA quantification was performed by a standard dot blot procedure.
RNA Extraction and Analysis.
Total cellular RNA was extracted by the Total SV RNA kit (Promega), fractionated on a 1.5% agarose gel and analyzed by standard Northern blot procedure (14). Control of the RNA amount transferred onto filters was performed after methylene blue staining.
Immunocytochemistry.
Infected and control HepaRG cells were fixed in 4% paraformaldehyde for 30 min at 4°C. Cell permeabilization was ensured by a 30-min treatment with a solution of 0.25% saponin. The primary antibody was a specific rabbit polyclonal antibody directed against the HBV core protein (used at a 1/400 dilution) (DAKO). The second antibody consisted of an FITC-labeled antibody directed against rabbit immunoglobulins.
Cytogenetic Analysis.
HepaRG cells were karyotyped eight passages after selection. The cells were grown for 24–48 h in RPMI medium 1640 supplemented with 10% FCS and blocked in metaphase by Colcemid exposure (10 μg/ml) for 45 min. The cells were then treated with hypotonic solution (0.1 M MgCl2), fixed with the Carnoy acetic solution, and stained for RHG bandings. Determination of the translocation and deletion breakpoints were determined by in situ hybridization.
cDNA Probes.
Human albumin (15), aldolase B (16), and α fetoprotein (AFP) and transferrin (17) cDNAs were kindly provided by A. Kahn (INSERM U567, Paris). Human CYP 2E1 (18) and GST α1 were kindly provided by A. Guillouzo (INSERM U456, Rennes, France). The human CYP 3A4-specific cDNA probe was constructed by I. de Wazier (INSERM U490, Paris) from a complete cDNA clone (19).
Results
Cell Line Isolation and Culturing.
Cells were isolated from a grade I differentiated liver tumor of a female patient suffering from hepatocarcinoma consecutive to chronic hepatitis C virus infection. After several weeks of cultivation, a spontaneous outgrowth of a cell population was observed. Initially, these cells had a hepatocyte-like morphology, but they acquired an undifferentiated morphology after few passages. To see whether these cells could again be committed to a differentiated phenotype, they were exposed to DMSO and hydrocortisone, two well known differentiation inducers. The combined use of these two agents has been shown efficient for maintaining the differentiation of normal hepatocytes in primary cultures, including those of human origin (1, 20). In the presence of 2% DMSO and 5 × 10−5 M hydrocortisone, the majority of cells died within 5 days, but after a 2-week exposure, some cells organized in small clusters and displayed a typical hepatocyte-like morphology. This indicated that it was possible to restore differentiation of a fraction of the original population. We took advantage of this observation and of the ability of these cells to actively proliferate after DMSO removal for selecting them from the other cells according to the procedure described in Methods. The resulting cell line, called HepaRG, has the potential to efficiently differentiate in the presence of DMSO and hydrocortisone with minor toxicity events. It was essential to continuously maintain this cell line in the presence of 5 × 10−5 M hydrocortisone to preserve an optimal differentiation capacity (data not shown). The presence of hepatitis C virus genome was not detectable by RT-PCR after the establishment of the HepaRG cell line in culture.
Morphology and Karyotype.
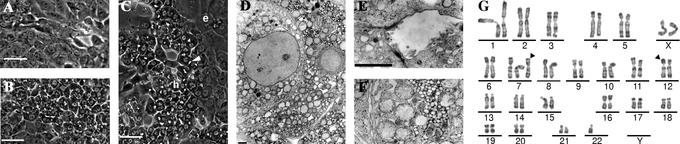
During the proliferation phase (doubling time of 24 h), HepaRG cells appeared as a homogeneous cell population with an epithelial phenotype showing no regular structural organization (Fig. 1A). Then, when reaching confluence, cells started to undergo progressive morphological changes, getting the appearance of granular hepatocyte-like cells (Fig. 1B). Addition of 2% DMSO to the medium, 2 weeks after plating, induced the granular hepatocyte-like cells to organize in well delineated trabeculae closely resembling those formed in primary human hepatocyte culture in which many bright canaliculi-like structures could be recognized. Few clear flat epithelium-like cells filled the empty spaces around. Trabeculae organization was completed within 2 weeks of DMSO exposure, while granular cell morphology closely resembled hepatocytes (Fig. 1C). Modulating the delay before applying DMSO, the length of the DMSO exposure, the DMSO, hydrocortisone, FCS, or insulin concentrations did not significantly enhance the differentiation of HepaRG cells. Therefore, 2 weeks in the presence of 5 × 10−5 M hydrocortisone followed by 2 weeks of exposure to both hydrocortisone and DMSO defined the conditions promoting a maximum morphological differentiation level.
Fig 1.
Morphology and karyotype of HepaRG cells. Phase contrast micrographs of HepaRG cells under proliferating conditions (A), maintained in culture for 30 days without DMSO (B), and maintained for 15 days without DMSO, then treated with 2% DMSO for 15 days (C). Hepatocyte-like cells and epithelium-like cells are indicated, respectively, by “h” and “e.” A bile canaliculus is indicated by a white triangle. (Bars = 50 μm.) Electron micrographs of HepaRG cells: low magnification view of HepaRG cells (D) and higher magnification views (E and F), showing a typical bile canaliculus-like structure and glycogen accumulation, respectively. (Bars = 2 μm.) RHG banding karyotype of a representative pseudodiploid metaphase of HepaRG cells (G). A karyotype of 46<2n>,XX,+del (7)(q11;q21)inv?(7)(q21q36),-der(22)t(12;22)(p11;q11) was interpreted. The two main anomalies are indicated by arrowheads.
Electron microscopy studies of these hepatocyte-like cells evidenced contacting cells tightly attached by desmosomes (Fig. 1D), with structures identical to bile canaliculi, delineated by typical junctional complexes and presenting several microvilli (Fig. 1E). We could also recognize numerous cytoplasmic glycogen granules (Fig. 1F).
In contrast to previously described hepatoma-derived cell lines, the karyotype was pseudodiploid. From the studied 40 mitoses, 65% contained 46 chromosomes and 22% had only one surnumerary chromosome. All exhibited the two following abnormalities: a surnumerary and remodeled chromosome 7 and a translocation t(12;22) with a loss of the 12p fragment leading to a monosomy 12p (Fig. 1G).
Expression of Liver-Specific Functions.
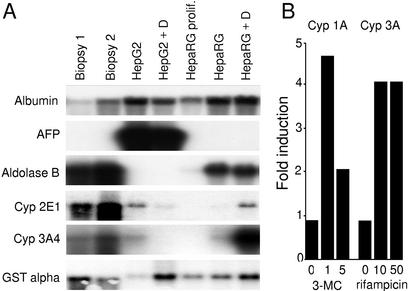
In addition to the morphological characterization, the differentiation state of HepaRG cells was monitored by analyzing a representative panel of liver-specific mRNAs, such as the mRNA for albumin and transferrin, the glycolytic enzyme aldolase B, and three enzymes involved in detoxification (CYP 2E1, CYP 3A4, and GST α), all being expressed in adult hepatocytes. We also looked for RNAs encoding α fetoprotein (AFP), a marker of fetal hepatocytes. RNA levels were examined in proliferating and differentiated cells and compared with human liver biopsies as well as to the differentiated human hepatoblastoma-derived HepG2 cell line (22). Low or even undetectable amounts of mRNAs were generally observed in proliferating HepaRG cells, whereas at confluence, they all accumulated except for AFP, which was never detected (Fig. 2A). Albumin and aldolase B mRNAs became highly expressed, whereas the cytochrome P450 CYP 2E1 and CYP 3A4 mRNAs remained weakly detected. In cells undergoing trabecular organization due to DMSO exposure, the amounts of all transcripts were high, particularly CYP 2E1 and CYP 3A4 mRNAs drastically increased, reaching levels close to those of liver biopsies. GST α transcripts, easily detectable in proliferating cells, slightly increased at confluence and were further induced in the presence of DMSO. In comparison, HepG2 cells evidenced a relatively high expression level of the studied specific functions, with a remarkable exception for aldolase B, which was undetectable. Another major difference is a high expression level of α fetoprotein, a protein normally expressed in fetal liver. These differences emphasize the unique features of HepaRG cells.
Fig 2.
Expression of liver-specific functions in HepaRG cells. (A) Northern blot analysis of liver-specific messenger RNA expression in two liver biopsy specimens, HepG2 cells, and HepaRG cells. Cultured cells were maintained either at confluence for 1 month (no indication) or under proliferating conditions (prolif.). A 2% DMSO treatment was performed for the last 15 days of the culture when indicated (+D). (B) Effect of inducers on CYP1A (phenacetine deethylase) and CYP3A (nifedipine oxidase) activities. The activities were measured after 72 h of treatment and are expressed as the ratio of treated cells versus corresponding control cells. The inducer concentrations (μM) are indicated below the graph.
We further quantitatively analyzed the ability of the cell line to express phase I and phase II drug metabolism enzyme activities, most of them being generally poorly expressed or not inducible in other hepatoma cell lines. These analyses were realized in confluent DMSO-treated HepaRG cells and compared with the activity observed in human hepatocyte primary cultures and HepG2 cells. Results, shown in Table 1, evidenced that most of the enzymatic activities, with the exception of dextromethorphan demethylase, the value of which was slightly lower, remained in the range observed with normal human hepatocytes, and were significantly higher than in HepG2 cells. The inducibility of phenacetin deethylase (cyp1A) by 3-methyl cholantrene and nifedipine oxidase (cyp3A) by rifampicin was also demonstrated (Fig. 2B).
Table 1.
Phase I and II activities measured in HepaRG, HepG2 cells, and primary human hepatocytes (PHH)
| Enzyme activity
|
HepaRG
|
HepG2
|
PHH | |
|---|---|---|---|---|
| Min/Max | Means | |||
| Phenacetin deethylase | 0.33/0.54 | 0.18/0.11 | 0.1–25 | 3.9 |
| Tolbutamid hydroxylase | 0.51/1.47 | <0.1/<0.1 | 0.2–2.1 | 0.95 |
| S-mephenytoin hydroxylase | 0.45/0.59 | <0.1/<0.1 | 0.1–2 | 0.7 |
| Dextromethorphan demethylase | 0.06/0.04 | <0.1/<0.1 | 0.1–2 | 0.5 |
| Nifedipine oxidase | 1 | <0.1 | 0.5–30 | 5.7 |
| Paracetamol glucuronyl transferase | 3.7 | <0.1 | 0.3–16 | 4.1 |
| Paracetamol sulfoconjugation | 0.7 | 0.3/0.43 | 0.1–14 | 3.6 |
| Procainimide N-acetyltransferase | 0.75 | 0.16/0.19 | ND | ND |
| GST (chrorodinitrobenzene as a substrate) | 0.04 | 0.01/0.01 | 0.03–0.5 | 0.12 |
When two independent assays were performed, the two values are indicated.
Activities are expressed in nanomoles of produced metabolites/h/mg protein.
ND, not determined.
Activities are expressed in units/mg protein.
HBV Infection.
HepaRG cells did therefore share many characteristics with normal human hepatocytes. We consequently examined their susceptibility to HBV infection. To this end, we applied the same conditions as validated previously for in vitro infection of primary human hepatocyte (3). These conditions, which include the use of PEG, have been shown to preserve the tissue and species specificity of the HBV infection process. They have also been successfully used to define the role of viral envelope protein in HBV infectivity and to determine the neutralization capacity of monoclonal antibodies directed against viral surface proteins (3, 21, 23–28).
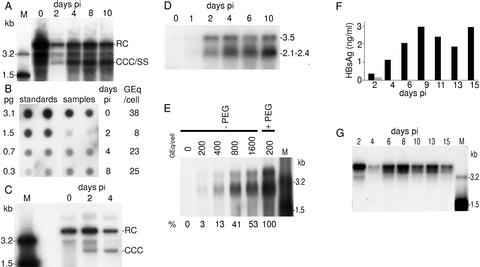
Viral DNA replicative forms were evidenced in HBV-infected cells (Fig. 3A). A strong signal was observed immediately after infection, likely corresponding to internalized viral particles. At day 2, the signal was reduced probably as a consequence of elimination of many viral particles, and only three discrete bands, with no intermediate forms, could be evidenced. They likely correspond to the input DNA which escaped the degradation pathway and was progressively converted into covalently closed circular DNA. The 2-kb band presumably represents this form. Thereafter, a signal was observed at intermediate positions and below the 2-kb position, corresponding to nascent viral DNA. Quantification of the HBV DNA signal by dot-blot analysis revealed that, at day 0, 2, 4, and 8 postinfection (pi), the number of genomes equivalent per cell was, respectively, 38, 8, 23, and 25 (Fig. 3B).
Fig 3.
HBV infection of HepaRG cells. (A) Southern blot kinetic analysis of intracellular HBV DNA after infection of HepaRG cells. Half of the DNA extracted from 106 infected cells was analyzed on a 1.5% agarose gel. The positions of the relaxed circular form (RC), covalently closed circular DNA (CCC), and single-stranded DNA (SS) are indicated on the right. Lane M corresponds to 3.5 pg of 3.2- and 1.5-kb HBV DNA restriction fragments (3.4 and 6.8 × 106 molecules, respectively). Autoradiographic exposure time was 24 h. (B) HBV DNA quantification by dot blot. Samples analyzed on Fig. 3A were quantified by comparison of the hybridization signal obtained from serial dilutions of known amounts of linear double-stranded HBV DNA. On the left is indicated the amount of standard HBV DNA spotted on the filter; on the right is the calculated HBV DNA copy number expressed in genome equivalents per cell (Geq/cell). Autoradiographic exposure time was 24 h. (C) Southern blot kinetic analysis of nonprotein-bound intracellular HBV DNA after infection of HepaRG cells. Half of the DNA extracted from 106 infected cells was analyzed on a 1.5% agarose gel. Positions of the relaxed circular form (RC) and covalently closed circular DNA (CCC) are indicated on the right. Lane M corresponds to 3.5 pg of 3.2- and 1.5-kb HBV DNA restriction fragments (3.4 and 6.8 × 106 molecules, respectively). Autoradiographic exposure time was 96 h. (D) Northern blot kinetic analysis of HBV RNA after infection of HepaRG cells. Half of the RNA extracted from 106 infected cells was analyzed on a 1.5% agarose gel. RNA sizes are indicated on the right (kb). Autoradiographic exposure time was 36 h. (E) Comparison of HBV RNA levels after infection of HepaRG cells in the presence or absence of PEG by Northern blot analysis. RNA were extracted from mock-infected cells or cells infected in the absence (−PEG) or in the presence (+PEG) of PEG. Cells were infected at a multiplicity of infection of 200, 400, 800, or 1,600 genome equivalents per cell (Geq/cell). Half of the RNA extracted from 106 infected cells was analyzed on a 1.5% agarose gel. Lane M corresponds to 3.5 pg of denatured 3.2- and 1.5-kb HBV DNA restriction fragments. RNA sizes are indicated on the right. The relative amounts of subgenomic RNA detected in infected cells are indicated below the figure. Autoradiographic exposure time was 96 h. (F) Kinetic analysis of HBsAg secretion in the supernatant of infected cells. Supernatants were collected during 15 days after infection of HepaRG cells (▪) or HepG2 cells ( ). (G) Southern blot kinetic analysis of extracellular HBV DNA in the supernatant of infected HepaRG cells. Complete viral particles were immunoprecipitated with an anti-HBsAg antibody from 1 ml of the supernatant of infected HepaRG cells. The viral DNA was extracted, and half of the preparation was analyzed by the Southern blot procedure. Lane M corresponds to 3.5 pg of 3.2- and 1.5-kb HBV DNA restriction fragments (3.4 and 6.8 × 106 molecules, respectively). Autoradiographic exposure time was 96 h.
). (G) Southern blot kinetic analysis of extracellular HBV DNA in the supernatant of infected HepaRG cells. Complete viral particles were immunoprecipitated with an anti-HBsAg antibody from 1 ml of the supernatant of infected HepaRG cells. The viral DNA was extracted, and half of the preparation was analyzed by the Southern blot procedure. Lane M corresponds to 3.5 pg of 3.2- and 1.5-kb HBV DNA restriction fragments (3.4 and 6.8 × 106 molecules, respectively). Autoradiographic exposure time was 96 h.
The presence of the covalently closed circular DNA form has been further established by a selective extraction procedure (Fig. 3C). Although this form, migrating at the 2-kb position, was absent immediately after infection, it became detectable from day 2.
The infection of HepaRG cells was ascertained by the kinetic analysis of viral RNA (Fig. 3D). Bands at 2.1- to 2.4- and 3.5-kb positions, which correspond to the main HBV RNA species, appeared 2 days pi and accumulated thereafter. By contrast, in an attempt to infect HepG2 cells cultured under identical conditions, no signal corresponding to viral RNA could be evidenced (data not shown).
To justify the use of PEG, we performed an infection experiment without the polymer. The infection efficiencies were evaluated by analyzing the quantities of HBV RNA present in cells 8 days pi. For an identical multiplicity of infection (200 genome equivalent per cell) when PEG was omitted, the detected amount of viral RNA only represented 3% of those obtained in the presence of PEG. However, a 53% infection level was reached by increasing 8-fold the multiplicity of infection (Fig. 3E).
To check if HepaRG cells support the entire HBV replication cycle, we looked for the production of viral particles in the supernatant by analyzing both the secretion of HBsAg, the dominant envelope protein that corresponds mainly to empty viral envelopes, and the production of virions (Fig. 3 F and G, respectively). The HBsAg concentration increased until day 9 pi and reached a high plateau level (2–3 ng/ml). In contrast, only traces of HBsAg were detectable in the supernatant of HepG2 cells, tentatively infected in parallel, presumably resulting from a gradual release of adsorbed viral particles issued from the inoculum. Repetitive analyses indicated a strict correlation between the long-term HBsAg secretion and the appearance of other viral infection markers, such as viral RNA and replicative DNA intermediates. The release of complete viral particles into the supernatant of infected HepaRG cells was also assessed (Fig. 3G). At day 2 pi, a strong HBV DNA signal was observed. Its intensity was reduced at day 4. Thereafter, HBV DNA remained permanently present during all of the culture period. These variations likely correspond to a release during the first 2 or 4 days followed by an active production of progeny virions after day 4.
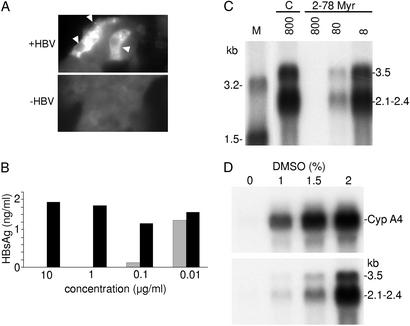
Finally, the percentage of infected cells was estimated by an immunostaining with an anticore antibody (Fig. 4A). The core antigen was preferentially detected in the cytoplasm of hepatocyte-like cells but was occasionally detected in their nucleus. These results revealed that at least 10% of HepaRG cells were infected.
Fig 4.
Specificity of the HBV infection. (A) Immunostaining of infected HepaRG cells for HBV core antigen. Cells were stained with an anticore antibody by immunofluorescence. Positive cells are indicated by a white arrow. (B) In vitro neutralization assay. Viral particles were incubated with serial dilutions of a monoclonal anti-HBs antibody (S 39–10;  ) or an unrelated monoclonal antibody (▪) and evaluated for their infectivity on HepaRG cells. Infectivity was evaluated by measuring the HBsAg secretion in the supernatant of infected cells at day 10 pi. (C) Peptide competition of HBV infection. HepaRG cells were preincubated with different concentrations (nM) of an HBV large protein-derived peptide (AA 2–78) bearing a myristic acid (myr) on gly 2 or, as a control, with a corresponding myristoylated duck hepatitis B virus-derived peptide (AA 2–41; C). The inoculum was then added for 20 h, without removing the peptide. Infectivity was evaluated by analyzing intracellular viral RNA at 10 days pi. Half of the RNA extracted from 106 infected cells was analyzed on a 1.5% agarose gel. Lane M corresponds to 3.5 pg of denatured 3.2- and 1.5-kb HBV DNA restriction fragments. Viral RNA sizes are indicated on the right. Autoradiographic exposure time was 48 h. (D) Efficiency of HBV infection and HepaRG cell differentiation. Northern blot analysis of CYP 3A4 (Upper) and HBV (Lower) RNAs after infection of HepaRG cells cultivated in presence of increasing concentrations of DMSO. RNA was extracted 10 days pi. Viral RNA sizes are indicated on the right.
) or an unrelated monoclonal antibody (▪) and evaluated for their infectivity on HepaRG cells. Infectivity was evaluated by measuring the HBsAg secretion in the supernatant of infected cells at day 10 pi. (C) Peptide competition of HBV infection. HepaRG cells were preincubated with different concentrations (nM) of an HBV large protein-derived peptide (AA 2–78) bearing a myristic acid (myr) on gly 2 or, as a control, with a corresponding myristoylated duck hepatitis B virus-derived peptide (AA 2–41; C). The inoculum was then added for 20 h, without removing the peptide. Infectivity was evaluated by analyzing intracellular viral RNA at 10 days pi. Half of the RNA extracted from 106 infected cells was analyzed on a 1.5% agarose gel. Lane M corresponds to 3.5 pg of denatured 3.2- and 1.5-kb HBV DNA restriction fragments. Viral RNA sizes are indicated on the right. Autoradiographic exposure time was 48 h. (D) Efficiency of HBV infection and HepaRG cell differentiation. Northern blot analysis of CYP 3A4 (Upper) and HBV (Lower) RNAs after infection of HepaRG cells cultivated in presence of increasing concentrations of DMSO. RNA was extracted 10 days pi. Viral RNA sizes are indicated on the right.
Specific Neutralization of HBV Infection.
To provide evidence that uptake of HBV into HepaRG cells follows the authentic entry pathway, we performed infection competition assays using either a monoclonal HBsAg-specific antibody or an HBV large surface protein-derived peptide. To this aim, viruses were incubated with various concentrations of the antibody before the incubation with cells. Residual infectivity was estimated by measuring the long-term HBsAg secretion. As shown in Fig. 4B, antibody concentrations of 10 and 1 μg/ml completely blocked viral infection, whereas lower concentrations induced a partial inhibition, the 50% inhibitory dose being 0.03 μg/ml.
In addition, we tested the inhibition capacity of myristoylated peptide representing the N-terminal 78 aa of the HBV large envelope protein. This region is required for HBV infectivity in primary human hepatocyte (24, 25). The corresponding peptide is therefore expected to address a cellular receptor(s). As depicted in Fig. 4C, an almost complete inhibition of HBV infection was obtained when this peptide was added during the infection process at a dose of 800 nM.
Influence of the Cellular Differentiation Status on HBV Infection.
To elicit the factors that determine the HBV susceptibility of HepaRG cells, we compared the infection efficiency on poorly and highly differentiated cells. To this purpose, we modulated the differentiation state of HepaRG cells by adding DMSO up to a final concentration of 2% and determined the accumulation of the CYP 3A4 transcripts (Fig. 4D). Two weeks after the infection, the detected amounts of the intracellular viral RNAs were correlated to DMSO doses. Our results showed a close correlation between the susceptibility to HBV infection and the cellular differentiation status (Fig. 4D). The possibility that the DMSO treatment may directly favor the viral transcription and not the earlier steps of viral infection was ruled out by adding DMSO after infection. In this case, only traces of viral RNA could be detected (data not shown).
Discussion
In this paper, we describe a human hepatocyte-derived cell line that supports the full replication cycle of HBV. Compared with other hepatoma cell lines, it is defined by two important hepatic functional features: (i) maintenance of an efficient proliferation/differentiation interplay accompanied by morphological changes leading to hepatocyte-like cells, and (ii) the ability to display optimal metabolic functions associated with a unique susceptibility to HBV infection.
The loss of hepatic phenotype during the proliferation phase is consistent with similar phenomenon observed for hepatocyte primary cultures that, when maintained in the absence of a reconstituted microenvironment or specific inducers, enter in G1 phase of the cell cycle and no longer express several characteristic hepatocyte traits (20, 29, 30). Although severe, this loss of function is not complete since albumin remains efficiently expressed. The other outstanding characteristic of HepaRG cells is their ability to differentiate in hepatocyte-like cells. This status is reached only after a prolonged quiescence of the cells maintained at confluence associated with a DMSO treatment. In the presence of hydrocortisone, confluence alone restores the expression of many hepatic functions. However, DMSO is absolutely required for high-level expression of cytochrome P450 and for the susceptibility to HBV infection.
The presence of only a few major chromosomal rearrangements constitutes a major advantage of this cell line, which is pseudodiploid and therefore much less likely profoundly altered than the previously described cell lines.
The most remarkable feature of HepaRG cells, however, is their susceptibility to HBV infection. Demonstration was provided by showing the kinetics of several markers of viral infections and, most important, the appearance of newly synthesized HBV transcripts. In addition, evidence that infection was blocked by both a specific anti-HBsAg monoclonal antibody and an HBV large envelope-derived peptide strongly argues that the incoming virus proceeds through the authentic HBV entry pathway. It is important to note that PEG is not required to infect HepaRG cells when an increased multiplicity of infection is used.
Interestingly, the susceptibility of HepaRG cells to HBV infection was strictly correlated to their differentiation as demonstrated by the efficient infection obtained only when cells were fully differentiated in the presence of DMSO and corticoids, emphasizing the specificity of this in vitro infection process. We may hypothesize that some HepaRG cells are less differentiated and therefore not susceptible to HBV infection. This might also explain the resistance of other hepatoma cell lines to HBV infection. A recent report (31) suggesting the successful infection of HepG2 cells in the presence of DMSO could not be confirmed in our laboratory.
The HepaRG hepatoma-derived cell line will provide a tool to improve our understanding of the early steps of the viral cycle, including identification of the yet unknown receptor molecules. It will enable us to functionally dissect the role of viral surface proteins in the entry process and consequently allow the development of types of antiviral drugs that interfere with early steps of infection. It will also be a powerful tool for the evaluation of other types of antiviral compounds, in the context of both a complete replication cycle and optimally differentiated human hepatocyte-like cells. This cell line is very promising for many other applications, including studies in pharmacology and toxicology.
Acknowledgments
We gratefully acknowledge F. V. Chisari for helpful comments and criticism of the manuscript. This work was supported by Institut National de la Santé et de la Recherche Médicale, Association pour la Recherche contre le Cancer, and Ligue Départementale Contre le Cancer d'Ille et Vilaine.
Abbreviations
pi, postinfection
HBV, hepatitis B virus
PEG, polyethylene glycol
HBsAg, hepatitis B surface antigen
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gripon P., Diot, C., Theze, N., Fourel, I., Loreal, O., Brechot, C. & Guguen-Guillouzo, C. (1988) J. Virol. 62 4136-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ochiya T., Tsurimoto, T., Ueda, K., Okubo, K., Shiozawa, M. & Matsubara, K. (1989) Proc. Natl. Acad. Sci. USA 86 1875-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gripon P., Diot, C. & Guguen-Guillouzo, C. (1993) Virology 192 534-540. [DOI] [PubMed] [Google Scholar]
- 4.Galle P. R., Hagelstein, J., Kommerell, B., Volkmann, M., Schranz, P. & Zentgraf, H. (1994) Gastroenterology 106 664-673. [DOI] [PubMed] [Google Scholar]
- 5.Sells M. A., Chen, M. L. & Acs, G. (1987) Proc. Natl. Acad. Sci. USA 84 1005-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang C. M., Jeng, K. S., Hu, C. P., Lo, S. J., Su, T. S., Ting, L. P., Chou, C. K., Han, S. H., Pfaff, E., Salfeld, J., et al. (1987) EMBO J. 6 675-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sureau C., Romet-Lemonne, J. L., Mullins, J. I. & Essex, M. (1986) Cell 47 37-47. [DOI] [PubMed] [Google Scholar]
- 8.Tsurimoto T., Fujiyama, A. & Matsubara, K. (1987) Proc. Natl. Acad. Sci. USA 84 444-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yaginuma K., Shirakata, Y., Kobayashi, M. & Koike, K. (1987) Proc. Natl. Acad. Sci. USA 84 2678-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urban S. & Gripon, P. (2002) J. Virol. 76 1986-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chesne C., Guyomard, C., Fautrel, A., Poullain, M. G., Fremond, B., De Jong, H. & Guillouzo, A. (1993) Hepatology 18 406-414. [DOI] [PubMed] [Google Scholar]
- 12.Guillouzo A. & Chesne, C., (1996) Xenobiotic Metabolism in Epithelial Cell Cultures (Oxford Univ. Press, Oxford).
- 13.Habig W. H. & Jakoby, W. B. (1981) Methods Enzymol. 77 398-405. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J., Fritsh, E. F. & Maniatis, T., (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 15.Dugaiczyk A., Law, S. W. & Dennison, O. E. (1982) Proc. Natl. Acad. Sci. USA 79 71-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besmond C., Dreyfus, J. C., Gregori, C., Frain, M., Zakin, M. M., Sala Trepat, J. & Kahn, A. (1983) Biochem. Biophys. Res. Commun. 117 601-609. [DOI] [PubMed] [Google Scholar]
- 17.Uzan G., Frain, M., Park, I., Besmond, C., Maessen, G., Trepat, J. S., Zakin, M. M. & Kahn, A. (1984) Biochem. Biophys. Res. Commun. 119 273-281. [DOI] [PubMed] [Google Scholar]
- 18.Urban P., Cullin, C. & Pompon, D. (1990) Biochimie 72 463-472. [DOI] [PubMed] [Google Scholar]
- 19.Guengerich F. P., Martin, M. V., Beaune, P. H., Kremers, P., Wolff, T. & Waxman, D. J. (1986) J. Biol. Chem. 261 5051-5060. [PubMed] [Google Scholar]
- 20.Isom I., Georgoff, I., Salditt-Georgieff, M. & Darnell, J. E., Jr. (1987) J. Cell Biol. 105 2877-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gripon P., Le Seyec, J., Rumin, S. & Guguen-Guillouzo, C. (1995) Virology 213 292-299. [DOI] [PubMed] [Google Scholar]
- 22.Knowles B. B., Howe, C. C. & Aden, D. P. (1980) Science 209 497-499. [DOI] [PubMed] [Google Scholar]
- 23.Ryu C. J., Gripon, P., Park, H. R., Park, S. S., Kim, Y. K., Guguen-Guillouzo, C., Yoo, O. J. & Hong, H. J. (1997) J. Med. Virol. 52 226-233. [DOI] [PubMed] [Google Scholar]
- 24.Le Seyec J., Chouteau, P., Cannie, I., Guguen-Guillouzo, C. & Gripon, P. (1998) J. Virol. 72 5573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Seyec J., Chouteau, P., Cannie, I., Guguen-Guillouzo, C. & Gripon, P. (1999) J. Virol. 73 2052-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chouteau P., Le Seyec, J., Cannie, I., Nassal, M., Guguen-Guillouzo, C. & Gripon, P. (2001) J. Virol. 75 11565-11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeng C. Y., Ryu, C. J., Gripon, P., Guguen-Guillouzo, C. & Hong, H. J. (2000) Virology 270 9-16. [DOI] [PubMed] [Google Scholar]
- 28.Park S. S., Ryu, C. J., Gripon, P., Guguen-Guillouzo, C. & Hong, H. J. (1996) Hybridoma 15 435-441. [DOI] [PubMed] [Google Scholar]
- 29.Loyer P., Cariou, S., Glaise, D., Bilodeau, M., Baffet, G. & Guguen-Guillouzo, C. (1996) J. Biol. Chem. 271 11484-11492. [DOI] [PubMed] [Google Scholar]
- 30.Corlu A., Kneip, B., Lhadi, C., Leray, G., Glaise, D., Baffet, G., Bourel, D. & Guguen-Guillouzo, C. (1991) J. Cell Biol. 115 505-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paran N., Geiger, B. & Shaul, Y. (2001) EMBO J. 20 4443-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]