Abstract
Immunophilins are intracellular receptors of the immunosuppressants cyclosporin A, FK506, and rapamycin. Although all immunophilins possess peptidyl-prolyl isomerase activity and are identified from a wide range of organisms, little is known about their cellular functions. We report the characterization and functional analysis of an FK506 and rapamycin-binding protein (AtFKBP13) from Arabidopsis. The AtFKBP13 protein is synthesized as a precursor that is imported into chloroplasts and processed to the mature form located in the thylakoid lumen, as shown by chloroplast import assays and Western blot analysis. Experiments show that AtFKBP13 is translocated across the thylakoid membrane by the ΔpH-dependent pathway. Yeast two-hybrid screening identified Rieske FeS protein, a subunit of the cytochrome bf complex in the photosynthetic electron transport chain, as an interacting partner for AtFKBP13. Both yeast two-hybrid and in vitro protein–protein interaction assays showed that the precursor, but not the mature form, of AtFKBP13 interacted with Rieske protein, suggesting that interaction between the two proteins occurs along the import pathway. When AtFKBP13 expression was suppressed by RNA interference method, the level of Rieske protein was significantly increased in the transgenic plants.
In the studies of immunosuppression by cyclosporin A, FK506, and rapamycin, receptor proteins for these drugs have been identified and characterized at the molecular level. Cyclosporin A binds to a group of proteins called cyclophilins, whereas FK506 and rapamycin both bind to FK506 and rapamycin-binding proteins (FKBPs). Cyclophilins and FKBPs are collectively referred to as immunophilins (1–3). Further studies have shown that the drug ligands bind to the receptors forming immunosuppressive complexes that target other components required for immune function (4, 5). Although the immunosuppression mechanism by cyclosporin A and FK506 has been determined, the function of their endogenous receptors in the absence of the drug ligands remains unknown. Studies have shown that all immunophilins have peptidylproline isomerase activity (1–3), suggesting a possible function for these proteins in protein-folding pathways. Evidence has accumulated in recent years to support this hypothesis. At least in several cases, immunophilins play a role as both rotamase and molecular chaperone (6, 7).
In higher plants, a family of cyclophilins with at least four members has been identified in Arabidopsis (8–10), and two major forms were found in Vicia faba (11, 12). The FKBP-type immunophilins from a higher plant were first purified by using an affinity chromatography approach (13). This biochemical approach suggests that at least four members are present in the FKBP family of V. faba, and these members are located in various compartments of plant cells (11). Two endoplasmic reticulum FKBP isoforms (FKBP15–1 and FKBP15–2) have been characterized in Arabidopsis (14). At least two high-molecular weight FKBP members have been identified from wheat (wFKBP73 and wFKBP77), two from Arabidopsis (ROF1/AtFKBP61 and PAS1/AtFKBP71) and one from maize (mzFKBP66; refs. 15–20). A cytosolic FKBP12 also has been characterized in Arabidopsis (21, 22). Analysis of the Arabidopsis genome sequence database indicates that there are at least 17 genes encoding distinct FKBP-like proteins in Arabidopsis (23).
Regarding the function of immunophilin proteins in plants, two recent reports suggest that they participate in important processes in plant development (19, 24). One study shows that an FKBP-like protein is essential for normal cell division and differentiation in Arabidopsis (19). The other report identified a cyclophilin as a critical regulator of normal development of leaf size and shape in Arabidopsis (24). Here, we report that a chloroplast-localized FKBP from Arabidopsis (AtFKBP13) regulates the accumulation of Rieske protein, an essential component of the photosynthetic electron transport chain.
Materials and Methods
Cloning and Characterization of a cDNA Encoding AtFKBP13.
A chloroplast-localized 13-kDa FKBP protein was purified from V. faba plants by affinity chromatography, as described (11). The N-terminal peptide sequence was obtained by microsequencing and used as a query to search the Arabidopsis sequence database. A genomic sequence (GenBank accession no. AB012245) was identified that encodes a protein that is highly homologous (74% identical) to V. faba FKBP13 protein over the sequenced N-terminal region of 42 aa. A 300-bp RT-PCR fragment was used to screen a cDNA library (25), resulting in isolation of a full-length AtFKBP13 cDNA.
RNA Gel Blot Analyses.
Arabidopsis plants (ecotype Columbia-gl) were grown in a greenhouse under long-day conditions (16-h light/8-h dark cycle). Total RNA from different organs was extracted by using Tripure reagent (Roche Molecular Biochemicals) according to the manufacturer's instructions. RNA gel blot analysis was performed as described (26).
Chloroplast Import Assays and Protein Localization.
Radiolabeled AtFKBP13 precursor was synthesized by a coupled transcription and translation procedure in a wheat germ extract, in the presence of [35S]methionine and [35S]cysteine. Chloroplasts were isolated from pea and incubated with the precursor protein, as described (27). Import assays contained intact chloroplasts (0.5 mg chlorophyll/ml), 5 mM methionine, 5 mM cysteine, and 10 mM MgATP in a final volume of 500 μl of import buffer (50 mM Hepes⋅KOH, pH 8.0/0.33 M sorbitol) with 45 μl of products from translation in vitro and were incubated in the light (100 μmol photons m−2 s−1) for 45 min. For protein import in the presence of nigericin or sodium azide, isolated chloroplasts were incubated with 5 μM nigericin or 10 mM sodium azide for 10 min on ice. 35S-labeled precursor protein was then added and samples were incubated at 25°C for 25 min in the light. After import, protein samples were analyzed by electrophoresis on 20% polyacrylamide gels in the presence of SDS followed by fluorography. For protein localization, Arabidopsis chloroplasts were isolated from protoplasts according to an earlier method (28). The stromal and luminal fractions were analyzed by Western blotting to determine the distribution of AtFKBP13 (and plastocyanin) in these two fractions.
Yeast Two-Hybrid Screening.
A Gal4p-based two-hybrid system (29) was used in this study. The coding region of the cDNA-encoding precursor of AtFKBP13 was amplified by PCR and cloned into pGBT9.BS in frame with the Gal4 DNA-binding domain. The yeast two-hybrid library screening was carried out essentially as described (30). The Arabidopsis ACT cDNA expression library CD4–22 was obtained from Arabidopsis Biological Resource Center (Columbus, OH). The β-galactosidase expression of the His+ colonies was analyzed by using a filter-lifting assay (30). For additional interaction assays between various domains of AtFKBP13 and Rieske protein, the DNA fragments encoding various domains were amplified with PCR by using flanking primers and cloned in frame with either activation or binding domain. All of the PCR reactions were performed by using pfu polymerase (Stratagene), and each fusion construct was confirmed by DNA sequencing. Assays for β-galactosidase activity were performed in triplicate by using chlorophenol red-β-d-galactopyranoside as substrate (30).
Expression and Purification of Recombinant Proteins in Escherichia coli.
To make GST-fusion constructs, cDNA fragments encoding various proteins or protein domains of AtFKBP13 and Rieske were amplified by PCR and cloned in frame with GST into pGEX-KG vector (Amersham Pharmacia). Fusion proteins were produced and purified as described (26).
Production and Purification of AtFKB13 and AtRieske Antibodies.
The mature form of AtFKBP13 and lumen domain of AtReiske protein (800 μg of each) were injected into rabbits (Cocalico Biological, Reamstown, PA), and antibodies were purified from anti-serum by using an affinity purification procedure (31).
In Vitro Protein–Protein Interaction Assays.
Purified precursor or mature form of AtFKBP13 protein was mixed with GST-Rieske fusion proteins (1:1 molar ratio), immobilized on the glutathione beads in a final volume of 500 μl containing binding buffer (50 mM Tris⋅HCl, pH 7.5/100 mM NaCl/0.1% Tween 20/1 mM PMSF). After gentle shaking for 1.5 h at room temperature, the beads were pelleted and washed three times with the binding buffer. Proteins were eluted by 10 mM glutathione, resolved by SDS/PAGE, and detected by Western blot analysis.
Generation of AtFKBP13 RNAi Construct and Arabidopsis Transformation.
To silence the AtFKBP13 gene, we used an RNA interference (RNAi) approach. The construct contained a partial GUS coding sequence in between the inverted repeat of partial AtFKBP13 cDNA sequence (nt 1–639 of cDNA) as described (32). The expression of RNAi cassette was driven by CaMV 35S promoter. This construct was named AtFKBP13 RNAi and transferred into Agrobacterium tumefaciens GV3101 strain for Arabidopsis transformation by the “floral dip” method (33). The putative transformants were rescued from kanamycin-containing medium and grown in green house under long-day conditions.
Protein Extraction and Western Blot Analysis.
Total proteins from leaves of 4-week old plants were extracted in a buffer (50 mM Tris, pH 7.5/100 mM NaCl/1 mM EDTA/0.1% Triton X-100/5 mM DTT/1 mM PMSF/1 mM benzamidine/5 μg/ml leupeptin/5 μg/ml aprotinin/5 μg/ml pepstatin A). The homogenate was mixed for 15 s, incubated on ice for 5 min, and centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was collected, and proteins were quantified by using the Bradford assay kit (Bio-Rad). For Western blot analysis, 30 μg of total proteins were separated in SDS/12% polyacrylamide gel and proteins were transferred onto nitrocellulose membrane. Bound antibodies were detected with a chemiluminescence kit (Amersham Pharmacia).
Results
Isolation and Characterization of a cDNA Encoding AtFKBP13.
We previously reported that a major FKBP in leaf tissues of V. faba is a 13-kDa protein (11). We purified a large quantity of FKBP13 protein from leaf extract of V. faba plants by using an FK506 affinity column and determined that the N-terminal amino acid sequence of the protein is AAEVAPCELTVAPSGLSFCDKVVGTGPQAVKGQLIKAHYVGRLEN.
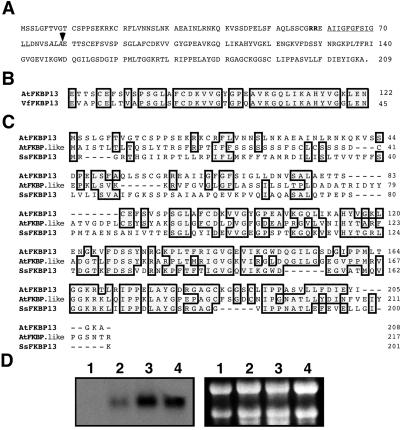
Based on this sequence information, we cloned a full-length cDNA encoding a homologue of V. faba FKBP13 protein from Arabidopsis, as described in Materials and Methods. This cDNA encodes a protein of 208-aa residues with a calculated molecular mass of 22 kDa, larger than the expected 13-kDa protein (Fig. 1A). Comparing the sequence of this gene product with the N-terminal sequence of mature V. faba FKBP13 revealed a 79-aa presequence in the Arabidopsis cDNA-deduced protein (Fig. 1B). The putative mature protein consists of 129-aa residues with a calculated molecular mass of 13.6 kDa. Based on the molecular weight and the high sequence identity between this protein and the V. faba FKBP13, we named this gene product AtFKBP13 (Arabidopsis thaliana FKBP13).
Fig 1.
Sequence and expression analysis of AtFKBP13 gene. (A) The amino acid sequence of the AtFKBP13 protein deduced from the cDNA. The double arginine motif and hydrophobic domain are shown as bold and underlined, respectively. The thylakoidal processing peptidase site (AxA) is shown in italics and the arrow indicates the putative cleavage site. (B) A comparison of N-terminal sequence of V. faba FKBP13 (VfFKBP13) and cDNA-deduced AtFKBP13 protein. (C) Amino acid sequence alignment of AtFKBP13, FKBP-like protein from Arabidopsis (AtFKBP.like, GenBank accession no. AJ242481) and a putative FKBP from Synechocystis sp. (SsFKBP13, GenBank accession no. S75144; ref. 35). In B and C, the shaded boxes indicate identical amino acid residues. The numbers indicate the amino acid positions in each protein. Dashes depict the gaps introduced to improve the alignments. (D) RNA gel blot analysis of the AtFKBP13 gene. (Left) RNA gel blot analysis of total RNA (10 μg) from roots (lane 1), stems (lane 2), leaves (lane 3), and developing flower buds (lane 4). (Right) Ethidium bromide-stained ribosomal RNAs in the corresponding agarose gel showing the relative amount of total RNA in each lane.
The AtFKBP13 protein contains one putative FKBP-like binding domain which spans from G92 to I205 (see Fig. 1A) including the 10 conserved amino acid residues (Y116, G118, F126, D127, V145, I146, W149, Y178, I192, and F200) that are essential for binding and maintaining the hydrophobic core of FK506 (34). Comparing AtFKBP13 to FKBPs from different organisms, we found that AtFKBP13 shares significant sequence homology with all other FKBPs. The highest sequence similarity was found among AtFKBP13 and another FKBP-like protein (GenBank accession no. AJ242481) from Arabidopsis and a putative FKBP (GenBank accession no. S75144) from Synechocystis sp. (Fig. 1C; ref. 35).
Genomic Southern blot analysis and search of Arabidopsis genome sequence found only one copy of the AtFKBP13 gene (data not shown). Stems, leaves, and developing flower buds contained green tissues and accumulated high levels of AtFKBP13 transcripts (Fig. 1D, lanes 2, 3, and 4), whereas roots did not show detectable AtFKBP13 mRNA (Fig. 1D, lane 1).
AtFKBP13 Is Targeted to the Thylakoid Lumen of Chloroplasts by the ΔpH-Dependent Pathway.
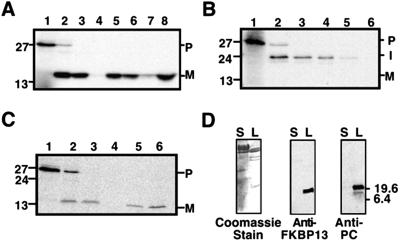
To determine whether AtFKBP13 is targeted to the chloroplast, and if so, what its suborganellar location is, protein import assays were performed with isolated chloroplasts and AtFKBP13 precursor as a substrate. The translated AtFKBP13 precursor was ≈27 kDa (estimated by mobility in SDS/PAGE). When isolated intact pea chloroplasts were incubated with the precursor protein in the presence of ATP, a 13-kDa protein was generated (Fig. 2A, lane 2). After incubations with the protease thermolysin (which under the conditions used does not penetrate the chloroplast envelope), intact chloroplasts were reisolated and fractionated. The resistance of the 13-kDa polypeptide to degradation by exogenously added thermolysin (Fig. 2A, lane 3) indicates that it is located within the chloroplast and is a product of precursor protein import. Further analysis revealed that AtFKBP13 protein was associated with the thylakoids (Fig. 2A, lane 5) and not in the stroma (Fig. 2A, lane 4). Sonication of the thylakoid fraction liberated the 13-kDa polypeptide in a soluble form (Fig. 2A, lane 8), indicating that the protein is located in the thylakoid lumen. From this experiment, we concluded that AtFKBP13 is a previously uncharacterized thylakoid lumen protein.
Fig 2.
AtFKBP13 is targeted to the chloroplast thylakoid lumen by the ΔpH-dependent pathway. (A) Chloroplast import assay of AtFKBP13. (B) Import assay of AtFKBP13 in the presence of 5 μM nigericin. (C) Import assay of AtFKBP13 in the presence of 10 mM sodium azide. (A–C) Translation products (lane 1); chloroplasts (lane 2); thermolysin-treated chloroplasts (lane 3); stromal fraction (lane 4); thylakoid fraction (lane 5); thermolysin-treated thylakoid fraction (lane 6). (A) Sonicated thylakoid membrane fraction (lane 7); soluble contents of thylakoid lumen (lane 8). The P, I, and M on the right side indicate precursor, intermediate, and mature forms of AtFKBP13, respectively. (D) Western blot analysis of stromal (lane S) and thylakoid lumen (lane L) fractions of Arabidopsis chloroplasts by Coomassie staining (Left), anti-AtFKBP13 (Middle), and anti-PC antibodies (Right). Approximate molecular masses (kDa) are shown on the sides.
The N-terminal extension of AtFKBP13 has the characteristic features of a thylakoid lumen protein presequence (Fig. 1A). It is bipartite, with the first region being hydrophilic in nature and enriched in basic and hydroxylated residues, which are features of a chloroplast envelope-transfer signal (36). As shown in Fig. 1A, the twin arginine motif is present in the presequence of AtFKBP13 and is followed by a highly hydrophobic domain ending with AXA. These features suggest that AtFKBP13 may be translocated into the thylakoid lumen by the ΔpH-dependent pathway (36). We tested this hypothesis by including nigericin, an inhibitor of ΔpH formation across the thylakoid membrane (36, 37).
As shown in Fig. 2B, nigericin completely inhibited the generation of the mature, 13-kDa polypeptide during import incubations, and an intermediate form of ≈24 kDa accumulated (Fig. 2B, lane 2). The intermediate form was resistant to degradation by thermolysin treatment of intact chloroplasts (Fig. 2B, lane 3), demonstrating that it was inside the chloroplast and was present in both the stromal and thylakoid fractions (Fig. 2B, lanes 4 and 5). The intermediate form in the thylakoid fraction was not protected from degradation by thermolysin (Fig. 2B, lane 6), indicating that the protein was on the stromal face of the thylakoid membrane. These results suggest that a transthylakoidal ΔpH is required for the efficient targeting of AtFKBP13 to the thylakoid lumen.
To support this conclusion, an import experiment was conducted by using 10 mM sodium azide, which is an inhibitor of Sec pathway (38). As shown in Fig. 2C, azide did not affect the targeting of AtFKBP13 into thylakoid lumen (Fig. 2C, lane 6). Thylakoid translocation was not dependent on ATP or stromal extract, further confirming that this protein is not targeted by the Sec pathway (data not shown). Control import assays were conducted with the nuclear-encoded thylakoid lumen proteins OEC23 (23-kDa component of the photosystem II oxygen-evolving complex) and plastocyanin, which are targeted to the lumen by ΔpH and Sec pathway, respectively (36, 38). We found that AtFKBP13 and OEC23 have exactly the same import behavior.
To confirm that native AtFKBP13 is localized in the thylakoid lumen, chloroplasts from Arabidopsis leaves were isolated and fractionated into stromal and lumen fractions. These two fractions were analyzed by Western blot analysis by using AtFKBP13 antibodies. AtFKBP13 was detected in the lumen fraction (Fig. 2D Center, lane L) but not in the stromal fraction (lane S). As a positive control, a Western blot was also performed with plastocyanin antibodies. Plastocyanin was present in the lumen fraction where AtFKBP13 was detected (Fig. 2D Right, lane L). The upper band in anti-PC Western blot (Fig. 2D Right, lane L) was most probably the result of dimerization of PC because this band disappeared in the urea-containing gel (data not shown).
AtFKBP13 Precursor Interacts with the Rieske Protein.
To function as chaperone or other regulatory entities, immunophilins often physically interact with their target proteins (3, 23). To identify a putative target for AtFKBP13, we used a yeast two-hybrid screening procedure. The precursor of AtFKBP13 was used as a bait in fusion with Gal4 DNA-binding domain to screen an Arabidopsis cDNA library in Gal4 activation-domain vector. Among the AtFKBP13 interacting clones that were sequenced, a number of them encoded the Rieske protein of various length fused in frame with the activation domain. The longest one had four amino acid truncations in the N terminus. Riesk protein is an essential subunit of cytochrome bf complex in the photosynthetic electron transport chain (39). It is located in the thylakoid with a transmembrane domain and a soluble region in the lumen (39). As AtFKBP13 is located in the lumen, Rieske protein may serve as a physiological target for AtFKBP13.
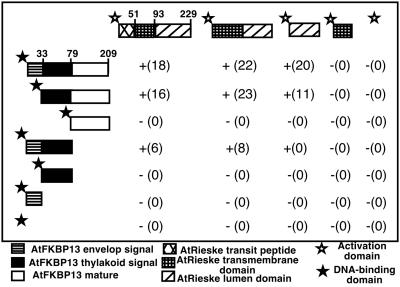
A search of Arabidopsis EST database identified an EST encoding full-length precursor of Rieske protein. This EST 39G11T7 (GenBank accession no. AJ243702) was sequenced on both strands to confirm its identity and was used in further experiments to determine the interacting domains of AtFKBP13 and Rieske proteins. A number of cDNA fragments encoding various domains of AtFKBP13 or Rieske protein were fused in frame with either DNA-binding or activation domain of Gal4 protein in the vectors. Different combinations of these constructs were cotransformed into yeast strain Y190, and interactions were studied by growth on selection medium and filter lift assay and quantified by the β-galactosidase activity. The precursor of AtFKBP13 interacts with the precursor, mature, and lumen domain but not with transit peptide of the Rieske protein (Fig. 3). When chloroplast envelope signal peptide was deleted from AtFKBP13, the truncated protein retained interaction with Rieske protein. However, the mature form of AtFKBP13 lacking the entire transit peptide did not interact with full-length or any domain of Rieske protein (Fig. 3). The transit peptide of AtFKBP13 interacted with precursor and mature form of Rieske, although the interactions were significantly weaker compared with the interaction between AtFKBP13 precursor and Rieske protein. The chloroplast envelope signal or thylakoid-targeting signal alone did not interact with full-length or any domain of Rieske protein (Fig. 3). We conclude that the transit peptide of AtFKBP13 is required and, to certain degree, sufficient for the interaction with Rieske protein. The mature region, more specifically, the lumen domain, of Rieske protein is sufficient for interaction.
Fig 3.
Interaction of Rieske and AtFKBP13 proteins assayed by yeast two-hybrid system. Interaction of various domains of AtFKBP13 and Rieske protein in yeast two-hybrid system. The symbols indicate interaction (+) and no interaction (−), whereas numbers in parentheses show the β-Gal activity in Miller units. The numbers associated with precursors of AtFKBP13 and Rieske protein indicate the positions of amino acid residues in these proteins. Results from one of the representative experiments of three duplicates are shown.
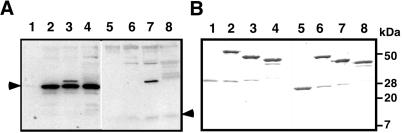
We also tested the interaction between AtFKBP13 and Rieske protein by in vitro protein–protein interaction assay. As shown in Fig. 4, precursor, mature, or lumen domain of Rieske protein copurified with the precursor but not with the mature form of AtFKBP13 (Fig. 4A). When GST was used as an affinity agent, neither precursor nor mature form of AtFKBP13 was copurified (Fig. 4A, lanes 1 and 5). The AtFKBP13 antibodies also reacted with an unknown 33-kDa protein associated only with the preparation of the GST-Rieske mature protein (Fig. 4A, lanes 3 and lane 7). The amount of GST and other bait proteins used in the experiment are shown in Fig. 4B. Some GST was detected in the preparations of Rieske precursor and mature protein fusions, suggesting that a small portion of GST fusion proteins were degraded (Fig. 4B, lanes 2, 3, 6, and 7).
Fig 4.
Interaction between AtFKBP13 and Rieske protein determined by in vitro protein interaction assays. GST (lanes 1 and 5), GST-Rieske precursor (lanes 2 and 6), GST-Rieske mature form (lanes 3 and 7), and GST-Rieske lumen domain (lanes 4 and 8) were immobilized on glutathione beads and used to “pull-down” purified precursor (lanes 1–4) or mature (lanes 5–8) form of AtFKBP13. The approximate molecular masses (kDa) are shown on the right side. (A) An immunoblot of copurified products probed with anti-AtFKBP13. The arrows on the left and right sides indicate the position of precursor and mature form of AtFKBP13, respectively. (B) A Coomassie blue-stained gel shows the amount of each bait protein used in the pull-down experiment.
Because the precursor, but not the mature form, of AtFKBP13 interacted with Rieske protein, and precursor of AtFKBP13 was not detectable by Western blot in Arabidopsis plants, it was difficult to study in vivo interaction of the two partner proteins by using immunoprecipitation. Indeed, we successfully precipitated AtFKBP13 mature protein by using AtFKBP13 antibody but did not detect any Rieske protein that was copurified. In a similar manner, Rieske antibody was used to precipitate Rieske protein but AtFKBP13 was not copurified (data not shown). This further supports the idea that the two mature proteins most probably do not interact with each other.
AtFKBP13 Down-Regulates Accumulation of Rieske Protein.
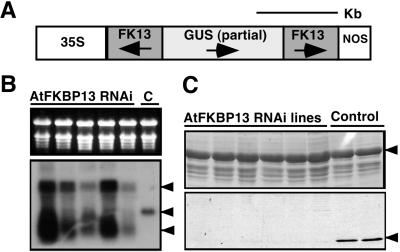
AtFKBP13 is localized in the thylakoid lumen, and Rieske protein is localized in the thylakoid membrane with a soluble domain in the lumen, making it physically possible for the two mature proteins to interact. However, our results suggest that these proteins probably interact before they are targeted to the final destination. What is the functional relevance of interaction between the two protein partners? To address this question, we took a “reverse genetics” approach to suppress the expression of AtFKBP13 in transgenic plants and examined the consequence of this manipulation to the Rieske protein. As described in Materials and Methods, AtFKBP13 gene was silenced by using RNAi procedure (32). The RNAi construct is illustrated in Fig. 5A. As expected endogenous AtFKBP13 mRNA (Fig. 5B Lower, lane C) was not detected in the RNAi plants (Fig. 5B Lower, middle arrow, lanes 1–5). Instead, the RNAi lines produced small RNA species hybridized to AtFKBP13 cDNA. In most of the RNAi plants, a larger RNA species of about 2 kb was detected, which corresponds to the full-length transcript from the RNAi cassette (Fig. 5B Lower, lanes 1–5). To examine the AtFKBP13 protein level in the RNAi plants, Western blot was performed by using AtFKBP13 antibody (Fig. 5C). In almost all of the transgenic lines harboring the RNAi construct, AtFKBP13 protein was not detectable (Fig. 5C, lanes 1–6), whereas control lines transformed with empty vector produced normal level of AtFKBP13 (Fig. 5C, lanes 7 and 8). These results indicated that the RNAi procedure effectively silenced the expression of AtFKBP13 in transgenic plants.
Fig 5.
Silencing of AtFKBP13 in transgenic Arabidopsis plants by RNAi. (A) Schematic representation of AtFKBP13 RNAi construct. Shaded boxes depict the inverted repeat of AtFKBP13 cDNA. (B) RNA gel blot of AtFKBP13 RNAi and control plant (lane C). (Upper) Ethidium bromide-stained ribosomal RNA showing the relative amount of total RNA in each lane. (Lower) AtFKBP13 level detected by AtFKBP13 cDNA probe. Arrows indicate RNAi cassette transcript, endogenous AtFKBP13 mRNA, and degraded form of AtFKBP13, respectively. (C) Western blot analysis of AtFKBP13 protein levels in the RNAi and control plants. (Upper) Coomassie blue-stained gel showing the large subunit of Rubisco (shown with arrow) as a loading control. (Lower) Anti-AtFKBP13 Western blot. The AtFKBP13 protein is indicated by an arrow on the right side.
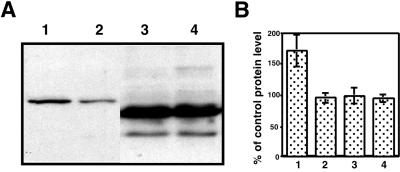
If AtFKBP13 interaction with Rieske protein is functionally relevant, it is expected that AtFKBP13 silencing would alter the level or function of Rieske protein. Because the interaction may occur before the proteins are imported to the final destination, we suspected that AtFKBP13 might regulate the accumulation of Rieske protein. As Rieske protein is an essential subunit of cytochrome bf complex in the photosynthetic electron-transport chain, changes in the Rieske protein may affect the photosynthetic electron transport. We assayed the relative PSII electron-transport rate of control and RNAi plants but did not observe any significant difference (data not shown). It is possible that the Rieske protein was modified in a more subtle manner. We examined the level of Rieske protein in the control and RNAi plants by Western blot analysis and found a significant difference in the accumulation of Rieske protein. As shown in Fig. 6, the RNAi plants produced higher levels of Rieske protein (Fig. 6A, lane 1) as compared with the control plants (Fig. 6A, lane 2). Statistical analysis on eight RNAi lines, eight empty vector control lines, and wild-type plants indicated that Rieske protein in the AtFKBP13-silenced plants produced ≈70% more Rieske (Fig. 6B, lanes 1 and 2), which was significant (P < 0.01). As a control, a Western blot analysis with plastocyanin antibody did not reveal significant difference (P > 0.05) in the accumulation of PC in the RNAi and control plants (Fig. 6, lanes 3 and 4). RNA gel blot analysis did not detect any significant difference in the Rieske mRNA levels among RNAi and control plants (data not shown), suggesting that AtFKBP13 affects the level of Rieske protein by a posttranscriptional process.
Fig 6.
Rieske protein accumulation in AtFKBP13 silenced plants. (A) Western blot analysis of AtFKBP13 RNAi (lanes 1 and 3) and control plants (lanes 2 and 4) with anti-Rieske (lanes 1 and 2) or anti-PC (lanes 3 and 4) antibody. (B) Statistical analysis of Rieske protein and PC levels in eight independent transgenic lines of AtFKBP13 RNAi (lane 1 and 3) or empty vector control lines (lane 2 and 4). Rieske (lanes 1 and 2) and PC (lanes 3 and 4) levels were measured by scanning the autoradiograph of Western blot of eight RNAi and eight vector lines. The band density average of vector lines is designated as 100%, and the density average of RNAi lines was compared and shown as a percentage of the control. (Bars = SD in eight samples.)
Discussion
Although immunophilins have been identified in a broad range of organisms from bacteria and plants to mammals, their physiological function is not well understood in any organism (6, 23). A yeast mutant disrupted in all known immunophilin genes has been constructed and shown to be viable (40), suggesting that immunophilins are not essential in yeast. In higher eukaryotes including plants and animals, rapidly accumulating evidence supports the hypothesis that each member in the immunophilin families plays a role in a specific cellular processes. For example, FKBP12 has been shown to interact with and regulate the function of ryanodine receptors, a family of calcium-release channels in the endoplasmic reticulum of muscle and other cell types (41). More recently, genetic disruption of FKBP12 gene in a transgenic mouse model clearly demonstrates that FKBP12 is essential to the normal function of calcium release channels: the mutant animals develop cardiovascular disorder (42). Another example is FKBP52 that interacts with the steroid-receptor complex in the cytoplasm (2, 3). The chaperone activity of FKBP52 is required for the regulation of steroid-receptor activity and nuclear translocation (3). A good example of cyclophilin function was presented by a genetic analysis that demonstrated a role of a cyclophilin protein in the trafficking of rhodopsin protein isoform in Drosophila eye (43).
In higher plants, a number of genes encoding immunophilins have been isolated in previous studies from several laboratories (2, 23). Completion of the Arabidopsis genome sequencing project revealed at least 51 genes that encode cyclophilins and FKBPs (S.L., unpublished results). The finding of these proteins has attracted much attention in plant biology. Several recent reports showed that some immunophilin members are required for the regulation of plant developmental processes. In one study, the authors found (19, 44) that a high molecular-weight FKBP-like protein, PAS1, is localized in the nucleus and is required for the control of cell division and differentiation in Arabidopsis. Loss of function mutant of PAS1 displays excessive activity of cell division, and a number of organs are transformed into callus-like structures (19). A homologue of cyclophilin 40, an immunophilin that interacts with heat-shock protein 90 in various organisms, is essential for normal development of vegetative organs in Arabidopsis (24).
Chloroplast is a green plant-specific organelle that not only serves as a “factory” for carbon fixation through photosynthesis but also plays an essential role in many other biosynthetic pathways (45). Despite the fact that chloroplasts contain their own genome, a large fraction of chloroplast proteins are encoded by nuclear genome, produced in the cytoplasm, and imported into various compartments of the organelle (36). Molecular chaperones, including HSP70, HSP60, and HSP100, play a vital role in the import and refolding of chloroplast proteins (3). Previous studies have shown that chloroplasts harbor several immunophilins (11, 12, 46). Based on their activity as rotamase and their possible function as chaperone, we speculated that chloroplast immunophilins are involved in protein import and/or refolding processes. In this study, we have characterized the first chloroplast FKBP from Arabidopsis and identified a component in the photosynthetic electron transport, the Rieske protein, as a putative target for this immunophilin member. The interaction between AtFKBP13 and the Rieske protein probably occurs before they are imported into the thylakoid, because the mature proteins do not interact with each other (Figs. 3 and 4). Both yeast two-hybrid and in vitro protein interaction assays demonstrated that the full-length precursor proteins (or cytoplasmic forms) interacted. In addition, the intermediate (or stromal) forms of the two protein partners also interacted well. These results suggest that AtFKBP13 associates with Rieske protein both before and after the import of the proteins into the chloroplast stroma. In addition, AtFKBP13 and Rieske intermediate forms also can interact after they are imported into the thylakoid lumen but before the thylakoid signal peptides are cleaved. After the cleavage of the transit peptide, the mature forms of the two proteins probably dissociate.
The Rieske protein has been found to associate with other chaperones including HSP70 and Cpn60 in the stroma (47). Interestingly, the previous study showed that only half of the Rieske protein is recovered from the protein complexes formed with HSP70 and Cpn60, suggesting that Rieske protein also may associate with other proteins (47). Our study has added another putative chaperone, AtFKBP13, into the regulatory circuit of Rieske protein during import process. It is generally believed that chaperones help imported proteins refold and should be positive regulators of the protein-accumulation processes. None of the studies has addressed the consequence of these interactions by genetic analysis. In this report, we used a gene-silencing method to assess the consequence of AtFKBP13–Rieske interaction. We have found that suppression of AtFKBP13 expression significantly increased the level of Rieske protein (Fig. 6). This finding suggests that AtFKBP13 interaction with Rieske may play a role in the down-regulation of Rieske protein accumulation. We speculate a possible mechanism based on this result. The AtFKBP13 protein may serve as an “anchor” chaperone that holds the Rieske protein in the cytoplasm or in the stroma so that excessive Rieske protein is not targeted to the thylakoid. Because Rieske is a subunit of cytochrome bf complex composed of a number of other protein subunits, coordination among the subunit proteins might be required for efficient assembly of the complex. Regulation of Rieske levels in the thylakoid could be important, and AtFKBP13 interaction may be responsible for this regulation. When AtFKBP13 level is suppressed, more Rieske protein would be targeted to the thylakoid because of reduced capacity of anchor proteins. Further study is required to test this hypothesis.
Acknowledgments
We thank Drs. John Gray, Colin Robinson, and Dick Malkin for providing PC, OEC23K clones, and PC antibodies, respectively. This work was supported by the National Institutes of Health and the U.S. Department of Energy. R.M.M. is supported by the Royal Society, U.K.
Abbreviations
RNAi, RNA interference
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ490171).
References
- 1.Schreiber S. L. (1991) Science 251 283-287. [DOI] [PubMed] [Google Scholar]
- 2.Luan S. (1998) Bot. Bull. Acad. Sin. (Taipei) 39 217-223. [Google Scholar]
- 3.Pratt W. B., Krishna, P. & Olsen, L. J. (2001) Trends Plant Sci. 6 54-58. [DOI] [PubMed] [Google Scholar]
- 4.Rao A., Luo, C. & Hogan, P. G. (1997) Annu. Rev. Immunol. 15 707-747. [DOI] [PubMed] [Google Scholar]
- 5.Schreiber S. L. & Crabtree, G. R. (1992) Immunol. Today 13 136-142. [DOI] [PubMed] [Google Scholar]
- 6.Freeman B. C., Toft, D. O. & Morimoto, R. I. (1996) Science 274 1718-1720. [DOI] [PubMed] [Google Scholar]
- 7.Freskgard P. O., Bergenhem, N., Jonsson, B. H., Svensson, M. & Carlsson, U. (1992) Science 258 466-468. [DOI] [PubMed] [Google Scholar]
- 8.Chou I. T. & Gasser, C. S. (1997) Plant Mol. Biol. 35 873-892. [DOI] [PubMed] [Google Scholar]
- 9.Fulgosi H., Vener, A. V., Altschmied, L., Herrmann, R. G. & Andersson, B. (1998) EMBO J. 17 1577-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasser C. S., Gunning, D. A., Budelier, K. A. & Brown, S. M. (1990) Proc. Natl. Acad. Sci. USA 87 9519-9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luan S., Albers, M. W. & Schreiber, S. L. (1994) Proc. Natl. Acad. Sci. USA 91 984-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luan S., Lane, W. S. & Schreiber, S. L. (1994) Plant Cell 6 885-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luan S., Li, W., Rusnak, F., Assmann, S. M. & Schreiber, S. L. (1993) Proc. Natl. Acad. Sci. USA 90 2202-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luan S., Kudla, J., Gruissem, W. & Schreiber, S. L. (1996) Proc. Natl. Acad. Sci. USA 93 6964-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blecher O., Erel, N., Callebaut, I., Aviezer, K. & Breiman, A. (1996) Plant Mol. Biol. 32 493-504. [DOI] [PubMed] [Google Scholar]
- 16.Hueros G., Rahfeld, J., Salamini, F. & Thompson, R. (1998) Planta 205 121-131. [DOI] [PubMed] [Google Scholar]
- 17.Kurek I., Aviezer, K., Erel, N., Herman, E. & Breiman, A. (1999) Plant Physiol. 119 693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurek I., Harvey, A. J., Lonsdale, D. M. & Breiman, A. (2000) Plant Mol. Biol. 42 489-497. [DOI] [PubMed] [Google Scholar]
- 19.Vittorioso P., Cowling, R., Faure, J.-D., Caboche, M. & Bellini, C. (1998) Mol. Cell. Biol. 18 3034-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vucich V. A. & Gasser, C. S. (1996) Mol. Gen. Genet. 252 510-517. [DOI] [PubMed] [Google Scholar]
- 21.Faure J. D., Gingerich, D. & Howell, S. H. (1998) Plant J. 15 783-789. [DOI] [PubMed] [Google Scholar]
- 22.Xu Q., Liang, S., Kudla, J. & Luan, S. (1998) Plant J. 15 511-519. [DOI] [PubMed] [Google Scholar]
- 23.Harrar Y., Bellini, C. & Faure, J. D. (2001) Trends Plant Sci. 6 426-431. [DOI] [PubMed] [Google Scholar]
- 24.Berardini T. Z., Bollman, K., Sun, H. & Poethig, R. S. (2001) Science 291 2405-2407. [DOI] [PubMed] [Google Scholar]
- 25.Gupta R., Webster, C. I. & Gray, J. C. (1998) Plant Mol. Biol. 36 897-907. [DOI] [PubMed] [Google Scholar]
- 26.Gupta R., Huang, Y., Kieber, J. & Luan, S. (1998) Plant J. 16 581-589. [DOI] [PubMed] [Google Scholar]
- 27.Mould R. M. & Gray, J. C. (1998) in Cell Biology: A Laboratory Handbook, ed. Celis, J. E. (Academic, New York), pp. 281–286.
- 28.Sommerville C. R., Sommerville, S. C. & Orgen, W. L. (1981) Plant Sci. Lett. 21 90-97. [Google Scholar]
- 29.Chien C. T., Bartel, P. L., Sternglanz, R. & Fields, S. (1991) Proc. Natl. Acad. Sci. USA 88 9578-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durfee T., Becherer, K., Chen, P. L., Yeh, S. H., Yang, Y., Kilburn, A. E., Lee, W. H. & Elledge, S. J. (1993) Genes Dev. 7 555-569. [DOI] [PubMed] [Google Scholar]
- 31.Yang Z. & Watson, J. C. (1993) Proc. Natl. Acad. Sci. USA 90 8732-8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chuang C.-F. & Meyerowitz, E. M. (2000) Proc. Natl. Acad. Sci. USA 97 4985-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clough S. J. & Bent, A. F. (1998) Plant J. 16 735-743. [DOI] [PubMed] [Google Scholar]
- 34.Van Duyne G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993) J. Mol. Biol. 229 105-124. [DOI] [PubMed] [Google Scholar]
- 35.Kaneko T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., Miyajima, N., Hirosawa, M., Sugiura, M., Sasamoto, S., et al. (1996) DNA Res. 3 109-136. [DOI] [PubMed] [Google Scholar]
- 36.Keegstra K. & Cline, K. (1999) Plant Cell 11 557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mould R. M. & Robinson, C. (1991) J. Biol. Chem. 266 12189-12193. [PubMed] [Google Scholar]
- 38.Mould R. M., Knight, J. S., Bogsch, E. & Gray, J. C. (1997) Plant J. 11 1051-1058. [DOI] [PubMed] [Google Scholar]
- 39.Hope A. B. H. (1993) Biochim. Biophys. Acta 1143 1-22. [DOI] [PubMed] [Google Scholar]
- 40.Dolinski K., Muir, S., Cardenas, M. & Heitman, J. (1997) Proc. Natl. Acad. Sci. USA 94 13093-13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brillantes A. B., Ondrias, K., Scott, A., Kobrinsky, E., Ondriasova, E., Moschella, M. C., Jayaraman, T., Landers, M., Ehrlich, B. E. & Marks, A. R. (1994) Cell 77 513-523. [DOI] [PubMed] [Google Scholar]
- 42.Shou W., Aghdasi, B., Armstrong, D. L., Guo, Q., Bao, S., Charng, M. J., Mathews, L. M., Schneider, M. D., Hamilton, S. L. & Matzuk, M. M. (1998) Nature 391 489-492. [DOI] [PubMed] [Google Scholar]
- 43.Stamnes M. A., Shieh, B. H., Chuman, L., Harris, G. L. & Zuker, C. S. (1991) Cell 65 219-227. [DOI] [PubMed] [Google Scholar]
- 44.Faure J. D., Vittorioso, P., Santoni, V., Fraisier, V., Prinsen, E., Barlier, I., Van Onckelen, H., Caboche, M. & Bellini, C. (1998) Development (Cambridge, U.K.) 125 909-918. [DOI] [PubMed] [Google Scholar]
- 45.Albertsson P.-A. (1995) Photosynth. Res. 46 141-149. [DOI] [PubMed] [Google Scholar]
- 46.Breiman A., Fawcett, T. W., Ghirardi, M. L. & Mattoo, A. K. (1992) J. Biol. Chem. 267 21293-21296. [PubMed] [Google Scholar]
- 47.Madueno F., Napier, J. A. & Gray, J. C. (1993) Plant Cell 5 1865-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]