Abstract
The detection of viral pathogens is of critical importance in biology, medicine, and agriculture. Unfortunately, existing techniques to screen for a broad spectrum of viruses suffer from severe limitations. To facilitate the comprehensive and unbiased analysis of viral prevalence in a given biological setting, we have developed a genomic strategy for highly parallel viral screening. The cornerstone of this approach is a long oligonucleotide (70-mer) DNA microarray capable of simultaneously detecting hundreds of viruses. Using virally infected cell cultures, we were able to efficiently detect and identify many diverse viruses. Related viral serotypes could be distinguished by the unique pattern of hybridization generated by each virus. Furthermore, by selecting microarray elements derived from highly conserved regions within viral families, individual viruses that were not explicitly represented on the microarray were still detected, raising the possibility that this approach could be used for virus discovery. Finally, by using a random PCR amplification strategy in conjunction with the microarray, we were able to detect multiple viruses in human respiratory specimens without the use of sequence-specific or degenerate primers. This method is versatile and greatly expands the spectrum of detectable viruses in a single assay while simultaneously providing the capability to discriminate among viral subtypes.
The rational diagnosis of viral diseases requires the identification of viral pathogens in clinical specimens and subsequent correlation between presence of the virus and the clinical syndrome. In some instances, where the disease is associated with a particular viral agent, the task is relatively straightforward, and a number of different methods can be used to determine the presence or absence of the virus. Historically, standard viral detection techniques have relied on isolation and in vitro viral culture or immunological assays such as shell vials, direct fluorescence antibody, or enzyme immunoassay (1). More recently, the emergence of PCR has revolutionized viral diagnostics (reviewed in ref. 2) by not only increasing detection sensitivity but also facilitating the detection of several viruses in parallel, either by multiplexing specific primers (for discrete viruses) or through careful design of degenerate primers (for members of a class).
However, in more complex biological situations, such as diseases where many different viruses are present or where no etiologic agent has been identified, the limitations of even the best current methodologies become readily apparent. Some viruses are completely refractory to in vitro culture (1), and immunoassays depend on the quality and availability of the antiserum. Furthermore, the complexity of the viral flora itself presents several problems. The existence of a large number of constantly evolving viral serotypes can render antibody-based detection nearly impossible. With PCR methods, because it is difficult to design compatible multiplex primer sets (3), the maximum number of viruses detectable in a single assay is relatively small (2). Moreover, unambiguous viral identification with degenerate PCR often is complicated by the existence of highly homologous relatives, and discrimination between viral subtypes or genera requires additional labor-intensive procedures such as restriction enzyme analysis, sequencing, or hybridization blotting of the PCR product (4–8). Perhaps most significantly, even the broadest multiplexing is inherently biased, requiring assumptions that ultimately restrict the possible outcome to the selected candidate viruses.
To address the limitations of existing viral detection methodologies, we have developed a genomic approach to virus identification. Using available sequence data from more than 140 sequenced viral genomes, we have designed a long oligonucleotide (70-mer) DNA microarray with the potential to simultaneously detect hundreds of viruses, including essentially all respiratory tract viruses. We describe here validation of this DNA microarray by using virally infected tissue culture cells as well as clinical specimens isolated from the human respiratory tract.
Materials and Methods
Microarray Design and Construction.
Viral sequence data were obtained primarily from the curated database of fully sequenced viral genomes in GenBank. For a given family of viruses, each fully sequenced genome was divided into overlapping 70-nt segments offset by 25 nt, and a pair-wise blastn (9) alignment was implemented between each 70-mer and each viral genome in the family. The results of these alignments were tabulated by using the best blast hit (if any) for each segment-viral genome pair. The 70-mers were then ranked by the number of viral genomes to which significant homology (>20-nt identity) was observed. In most cases, the five highest-ranking oligonucleotides for each virus and the corresponding reverse complement oligonucleotides were selected. In some cases additional steps were taken to distinguish between viral genera. For example, the family picornaviridae contains six genera, including the closely related rhinoviruses (RV) and enteroviruses, which share a similar genomic organization. To facilitate distinction between RVs and enteroviruses, the RV genus was considered as an independent category, and sequences with strong homology between the RV genus and the other picornaviruses were masked and removed from further analysis. Oligonucleotides (Illumina, San Diego) were suspended in 3× SSC at a concentration of 50 pmol/μl and printed on glass slides exactly as described for PCR products (10). In addition, ≈100 oligonucleotides derived from human gene sequences were printed both individually and in pools as controls for microarray scanning. (For a complete listing of viral oligonucleotide sequences represented on the current microarray, see http://derisilab.ucsf.edu/virochip.)
Viruses and Cell Culture.
Respiratory syncytial virus (RSV), parainfluenza 3, adenovirus 12, and human RVs 1b, 2, 14, 21, 62, 65, and 72 were obtained from the American Type Culture Collection. RV16 was obtained from W. Busse and E. Dick (University of Wisconsin, Madison). Poliovirus1 was kindly provided by R. Andino (University of California, San Francisco). All viral infections were performed by using HeLa cells, which were cultured in DMEM supplemented with 10% FCS and antibiotics. Viral infections were allowed to proceed until the onset of cytopathic effects (typically 24–72 h). The BCBL-1 cell line harbors Kaposi's sarcoma-associated herpes virus (KSHV), which was reactivated by treatment with tetradecanoyl phorbol acetate (11). RNA from virally infected and uninfected cell cultures was isolated by using RNAzol (Tel-Test, Friendswood, TX). After isopropanol precipitation, RNA was reverse-transcribed into cDNA in the presence of aminoallyl-dUTP (Sigma) as described (12).
Nasal Lavage Isolation, Amplification, and Labeling.
Nasal lavage was obtained from human subjects who participated in ongoing institutional review board-approved studies as described (13). In the first study, patients were deliberately inoculated with RV16. Before infection and at several time points postinfection, nasal lavage samples were isolated (13). Nasal lavage was also obtained from a cohort of patients who presented with natural colds. RNA was isolated from 250-μl samples of nasal lavage by using RNeasy (Qiagen, Chatsworth, CA). Samples were amplified with a modified version of a random PCR protocol (14). RNA was reverse-transcribed with PrimerD (5′-GTTTCCCAGTAGGTCTCNNNNNNNN), and second-strand DNA synthesis was carried out with Sequenase (United States Biochemical). Subsequently, this material was used as the template for 40 cycles of PCR with PrimerE (5′-GTTTCCCAGTAGGTCTC) by using the following profile: 30 s at 94°C, 30 s at 40°C, 30 s at 50°C, 60 s at 72°C. Reactions were supplemented with 2.5 units of Taq and amplified for 20 additional cycles. The resulting PCR product was random-primed with nonamers by using Klenow polymerase in the presence of aminoallyl-dUTP. The presence of RV in the clinical samples was independently analyzed by conventional RT-PCR using published primers RVF (5′-GAAACACGGACACCCAAAGTA) and RVR (5′-TCCTCCGGCCCCTGAATG) (15) or SEQF (5′-GCATCIGGYARYTTCCACCACCANCC; I = inosine; Y = T/C; R = G/A) and SEQR (5′-GGGACCAACTACTTTGGGTGTCCGTGT) (16). Similarly, RT-PCR for parainfluenza 1 was performed as described (17).
Microarray Hybridization and Data Visualization.
Microarray hybridization was performed as described (10, 12). For all de novo infections, virally infected HeLa cells were compared with uninfected HeLa cells. RNA from BCBL-1 was hybridized against BJAB, a KSHV-negative B cell line. For clinical samples, amplified nasal lavage from each patient was compared with amplified nasal lavage RNA from a healthy control subject without symptoms of upper respiratory infection or amplified HeLa RNA. All arrays were imaged by using an Axon Instruments (Foster City, CA) 4000B scanner and genepix pro software. Primary microarray data for all arrays are available at http://derisilab.ucsf.edu/virochip. Microarray data were converted to a color visualization in which the Cy5 intensity of each viral oligonucleotide was plotted by using a continuous linear color scale. The maximum intensity range was adjusted for each array individually to account for differences in overall hybridization signal. To minimize effects of nonspecific hybridization from human transcripts, a threshold ratio of Cy5/Cy3 >2.5 was implemented.
Results
Seventy-Mer Oligonucleotide Design.
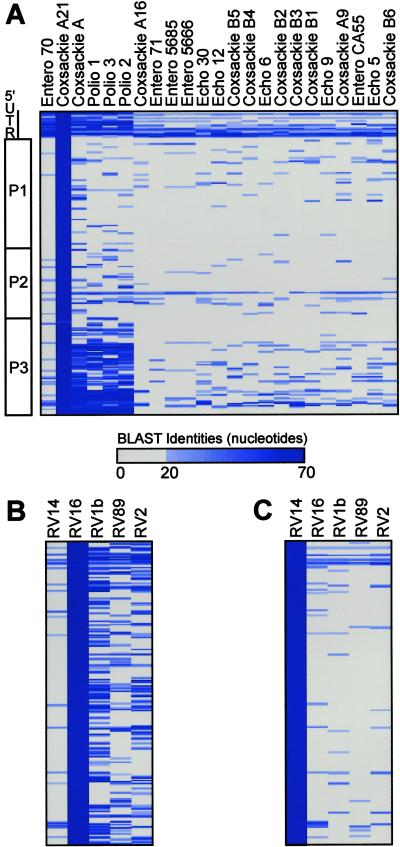
To maximize the spectrum of detectable viruses, the most highly conserved sequences within each viral family were selected for representation on the microarray as 70-mer oligonucleotides. The performance characteristics of long oligonucleotide microarrays have been documented (12). Homology between individual 70-nt viral fragments and each viral genome in the viral family (or genus) was assessed by the nucleotide identity score after blastn alignment. A simplified graphical representation of the regions of conservation between coxsackie virus A21 and other fully sequenced members of the enterovirus genus is shown in Fig. 1A as an example. Short regions of high nucleotide conservation, such as the sequences from the 5′ untranslated region, were evident throughout the genus. In most cases, these regions served as the primary source for array element selection. Similarly, analysis of RV16 (Fig. 1B) and RV14 (Fig. 1C) revealed the regions of conservation among the five fully sequenced members of the RV genus and recapitulated the known division of the RV genus into two taxonomic subgroups (18).
Fig 1.
(A) Graphical depiction of homology between coxsackie A21 and other enteroviruses. Picornavirus genomic organization is shown (Left). Seventy-nucleotide segments from the coxsackie A21 genome are ordered sequentially downward from the 5′ end of the genome. The number of nucleotides of identity between each 70-nt segment (rows) and each virus (columns) in the genus enterovirus is reflected by the intensity of the blue bar. Identities of <20 nt were plotted as gray. (B) Homology between RV16 and other RVs. (C) Homology between RV14 and other RVs.
Using the oligonucleotide selection strategy described above, we designed a first-generation viral detection microarray. The viral families represented on the microarray included double- and single-stranded DNA viruses, retroviruses, and both positive- and negative-stranded RNA viruses. Specifically, oligonucleotides were derived from potent human pathogens, including human T-lymphotropic virus, hepatitis B, hepatitis C, papillomaviruses, and all 20 fully sequenced human and animal herpes viruses. Five other viral families associated with respiratory tract infections (paramyxo-, orthomyxo-, nido-, adeno-, and picornavirus) were also extensively covered, using essentially every fully sequenced viral genome from these families. In total, the microarray harbors 1,600 unique viral oligonucleotides derived from ≈140 distinct viral genomes.
Detection of a Wide Range of Viruses from Cell Culture.
Initial validation of the microarray was accomplished by using RNA isolated from virally infected tissue culture cells. Viruses tested included KSHV, RSV, parainfluenza 3, poliovirus 1, adenovirus 12, and multiple serotypes of RV. In all cases, a two-color competitive hybridization was used to compare fluorescently labeled cDNA from virally infected cells to uninfected cells. Primary microarray data were converted to a color visualization scheme in which the Cy5 intensity was plotted as a linear yellow color scale (Fig. 2A). Hybridization results from de novo infections are shown in Fig. 2B, demonstrating the successful detection and classification of multiple viruses by family. In addition, endogenous viral infections, such as the presence of KSHV transcripts in the BCBL1 cell line (Fig. 2C), were readily detected.
Fig 2.
Detection of multiple viruses by DNA microarray. (A) One small region of a microarray is depicted with its corresponding bar graphic. Each detection oligonucleotide from the microarray is depicted as a vertical stripe. Detection oligonucleotides (stripes) are grouped by viral family of origin, and hybridization intensity on the microarray is reflected by the color of the stripes. Black indicates signal below threshold and Cy5 hybridization intensity above threshold is represented by a continuous color scale (yellow). The upper limit of the linear scale for each microarray was independently set to account for array-to-array hybridization variation. (B) Hybridization results from infection of HeLa cells with adenovirus 12, parainfluenza 3, RSV, polio1, and RV2. Detection oligonucleotides from the herpes and miscellaneous viruses are omitted for clarity (full plots are available in Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). (C) Hybridization results from the BCBL-1 cell line, which harbors KSHV. For clarity, only the herpes virus detection oligonucleotides are shown.
Detection of Multiple RV Serotypes.
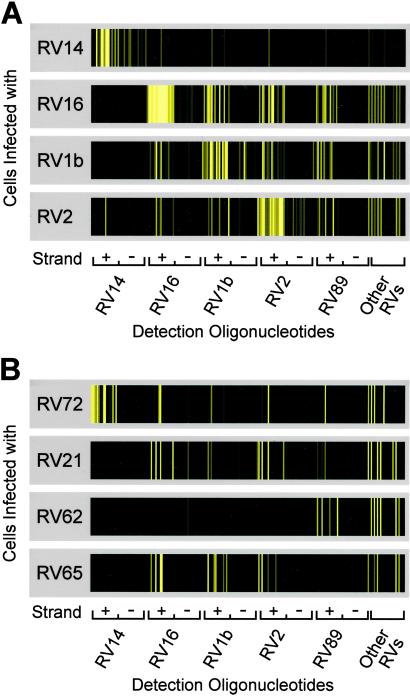
A total of 204 RV detection oligonucleotides designed to detect both the positive and negative strand of RV were present on the array. These oligonucleotides were derived primarily from the five fully sequenced RV genomes (of 102 serotypes of RV identified) in GenBank. Fig. 3 shows the hybridization patterns observed from RNA isolated from HeLa cells infected with eight different serotypes of RVs. The first four serotypes we examined (RV14, RV16, RV1b, and RV2) hybridized strongly to oligonucleotides derived from their respective genomic (positive-strand) sequences (Fig. 3A), making it possible to determine the virus subtype. In each of these cases the much less abundant negative-strand RNA was also detected (Fig. 3A). As predicted, cross-hybridization to spots derived from other RV serotypes was also observed, reflecting the successful representation on the array of conserved regions within the RV genus. Significantly, the presence of these conserved sequences on the microarray enabled detection of additional diverse serotypes (RV21, RV62, RV65, and RV72) (Fig. 3B), even though no sequence information from those serotypes was used in the oligonucleotide design process. This finding indicates that the chosen array elements are capable of broadly detecting many, if not all, RV serotypes. Moreover, a unique hybridization pattern for each serotype was observed, enabling discrimination between serotypes.
Fig 3.
Detection of multiple RV serotypes. Close-up views of the set of RV-derived detection oligonucleotides from hybridizations of different RV-infected HeLa cells. Cy5 hybridization intensities are plotted as a continuous color scale. Oligonucleotides are grouped by serotype of origin, with elements detecting the RV positive strand (+) followed by elements detecting the negative strand (−). The other RV category has 12 pairs of oligonucleotides (forward and reverse complement) derived from miscellaneous RV strains. (A) Four rhinoviruses used in the design of the detection oligonucleotides. (B) Detection of four additional RV serotypes.
The hybridization patterns also reflected the phylogenetic relationships between serotypes. As shown in Fig. 1 B and C and ref. 18, RV14 is in a subgroup distinct from RVs 1b, 2, 16, and 89. Correspondingly, only a minimal degree of cross-hybridization was observed between RV14 and array elements derived from the other sequenced RVs. Furthermore, RV72 is classified in the same subgroup as RV14 (18), and RV72 was the only tested serotype other than RV14 that hybridized strongly to the set of oligonucleotides derived from RV14.
Virus Detection in Clinical Samples.
To assess the performance of the microarray in a clinical setting, we examined nasal lavage fluid from a subject with a defined respiratory tract infection (deliberate inoculation with RV16) (13) as well as multiple patients with colds of undefined origin. As anticipated, whereas nasal lavage obtained before experimental infection with RV16 lacked appreciable RV signal, hybridization to many RV-derived oligonucleotides was detected in nasal lavage obtained from the same individual 2 days after infection (Fig. 4A). By visual inspection, the hybridization pattern from the clinical sample closely paralleled the pattern obtained after amplification of a reference sample of RV16 from infected HeLa cells (Fig. 4A). Note that after PCR amplification, the fluorescently labeled hybridization probes are double-stranded and thus amplified signatures differ from the single-stranded cDNA hybridization patterns in Fig. 3. Based on measured viral titers (TCID50, data not shown), the array detected RV in nasal lavage samples containing as few as 102 infectious RV particles.
Fig 4.
Detection of viruses in isolated nasal lavage. (A) Close-up of RV detection oligonucleotides: (Top) nasal lavage taken from patient before infection with RV16; (Middle) nasal lavage taken 2 days postinfection; (Bottom) amplified RV16-infected HeLa RNA. (B) Close-up of RV detection oligonucleotides from nasal lavage samples M18GV1 and J01LV1. (C) Close-up of family Paramyxoviridae-derived detection oligonucleotides, grouped by individual virus, after hybridization to sample H03AV1. Full plots for each hybridization are available in Fig. 6, which is published as supporting information on the PNAS web site.
We subsequently analyzed nasal lavage isolated from nine patients with natural colds. It is well known that RV infection is the leading cause of the common cold, but some cases result from infection with parainfluenza virus, corona virus, or RSV (1). In our initial microarray analysis, four samples gave clear indication of RV infection. Two examples of the RV-positive samples are shown in Fig. 4B. Distinct hybridization patterns were observed, suggesting that these patients were infected with different RV serotypes, thus underscoring our ability to detect a range of RV serotypes. The presence of RV in these clinical samples was independently confirmed by conventional RT-PCR and sequencing of the PCR products (data not shown). In addition, parainfluenza 1 was detected in sample H03AV1, as evidenced by hybridization primarily to the set of oligonucleotides derived from parainfluenza 1 (Fig. 4C). This observation was also confirmed by RT-PCR with parainfluenza 1-specific primers (data not shown). Cross hybridization was only observed to its close relatives parainfluenza 3 and Sendai virus (mouse parainfluenza 1).
Discussion
Existing methods to screen a broad range of viruses are inherently biased and thereby restricted to detecting a limited number of candidate viruses. To obviate this problem, we sought to develop a viral detection methodology based on a combination of viral genomics and long oligonucleotide DNA microarray technology. To achieve this goal, the most highly conserved 70-nt sequences within a viral family were chosen for representation on the microarray. By using the most conserved sequences, we hoped to maximize the probability that all members of each viral family, including unsequenced, unidentified, or newly evolved family members could be detected. As a secondary, but complementary, goal, we sought to take advantage of the high resolution of microarray hybridization to differentiate among viral subtypes, which is a difficult and problematic task with traditional methods.
Our initial studies used RNA isolated from tissue culture cells infected with a variety of viruses. These experiments demonstrated several powerful features of viral detection by using DNA microarrays. First, viruses that were explicitly represented on the microarray were readily detected and identified by specific hybridization to the appropriate oligonucleotides (Figs. 2 and 3A). This process was facilitated by internal redundancy; each virus was represented on the microarray by multiple oligonucleotides. One direct implication of these initial experiments is that any virus for which sequence information exists is capable of being detected by this approach. However, we have also demonstrated that detection is not limited to previously identified, sequenced viruses. By selecting highly conserved sequences within the RV genus as elements in our DNA microarray, several distinct serotypes of RV were still detected and identified as RVs in the absence of specific cognate oligoncleotides derived from those serotypes (Fig. 3B). These results demonstrate that maximizing potential cross-hybridization to conserved regions within a given viral family is a viable strategy for detecting unsequenced or uncharacterized viruses; this process may ultimately prove to be a useful approach to novel virus discovery.
In addition, distinct patterns of hybridization were obtained when different serotypes of RV were analyzed (Fig. 3). For example, there was almost no overlap between the hybridization patterns of some serotypes (RV14 and RV16), demonstrating the feasibility of subtype discrimination by microarray. One potential extension of these studies will be to establish a reference library of hybridization signatures, or “viral barcodes,” for hundreds of individual serotypes and to develop quantitative methods for comparing signatures to identify subtypes. Array-based genotyping of Mycobacteria (19) and, more recently, of rotaviruses has been reported (20). Classic serotyping of RVs (and other viruses) is tedious and limited by availability of antisera. As a consequence, field studies of many viruses have been severely hampered by a lack of serotype-specific data. Microarray-based viral detection may offer a powerful alternative for determination of viral subtypes.
Finally, we tested the efficacy of this diagnostic method in a clinical setting. Because the quantity of nucleic acid available for analysis from respiratory tract specimens is small, we used a random, sequence-independent PCR protocol to amplify material obtained from nasal lavage fluid. Diverse viruses, such as RV and parainfluenza, were detected on the microarray from clinical specimens after amplification in this fashion. The use of a random PCR step obviates the need for a priori knowledge of the infectious agent and thus identification is limited only by the spectrum of viral probes present on the array. This is in contrast to conventional RT-PCR-based detection schemes (2, 21), wherein the outcome is necessarily restricted by the initial selection of targets and corresponding primers. Even when degenerate multiplex RT-PCR is used, the range of target viruses remains narrow. A DNA microarray composed of carefully selected viral sequences, coupled to a random amplification step, bypasses these limitations and yields an extremely broad-reaching and unbiased detection strategy.
In conclusion, we have developed a genomic and microarray-based strategy for viral detection. Although our initial efforts were focused on only a few hundred viruses, efforts are now underway to include array elements derived from every sequenced human, animal, and plant virus. Such a diagnostic tool will undoubtedly have many uses in the study of viral pathogenesis and perhaps equally importantly has the potential to facilitate viral discovery and identification in diseases of unknown etiology as well as in instances of bioterrorism.
Supplementary Material
Acknowledgments
We thank L. Pereira (University of California, San Francisco) for cytomegalovirus oligonucleotides and D. Schnurr and S. Yagi (California Viral and Rickettsial Disease Laboratory, Richmond) for viral titer determination. We thank I. Herskowitz, M. Shuman, and members of the DeRisi laboratory for helpful discussions. This work was supported by an award from the Sandler Program in Asthma Research (to J.L.D.). The studies in which the clinical samples were collected were supported by Public Health Service Grants HL56385 and AI50496 (to H.A.B.).
Abbreviations
RV, rhinovirus
RSV, respiratory syncytial virus
KSHV, Kaposi's sarcoma-associated herpes virus
References
- 1.Storch G. A., (2000) Essentials of Diagnostic Virology (Churchill Livingstone, New York).
- 2.Elnifro E. M., Ashshi, A. M., Cooper, R. J. & Klapper, P. E. (2000) Clin. Microbiol. Rev. 13 559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broude N. E., Zhang, L., Woodward, K., Englert, D. & Cantor, C. R. (2001) Proc. Natl. Acad. Sci. USA 98 206-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saitoh-Inagawa W., Oshima, A., Aoki, K., Itoh, N., Isobe, K., Uchio, E., Ohno, S., Nakajima, H., Hata, K. & Ishiko, H. (1996) J. Clin. Microbiol. 34 2113-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeuchi S., Itoh, N., Uchio, E., Aoki, K. & Ohno, S. (1999) J. Clin. Microbiol. 37 1839-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liolios L., Jenney, A., Spelman, D., Kotsimbos, T., Catton, M. & Wesselingh, S. (2001) J. Clin. Microbiol. 39 2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreoletti L., Lesay, M., Deschildre, A., Lambert, V., Dewilde, A. & Wattre, P. (2000) J. Med. Virol. 61 341-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vinje J. & Koopmans, M. P. (2000) J. Clin. Microbiol. 38 2595-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215 403-410. [DOI] [PubMed] [Google Scholar]
- 10.Eisen M. B. & Brown, P. O. (1999) Methods Enzymol. 303 179-205. [DOI] [PubMed] [Google Scholar]
- 11.Renne R., Zhong, W., Herndier, B., McGrath, M., Abbey, N., Kedes, D. & Ganem, D. (1996) Nat. Med. 2 342-346. [DOI] [PubMed] [Google Scholar]
- 12.Hughes T. R., Mao, M., Jones, A. R., Burchard, J., Marton, M. J., Shannon, K. W., Lefkowitz, S. M., Ziman, M., Schelter, J. M., Meyer, M. R., et al. (2001) Nat. Biotechnol. 19 342-347. [DOI] [PubMed] [Google Scholar]
- 13.Fleming H. E., Little, F. F., Schnurr, D., Avila, P. C., Wong, H., Liu, J., Yagi, S. & Boushey, H. A. (1999) Am. J. Respir. Crit. Care Med. 160 100-108. [DOI] [PubMed] [Google Scholar]
- 14.Bohlander S. K., Espinosa, R., III, Le Beau, M. M., Rowley, J. D. & Diaz, M. O. (1992) Genomics 13 1322-1324. [DOI] [PubMed] [Google Scholar]
- 15.Blomqvist S., Skytta, A., Roivainen, M. & Hovi, T. (1999) J. Clin. Microbiol. 37 2813-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savolainen C., Blomqvist, S., Mulders, M. N. & Hovi, T. (2002) J. Gen. Virol. 83 333-340. [DOI] [PubMed] [Google Scholar]
- 17.Fan J. & Henrickson, K. J. (1996) J. Clin. Microbiol. 34 1914-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horsnell C., Gama, R. E., Hughes, P. J. & Stanway, G. (1995) J. Gen. Virol. 76 2549-2555. [DOI] [PubMed] [Google Scholar]
- 19.Gingeras T. R., Ghandour, G., Wang, E., Berno, A., Small, P. M., Drobniewski, F., Alland, D., Desmond, E., Holodniy, M. & Drenkow, J. (1998) Genome Res. 8 435-448. [DOI] [PubMed] [Google Scholar]
- 20.Chizhikov V., Wagner, M., Ivshina, A., Hoshino, Y., Kapikian, A. Z. & Chumakov, K. (2002) J. Clin. Microbiol. 40 2398-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson W. J., Strout, C. L., DeSantis, T. Z., Stilwell, J. L., Carrano, A. V. & Andersen, G. L. (2002) Mol. Cell. Probes 16 119-127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.