Abstract
Histone acetyltransferases (HATs) use acetyl CoA to acetylate target lysine residues within histones and other transcription factors, such as the p53 tumor suppressor, to promote gene activation. HAT enzymes fall into subfamilies with divergence in sequence and substrate preference. Several HAT proteins have been implicated in human cancer. We have previously reported on the preparation of peptide-CoA conjugate inhibitors with distinct specificities for the p300/CBP [cAMP response element binding protein (CREB)-binding protein] or GCN5 HAT subfamilies. Here we report on the crystal structure of the GCN5 HAT bound to a peptide-CoA conjugate containing CoA covalently attached through an isopropionyl linker to Lys-14 of a 20-aa N-terminal fragment of histone H3. Surprisingly, the structure reveals that the H3 portion of the inhibitor is bound outside of the binding site for the histone substrate and that only five of the 20 aa residues of the inhibitor are ordered. Rearrangements within the C-terminal region of the GCN5 protein appear to mediate this peptide displacement. Mutational and enzymatic data support the hypothesis that the observed structure corresponds to a late catalytic intermediate. The structure also provides a structural scaffold for the design of HAT-specific inhibitors that may have therapeutic applications for the treatment of HAT-mediated cancers.
It is now clear that enzymes that modify chromatin play particularly important roles in the regulation of gene expression (1). Many of these enzymes function by covalently modifying the N-terminal tail regions of histone proteins, which serve to package the DNA into chromatin. These enzymes include histone acetyltransferases (HATs), histone deacetylases (HDACs), methyltransferases, ubiquitinases, and kinases (1). Although histone acetylation and deacetylation are generally associated with gene activation and silencing, respectively, methylation and phosphorylation have been correlated with both transcriptional activation and repression, depending on the specific site and context of the modification (1, 2). Moreover, it now appears that many of these modifications act synergistically (3). In addition to their processing of histones, HATs have been found to catalyze acetyl transfer to many nonhistone cellular proteins, such as p53, MyoD, and E2F-1, to promote gene activation (4).
Many of the enzymes that regulate the histone acetylation balance have been correlated with human disease (5). For example, the cAMP response element binding protein (CREB)-binding protein (CBP) HAT forms translocation products with mixed lineage leukemia and monocytic leukemia zinc-finger protein, another HAT, in a subset of acute myeloid leukemias; and acute promyelocytic leukemias harbor retinoic acid receptor translocation products, which are thought to mediate their neoplastic phenotype through the aberrant recruitment of HDACs (5). In addition, the p300 HAT is mutated in a subset of colorectal and gastric cancers and the AIB1 HAT is gene-amplified or overexpressed in a significant subset of breast cancers (5).
As a result of the importance of acetylation in cellular function and human cancer, HATs and HDACs are attractive molecules for targeted inhibition. Indeed, the natural products trichostatin and trapoxin that induce tumor cell growth arrest, differentiation, and/or apoptosis are examples of potent HDAC inhibitors (6). In addition, several HDAC inhibitors have been shown to have impressive antitumor activity in vivo and are currently in phase I or II clinical trials (6). A structure determination of a bacterial HDAC homologue bound to the inhibitors trichostatin and suberoylanilide hydroxamic acid has further facilitated the structure-based design of HDAC-specific inhibitors and provided important insights into HDAC reaction mechanism (7).
Since their isolation in 1995–1996, the development of inhibitors for the HATs has progressed relatively slowly. We recently reported on the development of a series of peptide-CoA conjugates that displayed selectivity for the GCN5/p300/CBP-associating factor (PCAF) or CBP/p300 subfamily of HAT enzymes (8–10). In addition, we have reported on the crystal structure of the GCN5 HAT in various liganded forms (11). These crystal structures, together with additional mutational and biochemical data (12), reveal that catalysis proceeds through a ternary complex mechanism, whereby a glutamate residue located within a structurally conserved core domain functions as a general base for catalysis. We also show that N- and C-terminal domains, which diverge structurally from other N-acetyltransferases, contribute to histone H3-specific binding.
We now report on the crystal structure of the GCN5 HAT bound to a peptide-CoA conjugate, containing CoA covalently attached through an isopropionyl linker to the lysine ɛ-amino group of an N-terminal 20-aa fragment of histone H3 [H3-(Me)CoA-20]. We also report on biochemical analysis of the PCAF human homologue of GCN5, as well as analysis of a PCAF mutant, harboring a mutation inferred from the structure to affect inhibitor binding and catalytic turnover. Together, these studies provide insights into the mechanism of catalysis by the GCN5/PCAF HAT enzymes and suggest a structural scaffold for the design of improved HAT inhibitors that may have antineoplastic applications.
Materials and Methods
Protein Overexpression and Purification.
The HAT domain of Tetrahymena GCN5 (tGCN5) (residues 48–210) was overexpressed and purified as described (11). Purified protein was concentrated to ≈20 mg/ml in a buffer containing 20 mM sodium citrate (pH 6.0), 150 mM NaCl, and 10 mM β-mercaptoethanol, flash-frozen, and stored at −20°C. The production of WT hPCAFcat(His) was carried out as described (10, 13), except that the 6×His tag was not removed. The Y638A PCAFcat mutant was prepared by using the QuikChange procedure (Stratagene) and confirmed by DNA sequencing. The protein was overproduced and purified as described for the WT enzyme (10). The WT and Y638A PCAF proteins, which appeared >80% pure by SDS/PAGE, were concentrated to ≈2 mg/ml and stored at −80°C in 50 mM Na/Hepes (pH 7.0), 250 mM NaCl, 5 mM DTT, and 10% glycerol.
Preparation of HAT Inhibitors.
The peptide-CoA conjugates were synthesized by using a strategy as described (8), except that the bromopropionylated intermediate was prepared on resin and purified before solution-phase reaction with CoASH (detailed in Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org). In analogy to previous efforts in which bromopropionyl coupling to an amine resulted in stereochemical scrambling at the chiral center (18), we have likely obtained an HPLC-inseparable 1:1 epimeric mixture of both the H3-(Me)CoA-7 and H3-(Me)CoA-20 compounds. Purity of the peptide-CoA conjugates was confirmed to be >95% pure by HPLC. Structural identities were confirmed by matrix-assisted laser desorption ionization-MS.
Cocrystallization and Data Collection.
The tGCN5 HAT domain was cocrystallized with the H3-(Me)CoA-20 inhibitor by using hanging drop vapor diffusion at 20°C. A 2.0-μl complex solution, containing 1.0 μl of the protein-inhibitor complex (0.36 and 0.58 mM, respectively) and 1.0 μl of the reservoir solution [2.0 M (NH4)2SO4, 0.1 M Na cacodylate, pH 6.6, and 0.2 M NaCl], was equilibrated over a 1-ml reservoir. Crystals grew to average dimensions of 50 × 100 × 200 μm3 in 2–4 days and were harvested into reservoir solution supplemented with 25% glycerol and immediately flash-frozen in liquid propane. Diffraction data were collected on the 19-ID beamline at the Advanced Photon Source of Argonne National Laboratory, Argonne, IL at a single wavelength (λ = 1.0082 Å), and the data were processed with the HKL2000 program suite (14) (Table 1). The complex crystallized in the P212121 space group with cell dimensions of a = 67.42, b = 67.83, and c = 74.50 Å and two molecules per asymmetric unit.
Table 1.
Data and refinement statistics for the tGCN5/H3-(Me)CoA-20 complex
| Resolution range | 37.3–2.2Å |
| Rsymm | 5.9% (34.6%) |
| I/σ | 22.6 (5.0) |
| Completeness | 99.2% (97.7%) |
| I/σ cutoff | 2.0 |
| Final model | |
| Protein, inhibitor, water atoms | 1,369 (1,336), 93 (62), 108 |
| Rworking | 21.34 |
| Rfree | 26.52 |
| rms deviation values | |
| Bond lengths (Å), angles (°), B factors (Å2) | 0.0095, 1.75, 1.44 |
| Average B factors (Å2) | |
| Protein, inhibitor, water atoms | 24.14 (32.20), 41.06 (36.43), 30.64 |
Rworking = Σ | |Fo| − |Fc| |/Σ |Fo|.
Rfree = ΣT | |Fo| − |Fc| |/ΣT |Fo|, where T is a test data set of 10% of the total reflections randomly chosen and set aside before refinement.
The average value across the resolution range while that in parentheses is the value for the highest resolution bin (2.28–2.20 Å).
The value for complex A in the asymmetric unit while that in parentheses is the value for complex B.
Structure Determination and Refinement.
The structure of the tGCN5/H3-(Me)CoA-20 inhibitor complex was determined by molecular replacement with the program amore, using the tGCN5 ternary complex structure as an initial model (11, 15) (Protein Data Bank ID ). Structural refinement used simulated annealing (SA) and torsion angle dynamics protocols in the cns (16) program suite as well as iterative model adjustment with o (17), using Sigma A-weighted Fo − Fc and SA omit maps. Advanced stages of refinement incorporated individual atomic B factor, bulk solvent correction, introduction of the inhibitor model, and final addition of solvent. The final model was verified against a composite SA omit map. The final protein model has an excellent fit to the electron density except for residues 177–195 of complex B, which were modeled as polyalanine due to poor electron density. The histone H3 peptide portion of the bisubstrate analog includes residues 1–20. However, whereas residues 12–17 were modeled in complex A, only residue Lys-14 could be modeled in complex B. The final model was refined to 2.2 Å with excellent crystallographic statistics (Table 1), with only three residues within the β6–α6 loop of the complex B protein outside the allowed region of the Ramachandran plot.
Acetyltransferase Activity Assays.
The radioactive HAT assay was adapted from the methods in ref. 10. Briefly, unless otherwise noted, the concentrations of [14C]acetyl CoA and peptide substrate (H3–20) were fixed at 10 μM; the reaction buffer contained 50 mM Tris⋅HCl (pH 8), 1 mM DTT, 0.1 mM EDTA, and 50 μg/ml acetylated BSA. Reactions used purified enzyme at concentrations of 0.2–5 nM for the WT and 10–30 nM for the Y638A PCAFcat mutant, as needed. Assays were carried out at 30°C with reaction volumes of 30 μl. Reactions were initiated with enzyme after the other components equilibrated at 30°C and quenched after 3–6 min with 6 μl 6× Tris-tricine gel loading buffer (10). Mixtures were separated on 16% SDS Tris-tricine polyacrylamide gels and dried, and radioactivity was quantified by PhosphorImage analysis (Molecular Dynamics) by comparing to known quantities of 14C-labeled BSA standard (NEN Life Science Products). All assays were performed at least twice and agreed within 20%. Enzyme activities were demonstrated to be linear versus enzyme concentration and time in the concentration ranges used. Rate measurements were based on initial conditions (less than 10% consumption of the limiting substrate). In inhibition assays, the specified amount of inhibitors were added and in all cases were at least 5-fold greater than the enzyme concentration used (Figs. 6 and 7, which are published as supporting information on the PNAS web site). For Km (apparent) measurements, a range of, at least, five substrate concentrations was used at fixed concentration of the second substrate [10 μM acetyl CoA (AcCoA) or 180 μM H3–20]. The data were fitted to the Michaelis-Menten equation, v = E*kcat*S/(Km + S), using a nonlinear least-squares approach.
Results and Discussion
Design and Characterization of Peptide-CoA Conjugates.
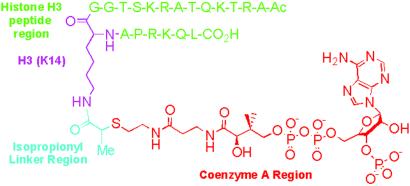
Our previous studies demonstrate that histone H3-derived peptide-CoA conjugates linked through an acetyl bridge between the peptide substrate and CoA could make potent inhibitors of the PCAF HAT. In analogy to enhanced inhibition of a weakly conserved GNAT superfamily member serotonin N-acetyltransferase by a bisubstrate analog containing an additional methyl group (18), we synthesized peptide-CoA conjugates containing an isopropionyl bridge (Fig. 1). This additional methyl group can serve to restrict rotational freedom and potentially provide enhanced hydrophobic interaction mediated by the methyl of AcCoA. We prepared two such molecules, whereby one inhibitor contained a seven-residue H3 peptide [H3-(Me)CoA-7] and the other contained a 20-residue H3 peptide [H3-(Me)CoA-20] (Fig. 1).
Fig 1.
Structure of the H3-(Me)CoA-20 inhibitor. The inhibitor is color-coded as follows: H3 peptide region (green), H3 Lys-14 residue (magenta), the isopropionyl linker (light blue), and CoA (red).
These compounds were evaluated as inhibitors of the human PCAF (hPCAF) HAT as well as tGCN5, using a standard HAT assay involving 14C-AcCoA and a 20-residue histone H3 peptide. A comparison was made between the H3-(Me)CoA-7 and H3-(Me)CoA-20 compounds and their nonmethylated counterparts (Table 2). As can be seen in summary form in Table 2, H3-(Me)CoA-20 was ≈4-fold more potent as a hPCAF and tGCN5 inhibitor compared with H3-CoA-20, assuming that only 50% of the epimeric mixture of H3-(Me)CoA-20 is responsible for the inhibition. The IC50 of the H3-(Me)CoA-20 inhibitor was 0.36 μM and 0.3 μM for hPCAF and tGCN5, respectively. The 20-residue methylated inhibitors were also about 10- or 30-fold more active than the corresponding seven-residue inhibitors for PCAF and tGCN5, respectively. Together, the H3-(Me)CoA-20 molecule is the most potent GCN5/PCAF inhibitor yet reported and these data reveal that the methyl moiety as well as residues outside the core 7 aa contribute to the potency of this compound. Based on previous kinetic characterization of H3-CoA-20 (10), it is likely that this compound is a competitive inhibitor versus AcCoA and a noncompetitive inhibitor versus peptide substrate, in accordance with an ordered BiBi kinetic mechanism (19).
Table 2.
Inhibitory properties of histone H3-CoA conjugates against PCAF and tGCN5
| Bisubstrate analog | IC50 (μM) PCAF | IC50 (μM) tGCN5 |
|---|---|---|
| H3-CoA-7 | >20 | >20 |
| H3-CoA-20 | 0.75 | 0.5 |
| H3-(Me)CoA-7 | 5.0 | 15 |
| H3-(Me)CoA-20 | 0.36 | 0.3 |
Previously reported data (8).
Measured with 50 μM H3-20 peptide substrate.
Data for the racemic mixture.
Structure of tGCN5 Bound to H3-(Me)CoA-20.
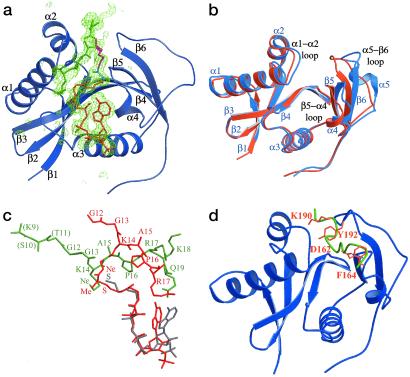
To gain further insight into the molecular basis of inhibition by H3-(Me)CoA-20, this compound was cocrystallized in complex with the HAT domain of tGCN5 (residues 48–210). The two complexes in the asymmetric unit are essentially superimposable with rms deviations of 1.54 Å for all protein atoms and 1.12 Å for the inhibitor regions modeled in complex B (Table 1). However, the atoms in complex B have higher average B factors than the corresponding atoms in complex A, reflecting more extensive crystal contacts in complex A and a higher degree of order. The protein molecules in the two complexes are essentially superimposable (with the exception of an inherently flexible C-terminal loop containing residues 184–198), consistent with an inherent flexibility within this region of the protein. In contrast, the peptide portion of the inhibitor shows significant differences in the two complexes. Whereas complex A reveals two ordered residues on each side of Lys-14 (residues 12–17) (Fig. 2a), complex B does not contain interpretable electron density outside of Lys-14 of the peptide. Inspection of crystal contacts reveal no specific intercomplex interactions that affect the relative positions of the inhibitors in the two complexes, and we infer that the greater apparent order of the inhibitor in complex A is related to the greater overall order of complex A in the crystal lattice. With the understanding that the peptide portion of the inhibitor is inherently more flexible than the CoA portion, we will therefore restrict further discussion of the protein/inhibitor structure to complex A, which contains more readily interpretable structural information for the tGcn5/H3-(Me)CoA-20 complex.
Fig 2.
Structure of the tGCN5/H3-(Me)CoA-20 inhibitor complex. (a) The protein is colored blue with secondary structural elements labeled. The inhibitor is color-coded as described in Fig. 1. A σA-weighted Fo − Fc omit map, omitting the entire inhibitor is shown. The map is contoured to 2.5 σ. (b) Superposition of the ternary tGCN5/CoA/histone H3 peptide complex protein (blue) and the tGCN5/H3-(Me)CoA-20 inhibitor complex protein (orange). (c) Superposition of the ternary (CoA in gray, H3 in green) and inhibitor (red) complex substrates. The respective proteins of the two complexes were used to align the superposition. (d) Superposition of the histone H3 peptide extracted from the ternary tGCN5/CoA/histone H3 complex (green) onto the protein component of the tGCN5/H3-(Me)-CoA-20 inhibitor complex (blue). The respective proteins of the two complexes were used for the superposition. Residues Asp-162 and Phe-164 of the β5−α4 loop and residues Lys-190 and Tyr-192 of the β6−α5 loops are in orange.
A superposition of the tGcn5/H3-(Me)CoA-20 complex with the ternary tGCN5 complex, containing CoA and a histone H3 peptide (11), reveals that the central core (β2-β3-β4-α3) and N-terminal (β1-α1-α2) domains are essentially superimposable, except for the α1–α2 loop within the N-terminal domain. In contrast, the C-terminal domain shows significantly more divergence (Fig. 2b). The C-terminal domain in the ternary complex contains a β5-α4-α5-β6 topology, whereas in the inhibitor complex the α5 helix adopts a random coil conformation and the entire C-terminal domain is shifted inward toward the protein core domain by about 9 Å relative to the corresponding C-terminal domain of the ternary complex. Correlated with this movement, the peptide-binding site, which normally resides over the central core domain and is flanked by the N- and C-terminal domains in the ternary complex, is occluded in the inhibitor complex. Consequently, the peptide portion of the inhibitor is bound along the edge of the N- and C-terminal domains almost orthogonal to the peptide in the ternary complex (Fig. 2c). In contrast to the divergent orientation of the peptide portion of the inhibitor, the pantetheine arm and pyrophosphate group of the CoA portion of the inhibitor are superimposable with the corresponding CoA region in the ternary complex and make very similar protein interactions in both complexes. The adenine base of the CoA and inhibitor adopt different conformations; however, this is unremarkable given that the adenine base does not make extensive contacts, in either the ternary or inhibitor complex structures, and adopts different conformations when compared with the structures of hPCAF (13), tGCN5 (11), and other GNAT superfamily members (20, 21). Taken together, the most dramatic differences between the tGCN5/H3-(Me)CoA-20 inhibitor and ternary complexes are largely restricted to the C-terminal region of the HAT domain and the position of the peptide portion of the substrate and inhibitor. Another unexpected feature of the inhibitor complex is that only five of the 20 residues within the peptide portion of the inhibitor are ordered in the crystal structure, despite the fact that the H3-(Me)CoA-20 molecule is a considerably better inhibitor than the seven-residue counterpart [H3-(Me)CoA-7]. These results suggest that residues outside the seven-residue core may be important for an initial binding event but do not contribute to the final state that is apparently captured in the crystal structure.
A superposition of the histone H3 peptide from the ternary complex onto the protein from the inhibitor complex, using the superimposable regions of the corresponding proteins for the overlay, reveals that the residues that play a particularly important role in blocking access of the peptide portion of the inhibitor to the normal substrate binding site are residues Asp-162 and Phe-164, of the β5–α4 loop, and residues Lys-190 and Tyr-192, of the large loop preceding β6 (Fig. 2d). Interestingly, each of these residues are mutationally sensitive for HAT activity (22) and all but Phe-164 are strictly conserved within the PCAF/GCN5 subfamily of HAT proteins. The structure of the ternary complex also reveals that all but Lys-190 contact the histone H3 peptide. The apparent importance of these residues in occluding the normal substrate binding site suggests that the inhibited complex observed in the crystal structure may represent a physiologically relevant catalytic intermediate in which the structural rearrangements observed in the inhibitor complex serve to mediate nucleophilic attack and/or peptide displacement by the enzyme.
Inhibitory and Enzymatic Analysis of hPCAF.
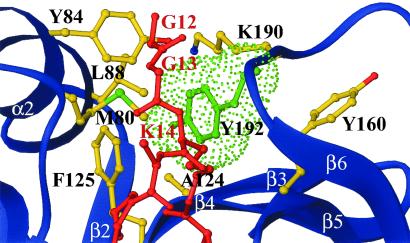
To test the hypothesis that the inhibitor complex that is observed in the crystal structure represents a catalytic intermediate, we mutated Tyr-638 of hPCAF, corresponding to residue Tyr-192 of tGCN5, to alanine and characterized the enzymatic and inhibitor binding properties of this mutant. Tyr-638 of PCAF is a particularly interesting residue to target for mutagenesis because it is strictly conserved within the GCN5/PCAF subfamily and shows mutational sensitivity (22), yet the structural significance of this residue had not been previously addressed. In the ternary tGCN5/CoA/histone H3 peptide complex, Tyr-192 showed only a few van der Waals interactions with the histone H3 backbone. However in the tGCN5/H3-(Me)CoA-20 inhibitor complex, this tyrosine residue is located within a hydrophobic pocket lined by the aliphatic arm of the Lys-14 side chain of the inhibitor and several protein residues in the active site, including Tyr-84, Leu-88, Ala-124, Phe-125, Tyr-160, and Lys-190 (Fig. 3). These observations suggest that this tyrosine may play a particularly important role in closing off the initial histone H3 binding site and facilitating acetyl transfer and/or product displacement. We reasoned that if the inhibitor complex conformation, which was observed in the crystal structure, did correspond to a catalytic intermediate, then substitution of Tyr-638 in hPCAF (Tyr-192 in tGCN5) with an alanine would have a significant impact on the acetyltransferase activity as well as on bisubstrate analog binding.
Fig 3.
tGCN5 H3-(Me)CoA-20 inhibitor interface proximal to Tyr-192. A van der Waals representation of Tyr-192 (green) is shown with the inhibitor (red), the tGCN5 backbone (blue), and relevant side chains. All protein residues shown are making van der Waals contact (<5 Å) with Tyr-192.
A comparison of the steady-state kinetic parameters (Table 3) for the WT and Y638A mutant of PCAF shows identical AcCoA Km values. In contrast, the Km for the histone H3 peptide substrate is elevated by 2.5-fold with the mutant and the kcat is 18-fold lower with the mutant. These results are nicely rationalized by the crystal structure where alignment of the substrate lysine nucleophile is modestly perturbed in the ground state (2.5-fold reduction in binding) but more severely affected in the transition state where the kcat/Km is reduced by 45-fold for the acetyltransferase reaction. The acetyltransferase inhibition studies (Table 3) show the IC50 values of the H3-(Me)CoA-20 for PCAF and PCAF(Y638A) to be 360 nM and 1700 nM, respectively. The nearly 5-fold reduction of inhibitor binding to the Y638A PCAF mutant clearly illustrates the importance of Tyr-638, and by extension the orientation of the C-terminal α5–β6 loop region harboring this residue, in facilitating the binding of the inhibitor to the protein. Taken together, these results are consistent with the importance of Tyr-638 in peptide substrate recognition and catalysis, and we propose that the observed reduction in catalytic rate, caused by the Y638A mutation, may result from the altered mobility of the Tyr-638-bearing loop during enzyme turnover, as well as substrate misalignment during acetyl transfer.
Table 3.
Binding and catalytic properties of hPCAF and hPCAF(Y638A)
| AcCoAKm, μM | H3-20 Km, μM | kcat, s−1 | H3-(Me)CoA-20 IC50, nM | |
|---|---|---|---|---|
| WT hPCAF | 1.6 | 50 | 3.5 | 360 |
| Y638A hPCAF | 1.6 | 125 | 0.19 | 1,700 |
Measured with 180 μM H3-20 peptide.
Given the correlation between effects on catalysis and bisubstrate analog inhibitor binding, we hypothesize that the bisubstrate inhibitor complex observed in the crystal structure corresponds to a late catalytic intermediate. This hypothesis is consistent with transition-state theory that correlates with the observations that the bisubstrate analog binds much tighter (≈10,000-fold) than the peptide to PCAF and GCN5, and that the change in binding to a catalytically defective mutant is more pronounced than the peptide substrate.
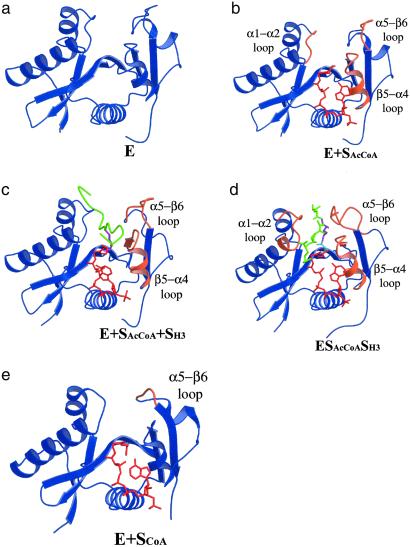
Model for Substrate Binding, Catalytic Turnover, and Product Release.
The structural and enzymatic information obtained in this study is complemented by a comparison with other known tGCN5 complexes, which provide the basis for a proposed model of the conformational change of the GCN5/PCAF enzyme as it relates to substrate binding, catalytic turnover, and product release. Specifically, we now have structural information for nascent tGCN5, tGCN5/AcCoA, tGCN5/CoA/histone H3 peptide (11) and the putative catalytic intermediate formed before product release, which is derived from this study. In addition, we have the previously determined structure of a PCAF/CoA (13) complex. The proposed model of the conformational change during catalysis is illustrated in Fig. 4. The nascent tGCN5 structure provides the starting point of the model and is referred to as the ground state of the enzyme, E. The enzymatic reaction proceeds first by binding AcCoA (19), denoted as E + SAcCoA, accompanied by a small widening of the active-site cleft, formed largely by reorientation of the α1–α2 loop in the N-terminal protein domain and the β5–α4 loop and the loop preceding β6, both in the C-terminal domain. In the next step, the second substrate, histone H3, binds in an ordered BiBi mechanism (19). Formation of the ternary complex, designated as E + SAcCoA + SH3, results in an expansion of the active-site cleft to accommodate the bulky residues in the histone H3 tail. This protein rearrangement largely involves reorientation of the β5–α4 loop and loop-β6 region of the C-terminal domain. We then propose that acetyl transfer from AcCoA to the Lys-14-Nɛ of histone H3 is accompanied by a movement of the histone H3 substrate away from the initial binding site. This histone rearrangement is associated with protein movements of the α1-loop region of the N-terminal protein domain, as well as significant movement in much of the C-terminal domain of the protein (β5−α4 loop and the loop preceding β6), forming the putative ESCoASAcH3 transition state intermediate. We propose that acetyl transfer may nucleate the protein movement, which in turn facilitates release of the acetylated histone H3 product. When the acetylated histone H3 product is finally released, it results in the generation of the E + SCoA state, which closely resembles the “closed” α5−β6 loop of the inhibitor complex, and the enzyme is primed for another round of AcCoA binding after release of the CoA substrate and return to the unbound E state.
Fig 4.
Model for the conformational changes during the GCN5/PCAF HAT catalytic cycle. (a) The nascent tGCN5 structure (blue). (b) The tGCN5/AcCoA complex (AcCoA red). Regions showing significant structural deviation (>0.5 Å) from the nascent structure (in a) are highlighted in orange. (c) The tGCN5/AcCoA/histone H3 complex (histone H3 11-mer peptide green). (d) The tGCN5/H3-(Me)-CoA-20 inhibitor complex. The catalytic Lys-14 shown in magenta and the methylated acetyl linker in light blue. Regions showing structural deviation (>0.5 Å and 0.8 Å in the N- and C-terminal loops, respectively) from the ternary complex (in c) are highlighted in orange. (e) The hPCAF/CoA complex. Color-coding as detailed in c. Regions showing structural deviation (>1 Å) from the inhibitor complex (in d) are highlighted in orange.
Model for Substrate Binding and Recognition.
In this study we show that the 20-residue H3-(Me)CoA-20 inhibitor is 10- to 30-fold more potent than the corresponding seven-residue inhibitor, despite the crystallographic observation that only five of the 20 residues of the H3-(Me)CoA-20 inhibitor are visible in the electron density map. We propose that residues outside of the seven-residue core may be important for an initial binding event of the histone substrate, within the normal substrate-binding pocket. However, after lysine acetylation, this binding mode is short-lived relative to the binding mode of the acetylated histone, which we propose closely mimics the tGCN5/H3-(Me)CoA-20 inhibitor complex reported here. It should also be mentioned that the apparent decrease in interactions between peptide substrate and enzyme might not be particularly energetically costly because of the relative increase in entropy of the disordered peptide residues in the bisubstrate inhibitor complex. It is also interesting to speculate that the structure of the bisubstrate analog complex may help explain altered PCAF and GCN5 sequence selectivity when present in multisubunit HAT complexes, such as SAGA and Ada in vivo (23, 24). Moreover, it is possible that the additional proteins present in these complexes may help bypass the initial recognition events observed in the ternary complex structure and lead to a catalytic state that resembles the bisubstrate inhibitor complex.
Implications for the Structure-Based Design of Improved GCN5/PCAF-Specific HAT Inhibitors.
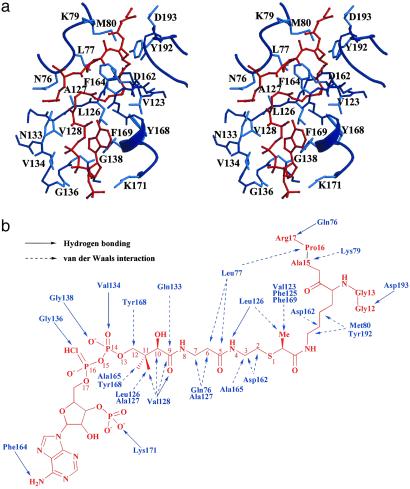
These studies indicate that the tGCN5/H3-(Me)CoA-20 inhibitor structure reported here represents a late catalytic intermediate of the GCN5/PCAF HAT enzymes. Unexpectedly, the structure reveals that the peptide portion of inhibitor is bound outside of the normal peptide-binding cleft and that only five of the 20 residues of the peptide portion have ordered structure. This result emphasizes the importance of determining the structure of an initial inhibitor complex in the design of second- and third-generation HAT inhibitors, rather than using the ternary complex as a model for inhibitor design. Not surprisingly, the pantetheine arm and pyrophosphate of CoA mediate extensive protein contacts (Fig. 5) consistent with the CoA contacts detailed in the binary and ternary HAT complexes (11, 13), as well as several other CoA-using GNAT enzymes (25). Indeed, these interactions likely provide a large degree of binding energy, but would be expected to exhibit a low degree of enzyme specificity. The introduction of the methyl group to the acetyl linker region of the inhibitor increases the potency of the compound against tGCN5 and hPCAF. The structure reveals that the bound linker has the (S)-configuration at the isopropionyl carbon (Fig. 5b), confirming the hypothesis that only 50% of the epimeric mixture is contributing to inhibition. The linker region is located in a highly hydrophobic cavity, formed by residues Val-123, Phe-125, Leu-126, and Phe-169 of the protein, and we infer that the methyl group helps orient and enhance the binding of the target lysine within this cavity. The lysine side chain and the isopropionyl linker make extensive van der Waals interaction with Met-80 and Asp-162, but most notably with Tyr-192. Despite its apparent significance for initial binding, the peptide region of the inhibitor appears to make only a handful of interactions with the protein. The inhibitor peptide backbone makes two hydrogen bonds with the protein backbone via Asp-193 and Gln-76 (Fig. 5). The peptide also makes van der Waals contact to two protein residues, Lys-79 and Asp-193, beyond those centered on the Lys-14 side chain discussed previously, which contribute to peptide binding. Surprisingly, the “closed conformation” of the α5–β6 loop is not characterized by specific interactions with the inhibitor. Taken together, the structure of the tGCN5/H3-(Me)CoA-20 complex reveals that the majority of protein–inhibitor interactions are concentrated around the pantetheine arm of CoA and the hydrophobic cavity surrounding the lysine side chain and ispropionyl linker region of the inhibitor.
Fig 5.
Detailed interactions between tGCN5 and the H3-(Me)CoA-20 inhibitor in the complex. (a) Detailed stereo diagram of the tGCN5/H3-(Me)-CoA-20 inhibitor interface. Protein residues making van der Waals or hydrogen bonding interactions with the inhibitor are shown with side chains (main chain for glycines) in light blue. The inhibitor is shown in red. (b) Schematic diagram of tGCN5/H3-(Me)-CoA-20 inhibitor interactions. Protein residues making hydrogen bonding interactions are shown with a solid line arrow and van der Waals contacts with the inhibitor are shown with a dashed line arrow.
We have demonstrated that the analogous H3-CoA-20 inhibitor works poorly against p300, a member of the CBP/p300 family of HATs (8, 10), and also poorly against the Esa1 member of the MYST family of HAT enzymes (data not shown). We propose that, at least, part of the reason for this HAT selectivity is that the catalytic intermediate seen here is unique to the GCN5/PCAF family of enzymes. This notion is consistent with recent enzymology studies showing that both p300 (9) and Esa1 (28) have catalytic mechanisms that differ from that of the GCN5/PCAF family of HAT enzymes. Because the interaction of the CoA and lysine would be expected to be, at least, moderately conserved, substrate specificity would be derived from the interactions mediated by the peptide portion of the inhibitor with the protein. However, in this intermediate state, the reoriented inhibitor mediates modest peptide–protein interactions in this region. Optimization of the interactions in the lysine and acetyl region, combined with enhanced interactions at the peptide–protein surface, would provide a reasonable starting point for elaborating analogs of H3-(Me)CoA-20 with enhanced GCN5/PCAF HAT specificity and potency.
A remaining challenge is to elucidate the specific roles of the disordered residues in binding affinity and specificity. Although both Lys-CoA and H3-CoA-20 are expected to show suboptimal pharmacokinetic properties because of their polarity and charge, effective use of even the original compounds has been made in cell culture systems by using microinjection, cell permeabilizing phospholipid, and appending Tat sequence (26, 27) (M.C., V. Sartorelli, and P.A.C., unpublished data). Based on the crystal structure, it should be possible to delete the adenosine-monophosphate of the CoA moiety of the bisubstrate analogs, which should further enhance bioavailability. Ultimately, it would be preferable to use the structural information obtained in this and subsequent iterations of peptide-CoA conjugates to move toward HAT inhibitors with inherently favorable pharmacokinetic properties.
In summary, the structure presented here provides insights into the catalytic mechanism of the GCN5/PCAF HAT enzymes and the structural scaffold for the design of HAT-specific inhibitors. Because of the important role that HATs play in gene regulation and the involvement of HATs in human cancer, the further development of HAT inhibitors should provide important insights into the unique role played by different HAT families in gene expression and provide a therapeutic avenue for the treatment of cancer.
Supplementary Material
Acknowledgments
We thank A. Clements, J. Rojas, and B. Poux for useful discussions and A. Joachimiak, R. Zhang, N. Duke, and staff for access to and assistance with the 19-ID beamline at the Advanced Photon Source of Argonne National Laboratory. This work was supported by National Institutes of Health grants (to R.M. and P.A.C.). A.N.P. was supported by a National Institutes of Health training grant, awarded to The Wistar Institute.
Abbreviations
HAT, histone acetyltransferase
HDAC, histone deacetylase
tGCN5, Tetrahymena GCN5
PCAF, p300/CBP-associating factor
hPCAF, human PCAF
AcCoA, acetyl CoA
CBP, cAMP response element binding protein (CREB) binding protein
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code ).
References
- 1.Berger S. L. (2002) Curr. Opin. Genet. Dev. 12, 142-148. [DOI] [PubMed] [Google Scholar]
- 2.Richards E. J. & Elgin, S. C. (2002) Cell 108, 489-500. [DOI] [PubMed] [Google Scholar]
- 3.Strahl B. D. & Allis, D. C. (2000) Nature 403, 41-45. [DOI] [PubMed] [Google Scholar]
- 4.Sterner D. E. & Berger, S. L. (2000) Microbiol. Mol. Biol. Rev. 64, 435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timmermann S., Lehrmann, H., Polesskaya, A. & Harel-Bellan, A. (2001) Cell. Mol. Life Sci. 58, 728-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marks P., Rifkind, R., Richon, V., Breslow, R., Miller, T. & Kelly, W. (2001) Nat. Rev. Cancer 3, 194-202. [DOI] [PubMed] [Google Scholar]
- 7.Finnin M., Donigian, J., Cohen, A., Richon, V., Rifkind, R., Marks, P., Breslow, R. & Pavletich, N. (1999) Nature 401, 188-193. [DOI] [PubMed] [Google Scholar]
- 8.Lau O. D., Kundu, T. K., Soccio, R. E., Ait-Si-Ali, S., Khalil, E. M., Vassilev, A., Wolffe, A. P., Nakatani, Y., Roeder, R. G. & Cole, P. A. (2000) Mol. Cell 5, 589-595. [DOI] [PubMed] [Google Scholar]
- 9.Thompson P. & Cole, P. A. (2001) J. Biol. Chem. 276, 33721-33729. [DOI] [PubMed] [Google Scholar]
- 10.Lau O. D., Courtney, A., Vassilev, A., Marzilli, L. A., Cotter, R. J., Nakatani, Y. & Cole, P. A. (2000) J. Biol. Chem. 275, 21953-21959. [DOI] [PubMed] [Google Scholar]
- 11.Rojas J. R., Trievel, R. C., Zhou, J., Mo, Y., Li, X., Berger, S. L., Allis, D. C. & Marmorstein, R. (1999) Nature 401, 93-98. [DOI] [PubMed] [Google Scholar]
- 12.Tanner K. G., Trievel, R. C., Kuo, M.-H., Howard, R. M., Berger, S. L., Allis, C. D., Marmorstein, R. & Denu, J. M. (1999) J. Biol. Chem. 274, 18157-18160. [DOI] [PubMed] [Google Scholar]
- 13.Clements A., Rojas, J. R., Trievel, R. C., Wang, L., Berger, S. L. & Marmorstein, R. (1999) EMBO J. 18, 3521-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otwinowski Z. & Minor, W. (1997) in Macromolecular Crystallography: Part A, eds. Carter, C. W., Jr. & Sweet, R. (Academic, New York), Vol. 276, pp. 307–326. [Google Scholar]
- 15.Navaza J. (2001) Acta Crystallogr. D 57, 1367-1372. [DOI] [PubMed] [Google Scholar]
- 16.Brunger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 17.Jones T. A., Zou, J. Y. & Cowen, S. W. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 18.Khalil E. M., De Angelis, J., Ishii, M. & Cole, P. A. (1999) Proc. Natl. Acad. Sci. USA 96, 12418-12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanner K. G., Langer, M. R., Kim, Y. & Denu, J. M. (2000) J. Biol. Chem. 275, 22048-22055. [DOI] [PubMed] [Google Scholar]
- 20.Dutnall R. N., Tafrov, S. T., Sternglanz, R. & Ramakrishnan, V. (1998) Cell 94, 427-438. [DOI] [PubMed] [Google Scholar]
- 21.Wolf E., De Angelis, J., Khalil, E. M., Cole, P. A. & Burley, S. K. (2002) J. Mol. Biol. 317, 215-224. [DOI] [PubMed] [Google Scholar]
- 22.Wang L., Liu, L. & Berger, S. L. (1998) Genes Dev. 12, 640-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balasubramanian R., Pray-Grant, M. G., Selleck, W., Grant, P. A. & Tan, S. (2002) J. Biol. Chem. 27, 7989-7995. [DOI] [PubMed] [Google Scholar]
- 24.Grant P. A., Eberharter, A., John, S., Cook, R. G., Turner, B. M. & Workman, J. L. (1999) J. Biol. Chem. 274, 5895-5900. [DOI] [PubMed] [Google Scholar]
- 25.Yan Y., Barlev, N. A., Haley, R. H., Berger, S. L. & Marmorstein, R. (2000) Mol. Cell 6, 1195-1205. [DOI] [PubMed] [Google Scholar]
- 26.Polesskaya A., Naguibneva, I., Fritsch, L., Duquet, A., Ait-Si-Ali, S., Robin, P., Vervisch, A., Pritchard, L. L., Cole, P. & Harel-Bellan, A. (2001) EMBO J. 20, 6816-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Costanzo A., Merlo, P., Pediconi, N., Fulco, M., Sartorelli, V., Cole, P. A., Fontemaggi, G., Fanciulli, M., Schiltz, L., Blandino, G., et al. (2002) Mol. Cell 9, 175-186. [DOI] [PubMed] [Google Scholar]
- 28.Yan, Y., Harper, S., Speicher, D. & Marmorstein, R. (2002) Nat. Struct. Biol., in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.