Abstract
The use of live bacteria in the treatment of cancer has a long and interesting history. We report the use of a purified bacterial redox protein, azurin, that enters human cancer (melanoma UISO-Mel-2) cells and induces apoptosis. The induction of apoptosis occurs readily in melanoma cells harboring a functional tumor suppressor protein p53, but much less efficiently in p53-null mutant melanoma (UISO-Mel-6) cells. A redox-negative mutant form of azurin (M44K/M64E) demonstrates much less cytotoxicity to the UISO-Mel-2 cells than the wild-type protein. Azurin has been shown to be internalized in UISO-Mel-2 cells and is localized predominantly in the cytosol and in the nuclear fraction. In the p53-null UISO-Mel-6 cells, azurin is localized only in the cytosol. Thus, intracellular trafficking of azurin to the nucleus is p53-dependent. Azurin forms a complex with p53, thereby stabilizing it and raising its intracellular level in cytosolic, mitochondrial, and nuclear fractions. Corresponding to an increasing level of p53, an inducer of apoptosis, the level of Bax also increases in mitochondria, allowing significant release of mitochondrial cytochrome c into the cytosol, thus initiating the onset of apoptosis. The M44K/M64E mutant form of azurin, deficient in cytotoxicity, is also deficient in forming a complex with p53 and is less efficient in stabilizing p53 than wild-type azurin. Azurin has been shown to allow regression of human UISO-Mel-2 tumors xenotransplanted in nude mice and may potentially be used in cancer treatment.
Reports on regression of cancer in humans and animals infected with microbial pathogens date back more than 100 years, originating with the initial report by Coley (1). Several subsequent reports (2–4) have shown that microbial pathogens replicate at tumor sites under hypoxic conditions and also stimulate the host's immune system during infection, leading to an inhibition of cancer progression. For example, the vaccine strain Mycobacterium bovis bacillus Calmette–Guérin is widely used for the topical treatment of superficial bladder cancer (2) and has also been used in other forms of cancer (2). In addition to M. bovis bacillus Calmette–Guérin, bacterial pathogens such as Listeria monocytogenes or Salmonella species are often used as vaccine vectors for cancer prevention because of their ability to target antigen-presenting cells and mount vigorous immune response against tumor cells (3). Infection of tumor-bearing mice with live attenuated cells of Salmonella typhimurium has also been reported to enhance tumor regression (4). It has generally been believed that infection with microbial pathogens leads to activation of macrophages and lymphocytes, resulting in the production of cytotoxic agents with anticancer activity. Anaerobic bacteria such as Salmonella or Clostridia are believed to grow preferentially in the anaerobic core of the tumors (4). More recently, the use of anaerobic bacteria with selected chemotherapeutic and antivascular agents has resulted in widespread and significant regression of s.c. tumors in mice, a treatment called combination bacteriolytic therapy (COBALT) (5). The studies above (5), as well as the publication by Jain and Forbes (6), have reviewed the long history of the use of anaerobic bacteria in the treatment of cancer. Live bacteria, however, produce significant toxicity and side reactions (5, 6), thus limiting the use of bacteria in the treatment of human cancer (2).
Bacterial pathogens are not the only infectious agents that are known to induce tumor regression in infected hosts. Protozoa such as Toxoplasma gondii or Besnoitia jellisoni are also known to activate macrophages to induce tumor shrinkage. The idea that infection with bacterial or protozoan pathogens may trigger cancer regression through activation of the innate or adaptive immunity has recently been questioned when it was shown (7) that infection of melanoma-harboring mice with T. gondii resulted in tumor shrinkage without concomitant activation of cytotoxic T or NK cells, production of nitric oxide by macrophages or the function of the cytokines IL-12 and tumor necrosis factor α. A major finding of these authors (7) was that T. gondii-infected tissues produce some factors that prevented formation of blood vessels in the tumors, thereby creating hypoxic conditions that then triggered necrotic death of the tumor cells. These authors surmised that inhibition of angiogenesis during infection might be due to synthesis of soluble antiangiogenic factors by the infected tissues which could be of potential therapeutic interest, similar to endostatin, an endogenous inhibitor of angiogenesis (8).
Although various bacteria or protozoa are believed to mediate tumor regression through selective proliferation in the anaerobic zone of the tumors or through inhibition of angiogenesis, very little is known about the detailed mechanism of tumor regression by such pathogens. In particular, little is known about the production of soluble or secreted factors by bacteria that may specifically act on cancer cells, resulting in their death and regression. In this article, we present evidence that a purified redox protein, azurin, which has been reported to be secreted by Pseudomonas aeruginosa and induce apoptosis in macrophages (9), also triggers apoptosis in various human cancer cells. These in vitro findings can be confirmed in vivo in human cancer xenotransplanted in nude mice.
Materials and Methods
Preparation of Azurin.
The azurin-encoding gene was amplified by PCR with genomic DNA of P. aeruginosa strain PAO1 as a template DNA. The forward and reverse primers used were 5′-GCCCAAGCTTACCTAGGAGGCTGCTCCATGCTA-3′ and 5′-TGAGCCCCTGCAGGCGCCCATGAAAAAGCCCGGC-3′. The azurin gene was placed downstream of the lac promoter in the vector pUC19. Escherichia coli JM109 was used as a host strain for expression of the azurin gene. Wild-type (wt) azurin and mutant azurin were purified by Q-Sepharose FF and Superdex 75 chromatography from periplasmic fractions of the recombinant E. coli cells according to the method described by Kukimoto et al. (10).
Cytotoxicity Assay.
The human melanoma cells of UISO-Mel-2 and UISO-Mel-6 (11) were cultivated either in MEM with Hanks' medium supplemented with 10% FBS, or MEM with Eagle's supplemented with 10% heat-inactivated FBS and 1 mM glutamic acid, respectively. The 3-(4,5-dimethylthiazol-2-yl-2,5-diphenyl tetrazolium bromide) (MTT) assay (12) was performed to determine the cytotoxicity of azurin toward cancer cells. Cells (5 × 103 per well) were seeded into 96-well culture plates in 200 μl of the medium at 37°C with 5% CO2. After 24 h, the supernatant was removed and new media containing azurin at various concentrations as specified were added to the attached cells. These cells were incubated for 24 h before the number of live cells was determined by MTT assay by adding 10 μl of 5 mg/ml MTT (Sigma) solution to the culture, incubating for 3 h at 37°C, and measuring the formation of MTT formazan spectrophotometrically (12).
Preparation of Whole Cell Lysate.
UISO-Mel-2 cells (107 cells) were treated with azurin (500 μg/ml) for the times indicated. Cell lysate was prepared as described by Asher et al. (13).
Subcellular Fractionation.
UISO-Mel-2 and UISO-Mel-6 cells (107 cells) were treated with azurin (500 μg/ml) for the indicated times. After treatment, mitochondrial and cytosolic fractions were prepared as described by Han et al. (14). Nuclear extracts were prepared as described (15). Monoclonal antibodies raised against p53, Bax, and mitochondrial cytochrome c (Santa Cruz Biotechnology) were used for Western blotting. Blots were also probed for actin by using monoclonal anti-actin (Sigma) and mitochondrial membrane protein cytochrome c oxidase subunit IV (COX IV) by using monoclonal anti-COX IV antibody (Molecular Probes) for checking cross-contamination and as internal controls. Protein bands were visualized by using enhanced chemiluminescence reagents (Amersham Pharmacia).
Intracellular Localization of Azurin.
UISO-Mel-2 and UISO-Mel-6 cells (107 cells) were used for subcellular fractionation after azurin treatment (50 μg/ml) for 0, 3, 6, and 12 h. Each fraction (10 μg protein) was loaded on SDS/PAGE and blotted on Immuno-P membrane (Millipore). The presence of azurin was confirmed by using anti-azurin antibody.
Microinjection of Azurin in UISO-Mel-2 and UISO-Mel-6 Cells.
UISO-Mel-2 and UISO-Mel-6 cells were cultured overnight on 22-mm glass coverslips coated with poly-L-lysine. Azurin was labeled with Alexa Fluor 568 (Molecular Probes) having absorption and emission fluorescence maxima of 577 and 603 nm, respectively. Chemically labeled azurin (0.36 μM; fluorescing red) was microinjected into the cytoplasm of single cells by using an Eppendorf pressure injector and micromanipulator. All microinjection experiments were performed with a 0.5-s injection time and 100 hPa pressure, with the aid of LSM 5 Pascal microscope (16). Approximately 70–100 cells were injected in each dish. After injection, cells were incubated at 37°C in 5% CO2 for different times varying from 15 min to 3 h, as noted in the text, and fixed; nuclear DNA was stained with 4,6-diamidino-2-phenylindole. Fluorescent images were collected with a Zeiss LSM 510 confocal laser microscope. Only blue (4,6-diamidino-2-phenylindole) and red (Alexa Fluor-labeled azurin) fluorescence were recorded.
Complex Formation of Azurin with p53.
Complex formation between wt azurin, mutant azurin, and p53 was confirmed by glycerol gradient centrifugation analysis as described (17).
In Vivo Xenotransplanted Tumor Regression by Azurin: Statistical Analysis.
Two groups of male nude mice (average weight, 20 g) received 1 × 106 UISO-Mel-2 cells s.c. in the flank. When the s.c. tumor grew to the size of 0.5 mm (in one of the three diameters), the animals were clustered into two groups of five mice each. One group of five mice comprised the control, whereas the treated group of five mice received 0.5 mg of azurin in saline (0.1 ml) i.p. daily for 3 weeks. The tumor volumes were measured before each injection. To determine differences in tumor volumes between the treated and control groups, both univariate and multivariate statistical analyses were used. The univariate method consisted of computing summary statistics for tumor volume for each group (treated and control) for each of the 12 days for which measurements were taken. The multivariate method was based on the 10 animals with 12 observations on each, using random coefficients mixed models for the treated and control groups, to study the growth of tumor over time.
Results
Cytotoxic Activity of Purified Azurin and Cytochrome c551 Toward Various Cancer Cells.
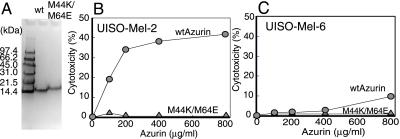
We recently reported that two redox proteins, azurin and cytochrome c551, elaborated by P. aeruginosa in its growth medium, induce apoptotic cell death in phagocytic cells such as macrophages and mast cells (9). Azurin and cytochrome c551 are involved in electron transfer during denitrification (18, 19) and are not known for their cytotoxic activity. To examine whether the cytotoxic activity is restricted to phagocytic cells, we purified azurin by cloning the gene from the chromosome of P. aeruginosa into an expression vector of E. coli and hyperproduced the protein as described under Materials and Methods. The purified azurin produced a single band on SDS-polyacrylamide gel with a molecular size of 14 kDa, as expected. As reported for macrophages (9), a combination of azurin and cytochrome c551 (50:25 μg/ml) induced apoptosis in the human melanoma (UISO-Mel-2) cells, as determined by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) (Fig. 1A) and caspase assays (Fig. 1 B and C). A great deal is known about the structure of azurin and cytochrome c551 (19), and mutations in two critical amino acids Met-44 and Met-64 have been shown to lead to a loss of >95% of the azurin electron transfer activity (20). To determine the mode of action of a single redox protein such as azurin, we isolated a double mutant Met44Lys/Met64Glu (M44K/M64E) defective in redox activity and purified the wt azurin and the M44K/M64E mutant protein (Fig. 2A). We then examined the cytotoxicity of the wt and the mutant azurin against a human melanoma cell line UISO-Mel-2 (11). Rauth et al. (11) demonstrated that the UISO-Mel-2 cell line was p53-positive, whereas another melanoma cell line (UISO-Mel-6) was p53-negative. Wt azurin demonstrated significant cytotoxicity toward UISO-Mel-2 cells (Fig. 2B), whereas the redox-negative M44K/M64E mutant azurin protein had very little cytotoxicity (Fig. 2B). Wt azurin had comparatively less cytotoxicity toward p53-null UISO-Mel-6 cells (Fig. 2C), suggesting that the presence of p53 might be important for azurin-induced cytotoxicity.
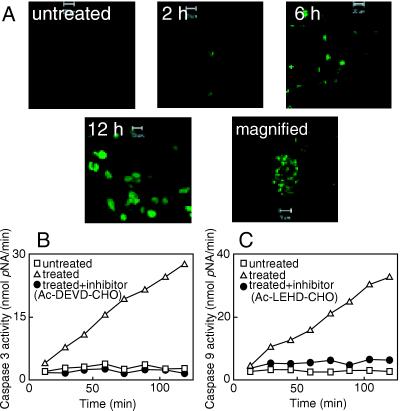
Fig 1.
(A) Confocal microscopic images for the detection of DNA fragmentation in UISO-Mel-2 cells by TUNEL assay. The incorporation of fluorescein-dUTP in the 3′-OH ends of the fragmented nuclear DNA generates the green fluorescence that is a measure of nuclear DNA fragmentation. A magnified view of a treated UISO-Mel-2 cell undergoing nuclear DNA fragmentation is also shown. (B and C) Activation of caspase-3 (B) and caspase-9 (C) in cell extracts of UISO-Mel-2 cells in response to either no treatment (untreated) or treatment with azurin and cytochrome c551 (treated) in absence or in presence of specific inhibitors. Details of the assay are described in ref. 9.
Fig 2.
(A) SDS/PAGE mobility of wild-type (wt) and M44K/M64E double-mutant azurin proteins obtained after hyperexpression in E. coli. The cytotoxicity exhibited by wt azurin and M44K/M64E mutant azurin toward UISO-Mel-2 (B) and UISO-Mel-6 (C) cells was determined by the MTT assay as described under Materials and Methods.
Treatment of UISO-Mel-2 Cells with Azurin Leads to Elevated Levels of p53 in Cytosolic and Nuclear Fractions.
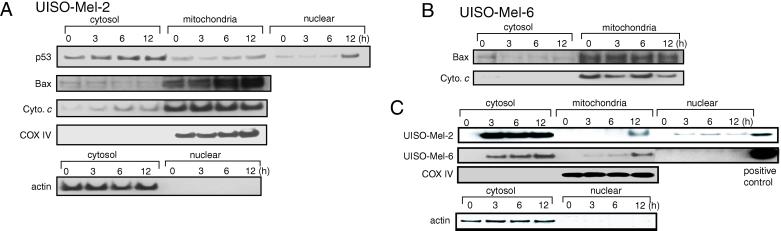
Cytotoxicity assays (Fig. 2 B and C) demonstrated a putative role of p53 in the induction of azurin-mediated apoptosis in UISO-Mel-2 cells. Because p53 is an inducer of mammalian cell apoptosis (21–23), it was of interest to examine whether azurin treatment might enhance the intracellular level of p53, which in turn might induce apoptosis in UISO-Mel-2 cells. We therefore treated UISO-Mel-2 cells with azurin (500 μg/ml) for 0 (no treatment), 3, 6, and 12 h, made extracts, gel-electrophoresed equal amounts of protein from each extract (50 μg), and determined the level of p53 by Western blotting. We also determined the level of actin as an internal control. Although the actin level remained more or less the same during the 12-h treatment period, the level of p53 increased substantially in the cell extract during the 12-h period (data not shown). To localize the p53, the extracts of the untreated and azurin-treated UISO-Mel-2 cells were fractionated to obtain cytosolic, mitochondrial, and nuclear fractions, as described under Materials and Methods. The enhanced p53 levels during the 3- to 12-h treatment were mostly found in the nuclear and the cytosolic fractions, particularly at 12 h. The level of p53 in mitochondria also rose slightly in 6–12 h (Fig. 3A). In some human cell lines, such as the myeloid leukemia line ML-1 or the colorectal carcinoma line RKO, a portion of p53 rapidly localized to the mitochondrial fraction of cells during induction of apoptosis by DNA-damaging agents such as camptothecin or hypoxic stress (24). Mitochondrial p53 localization was shown to be specific for p53-dependent apoptosis and did not occur in p53-independent apoptosis. These data, as well as the data of Ding et al. (25) that p53 protein from cell-free postnuclear extracts of irradiated transformed fibroblast cells (containing mitochondria) directly activated caspase 3 through an unknown mechanism, led to the postulation of a model where p53 localization in mitochondria caused oxidative damage to initiate apoptosis that is independent of the nuclear transcriptional activation by p53 (26, 27). Azurin-induced enhanced mitochondrial p53 level may thus contribute, at least in part, to the initiation of apoptosis in UISO-Mel-2 cells.
Fig 3.
The effect of the treatment of UISO-Mel-2 cells with azurin (500 μg/ml) for various times (0, 3, 6, and 12 h) on the levels of p53, Bax, mitochondrial cytochrome c (Cyto. c), cytochrome oxidase subunit IV (COX IV), and actin in various subcellular fractions (A). The levels of Bax and mitochondrial cytochrome c in the cytosolic and mitochondrial fractions of the untreated or azurin-treated UISO-Mel-6 cells are shown in B. The subcellular localization of azurin in UISO-Mel-2 and UISO-Mel-6 cells, as well as the localization of actin and COX IV in cytosolic and mitochondrial fractions of the UISO-Mel-2 cells, are shown in C. A positive control for azurin is maintained for both UISO-Mel-2 and UISO-Mel-6 cells (C).
Azurin Treatment Allows Enhanced Accumulation of Bax in Mitochondria and Release of Mitochondrial Cytochrome c to the Cytosol.
The localization of p53 to mitochondria has been suggested to be reminiscent of the proapoptotic protein Bax (24), which is also known to accumulate in mitochondria during apoptosis. In apoptotic cells, the cytosolic Bax undergoes a conformational change leading to its relocalization in the mitochondria (28). Similarly, the BH3-domain-containing protein Bid, during staurosporine-induced apoptosis in HeLa cells, translocates from the cytosol to the mitochondria, leading to a change in the conformation of Bax and resulting in the release of cytochrome c from mitochondria (29). Therefore, we examined the subcellular level of Bax and mitochondrial cytochrome c during treatment of UISO-Mel-2 cells with azurin. The Bax level was low in the cytosol but increased steadily in the mitochondria upto 12 h (Fig. 3A). Persuant to the enhanced mitochondrial accumulation of Bax, the cytosolic level of mitochondrial cytochrome c increased substantially during the period of treatment (Fig. 3A). The level of actin in the cytosol or COX IV in the mitochondria remained basically unchanged and were confined to their respective organelles, suggesting that the cytosolic fraction was virtually free of mitochondria or the nuclear fraction was free of cytosolic contamination. When the Bax or mitochondrial cytochrome c levels were checked as a function of azurin treatment in the p53-null UISO-Mel-6 cells (11), where azurin demonstrated very little cytotoxicity (Fig. 2C), very little increase of Bax in mitochondria or release of cytochrome c from mitochondria to cytosol was observed (Fig. 3B), suggesting a role of p53 in such a process.
Role of p53 in the Nuclear Transport of Azurin.
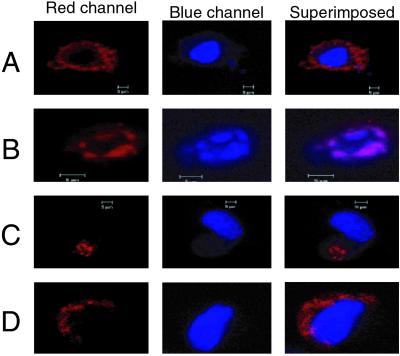
We also used anti-azurin antibodies to localize azurin in various subcellular fractions of UISO-Mel-2 cells. The bulk of azurin was found in the cytosol, but azurin was also found to be in the nuclear fraction (Fig. 3C). Azurin was also found in the mitochondria, but not during the earlier period. In the p53-null UISO-Mel-6 cells, azurin was located in the cytosol and in mitochondria, but not in the nucleus, demonstrating a role of p53 in the nuclear transport of azurin (Fig. 3C). A possible role of p53 in the nuclear transport of azurin raised the question whether a putative receptor-binding of azurin might be important for its subsequent trafficking to the nucleus, likely mediated through an association with p53. p53 is known to traffic freely from the cytosol to the nucleus and vice versa (21) and can therefore help in the transport of azurin to the nucleus. To determine whether microinjected azurin, which did not have to go through cell surface-associated receptor binding, can be transported to the nucleus in UISO-Mel-2 cells but not in UISO-Mel-6 cells, we microinjected wt azurin in these two cell types and determined the localization of azurin soon after microinjection (15 min) and after 2 or 3 h. The results demonstrate that, when microinjected in p53+-UISO-Mel-2 cells, azurin is found primarily in the cytosol at 15 min (Fig. 4A). At 2 h after microinjection, however, azurin is found in the nucleus of UISO-Mel-2 cells, which also seems to undergo structural deformity, perhaps because of the onset of apoptosis (Fig. 4B). In the p53-null UISO-Mel-6 cells, microinjected azurin is found localized at one end of the cytosol in 15 min (Fig. 4C); 3 h after microinjection, azurin is still found only in parts of the cytosol surrounding the nucleus, but not within the nucleus itself (Fig. 4D), confirming the Western blotting data (Fig. 3C) that p53 is important for the transport of azurin to the nucleus. No condensation or fragmentation of the nucleus was observed under these conditions (Fig. 4D).
Fig 4.
Representative confocal microscopy images for localization of azurin in the cytosol and subsequent trafficking to nucleus. Microinjection of Alexa Fluor 568-labeled azurin to UISO-Mel-2 and UISO-Mel-6 cells is described under Materials and Methods. (A) UISO-Mel-2 cells 15 min after injection. (B) UISO-Mel-2 cells 2 h after injection. The induction of apoptosis is evident from the shrinkage and fragmentation of nuclear DNA. (C) UISO-Mel-6 cells 15 min after injection of azurin in the cytosol. (D) UISO-Mel-6 cells 3 h after injection of azurin.
Azurin Forms a Complex with p53 Leading to Its Stabilization.
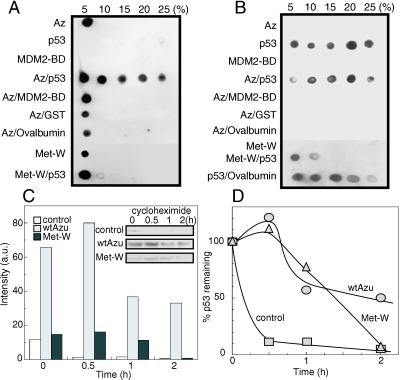
The enhanced intracellular level of p53 during treatment of the UISO-Mel-2 cells with azurin and the apparent role of p53 in the transport of azurin to the nucleus, raised the question whether azurin might stabilize p53. p53 is a highly labile protein that can be stabilized or destabilized by protein–protein interaction (21, 30, 31). Of particular interest is a recent report that a mammalian oxidoreductase, NADH quinone oxidoreductase 1 (NQO1), stabilized p53 by inhibiting its degradation in a distinct pathway (13, 32). Another oxidoreductase WOX1, a mitochondrial apoptogenic protein, has been shown to bind to the proline-rich region of p53 and is an important partner in p53-mediated cell death (33). We thus examined any putative complex formation between azurin and p53 to determine whether this bacterial redox protein can bind p53, perhaps to stabilize it and raise its intracellular level for ultimate induction of apoptosis and growth arrest of the cancer cells. To determine complex formation between azurin and p53, we used the glycerol gradient centrifugation method used by us to study σ factor–anti-σ factor interaction (34) or complex formation between nucleoside diphosphate kinase and various other proteins such as elongation factor Tu (35), Ras-like protein Pra (36), pyruvate kinase (37), etc. We prepared glycerol gradients comprising of 5–25% glycerol as described (17, 37) and azurin, p53 (we used a GST-p53 fusion protein), the MDM2-binding domain (MDM2-BD) of p53 (38), and other proteins, singly or in combination, were sedimented through this gradient by centrifugation. After centrifugation, various fractions were collected from different glycerol gradients and the presence of azurin (Fig. 5A) or p53 (Fig. 5B) was determined in various gradient fractions by Western blotting with anti-azurin or anti-p53 monoclonal antibodies. Azurin, as a low molecular mass (14 kDa) protein, sediments at 5% glycerol fraction and not in others (Az, Fig. 5A). However, when preincubated with p53, it is found in the higher glycerol fractions as well (Fig. 5A, Az/p53). Preincubation of azurin with the MDM2-BD of p53, in contrast to p53 itself, does not allow a higher sedimentation profile, suggesting either a lack of complex formation or that the binding of this low-molecular-weight fragment (4 kDa) does not significantly affect the molecular size of the complex. Other proteins such as GST, ovalbumin, and BSA (not shown) did not allow sedimentation of azurin at higher glycerol gradients, suggesting that complex formation between azurin and p53 is specific. The lack of complex formation between azurin and GST strongly implies that complex formation between azurin and GST-p53 fusion protein is due to p53 and not due to GST. p53 by itself forms a polydisperse aggregate (Fig. 5B), presumably because of its ability for oligomerization (39). The M44K/M64E double mutant (termed Met-W; Fig. 5 A and B), deficient in cytotoxicity toward UISO-Mel-2 and UISO-Mel-6 cells (Fig. 2 B and C), was also deficient in forming a complex with p53 (Fig. 5A).
Fig 5.
Glycerol gradient centrifugation to detect complex formation between wt azurin and p53, leading to stabilization of p53. Azurin and p53 were detected in various glycerol gradient fractions by Western blotting with anti-azurin (A) and anti-p53 (B) monoclonal antibodies. Azurin was found in various glycerol gradient fractions only when preincubated with p53, but not with GST, MDM2-BD, or ovalbumin (A). The double mutant Met-W, in the absence or in the presence of p53, demonstrated its sedimentation at 5% glycerol fraction. The stability of p53 was determined in UISO-Mel-2 cells incubated for 12 h with buffer (control), wt azurin or mutant azurin Met-W; after 12 h of incubation, cycloheximide was added to prevent protein synthesis, and the level of p53 was determined at 0.5, 1.0, and 2.0 h after cycloheximide addition by Western blotting using anti-p53 monoclonal antibodies (C and D).
The ability of azurin to form complexes with p53 raised the important question whether such complex formation conferred stability to p53, as was shown for NQO1 (13, 32). We therefore incubated UISO-Mel-2 cells with azurin for 12 h. A control UISO-Mel-2 cell suspension without azurin, but only treated with buffer for 12 h, was maintained. After 12 h, 20 μg/ml cycloheximide was added to the cell suspensions to prevent protein synthesis, and the amount of residual p53 was determined in the cell extracts (50 μg protein each) for the next 2 h by Western blotting (Fig. 5C). The amount of p53 was quantitated during 0 (time of addition of cycloheximide), 0.5, 1, and 2 h and depicted as percent of residual p53 remaining with the 0-h value as 100% in each case (Fig. 5D). Very little p53 was seen 2 h after cycloheximide addition in the extracts of untreated control or mutant Met-W azurin-treated UISO-Mel-2 cells (Fig. 5 C and D), whereas substantial p53 was still present in the extracts of wt azurin-treated cells, clearly suggesting that wt azurin treatment allows stabilization of p53, thereby raising the intracellular level of this apoptosis-inducing protein. The Met-W mutant azurin, deficient in forming a complex with p53 (Fig. 5A) was also deficient in stabilizing p53 in UISO-Mel-2 cells.
Azurin Induces Tumor Regression in Vivo.
On the basis of the observation of specificity of azurin in exerting its cytotoxicity to p53+ human melanoma (UISO-Mel-2) cells in vitro, it was thought that azurin might produce an inhibitory effect on the growth and progression of UISO-Mel-2 xenotransplants in nude mice. If indeed this is the case, then azurin, singly or in combination with other redox proteins and/or known chemotherapeutic agents, can be used in the treatment of human neoplasia. Thus, initially, an in vivo study using only azurin was performed.
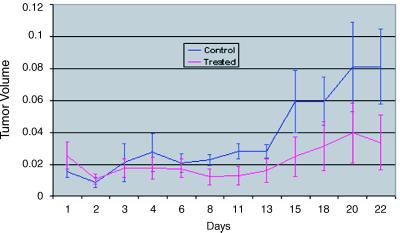
UISO-Mel-2 cells (1 × 106) were injected s.c. into the right flanks of nude (athymic) mice. After 2 weeks when small tumors appeared, the animals were divided into an untreated control group (n = 5) and treated group (n = 5). The treated group received i.p. 0.5 mg of wt azurin daily for 22 days. Tumor volume was measured for each mouse on various days as shown in Fig. 6. Azurin-treated mice showed tumor growth inhibition. At the conclusion of the study, the mean tumor volume in azurin-treated mice was 59% lower than that in the controls (Fig. 6). Multivariate analysis based on a random coefficient model showed that the difference in the tumor volume between treated and control animals was significant (P = 0.0116).
Fig 6.
Regression of melanoma (UISO-Mel-2) in nude mice as a function of treatment time with azurin. Tumor volume is in cubic centimeters.
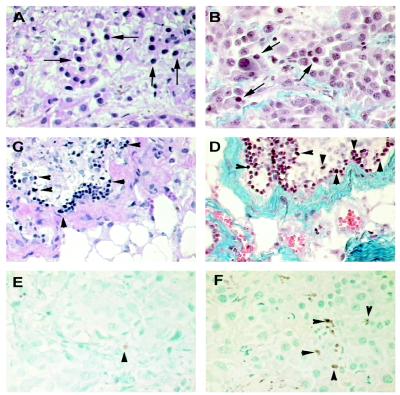
Histologic and immunocytochemical studies showed that azurin-induced tumor regression in UISO-Mel-2 xenotransplants mimics the morphologic features that characterize regression of primary human cutaneous melanoma (43, 44). Although the mechanism of regression of human cutaneous melanoma has not yet been fully explained, two signal microscopic features are commonly observed in regressing melanoma. These are excessive proliferation of stromal fibrous tissue encompassing melanoma cells and induction of apoptosis (45, 46). Both of these were observed in all of the azurin-treated xenotransplants as compared with untreated controls (Fig. 7). Furthermore, at the time of conclusion of the study, complete lack of toxicity, as evidenced by any changes in body weight or other form of toxicity, was noted at this dose level and for this period. At necropsy, none of the treated mice showed any histologic evidence of apparent toxicity and all of the viscera were within normal limits.
Fig 7.
Histological and immunocytochemical analysis of tumors from nude mice. (A) Untreated control UISO-Mel-2 flank tumor 3 weeks after inoculation in nude mice. Abundance of viable malignant cells as shown by arrows. [Hematoxylin/eosin (H&E) stain, ×40.] (B) Untreated control UISO-Mel-2 tumor 3 weeks after inoculation. Masson's trichrome selectively stains fibrous tissues blue; a normal distribution of blue-stained fibrous tissues with similar numbers of malignant cells as in A is seen (shown by arrows). (×40.) (C) Azurin-treated UISO-Mel-2 tumor after 3 weeks of treatment. Apparent is a paucity of viable malignant cells but with many nonviable (apoptotic) cells (arrowhead) with areas of regression and fibrosis, stained deep purple with H&E stain. (×40.) (D) Same azurin-treated UISO-Mel-2 tumor after 3 weeks of treatment. Stained with Masson's trichrome stain. Many nonviable (apoptotic) cells (shown by arrowhead) are seen. The extensive blue areas of fibrosis are pronounced. Similar to C but in contrast to B, very few viable melanoma cells are observed. (×40.) (E) Untreated control 3 weeks after inoculation in nude mice. TUNEL stain shows scanty apoptotic cells. In this section, only one is easily recognizable (shown by arrowhead). (×40.) (F) Azurin-treated tumor cells, 3 weeks later. TUNEL stain shows a multitude of apoptotic cells (shown by arrowheads). (×40.)
Discussion
Much effort has been spent over the years in developing wild-type or attenuated bacterial and viral strains for the treatment of cancer (2, 4–6, 40). The results have been mixed, given significant toxicity associated with the administration of live cells and the consequent immune response to such cells. The use of purified low molecular weight bacterial proteins as potential anticancer agents may thus allow us to bypass these problems, although the proteins themselves may trigger immune response leading to their elimination from the body. It might be possible to isolate site-directed mutants of azurin in the antigenic epitope regions that may reduce the effect of such immune response without affecting its cytotoxicity. It might also be possible to use truncated versions of this redox protein, which may possess cytotoxicity, but little immunogenicity. Preparation and testing of such mutant and truncated derivatives are currently under progress in our laboratory.
A lack of p53 in the UISO-Mel-6 cells leads to a comparative loss of cytotoxicity of azurin toward these cells. The inability of the M44K/M64E double mutant to form a stable complex with p53 in UISO-Mel-2 cells and its lack of cytotoxicity indicates that complex formation may be the primary reason for azurin-mediated cytotoxicity. Such complex formation may also account for the transport of azurin to the nucleus where p53 may be stabilized and may allow a higher level of synthesis of Bax and other proapoptotic proteins (41, 42). Bax levels are indeed higher in the mitochondria of UISO-Mel-2 cells during 6–12 h of incubation with azurin (Fig. 3A), but not in the mitochondria of the p53-null UISO-Mel-6 cells (Fig. 3B). Higher levels of Bax may then trigger apoptosis in the cancer cells by lowering the mitochondrial membrane permeability, thus enhancing the release of mitochondrial cytochrome c to the cytosol in UISO-Mel-2 cells (Fig. 3A) but not in UISO-Mel-6 cells (Fig. 3B). To our knowledge, this is the first report of a bacterial redox protein that induces apoptosis in cancer cells by forming a complex with p53 and allowing subsequent apoptosis and regression of cancer cells.
Acknowledgments
We thank Albert Green and Laura Bratescu for their expert technical assistance. M.G. is on a leave of absence from Kyushu University, Japan, and is supported by a fellowship from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. T.Y. is a visiting scholar from the Department of Built Environment, Tokyo Institute of Technology, Tokyo. This work was supported by Public Health Service Grant ES-04050-17 from the National Institute of Environmental Health Sciences AI 16790 (to A.M.C.) and by National Cancer Institute Grant T-32 CA 09432 (to T.K.D.G.).
Abbreviations
TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
MTT, 3-(4,5-dimethylthiazol-2-yl-2,5-diphenyl tetrazolium bromide)
wt, wild type
References
- 1.Coley W. B. (1891) Clin. Orthop. Relat. Res. 262, 3-12. [PubMed] [Google Scholar]
- 2.Alexandrof A. B., Jackson, A. M., O'Donnell, M. A. & James, K. (1999) Lancet 353, 1689-1694. [DOI] [PubMed] [Google Scholar]
- 3.Paglia P. & Guzman, C. A. (1998) Cancer Immunol. Immunother. 46, 88-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pawelek J., Low, K. & Bermuda, D. (1997) Cancer Res. 57, 4537-4544. [PubMed] [Google Scholar]
- 5.Dang L. H., Bettegowda, C., Huso, D. L., Kinzler, K. & Vogelstein, B. (2001) Proc. Natl. Acad. Sci. USA 98, 15155-15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain R. K. & Forbes, N. S. (2001) Proc. Natl. Acad. Sci. USA 98, 14748-14750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter C. A., Yu, D., Gee, M., Ngo, C. V., Sevignani, C., Goldschmidt, M., Golovkina, T. V., Evans, S., Lee, W. F. & Thomas-Tekhonenko, A. (2001) J. Immunol. 166, 5878-5881. [DOI] [PubMed] [Google Scholar]
- 8.O'Reilly M. S., Boehm, T., Shing, Y., Fukai, N., Vasios, G., Lane, W. S., Flynn, E., Birkhead, J. R., Olsen, B. R. & Folkman, J. (1997) Cell 88, 277-285. [DOI] [PubMed] [Google Scholar]
- 9.Zaborina O., Dhiman, N., Chen, M. L., Kostal, J., Holder, I. A. & Chakrabarty, A. M. (2000) Microbiology 146, 2521-2530. [DOI] [PubMed] [Google Scholar]
- 10.Kukimoto M., Nishiyama, M., Tanokura, M., Murphy, M. E., Adman, E. T. & Horinouchi, S. (1996) FEBS Lett. 394, 87-90. [DOI] [PubMed] [Google Scholar]
- 11.Rauth S., Kichina, J., Green, A., Bratescu, L. & Das Gupta, T. K. (1994) Anticancer Res. 14, 2457-2463. [PubMed] [Google Scholar]
- 12.Mosmann T. (1983) J. Immunol. Methods 65, 55-63. [DOI] [PubMed] [Google Scholar]
- 13.Asher G., Lotem, J., Cohen, B., Sachs, L. & Shaul, Y. (2001) Proc. Natl. Acad. Sci. USA 98, 1188-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Z., Bhalla, K., Pantazis, P., Hendrickson, E. A. & Wyche, J. H. (1999) Mol. Cell. Biol. 19, 1381-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raffo A. J., Kim, A. L. & Fine, R. L. (2000) Oncogene 19, 6216-6228. [DOI] [PubMed] [Google Scholar]
- 16.Marzo I., Brenner, C., Zamzami, N., Jurgensmeier, J., Susin, S. A., Vieira, H. L. A., Prevost, M. C., Xie, Z., Mutsuyama, S., Reed, J. C. & Kroemer, G. (1998) Science 281, 2027-2031. [DOI] [PubMed] [Google Scholar]
- 17.Shankar S., Kapatral, V. & Chakrabarty, A. M. (1997) Mol. Microbiol. 26, 607-618. [DOI] [PubMed] [Google Scholar]
- 18.van de Kamp M., Silvestrini, M. C., Brunori, M., van Beeumen, J., Hali, F. C. & Canters, G. W. (1990) Eur. J. Biochem. 194, 109-118. [DOI] [PubMed] [Google Scholar]
- 19.Cutruzzola F., Arese, M., Ranghino, G., van Pouderoyen, G., Canters, G. & Brunori, M. (2002) J. Inorg. Biochem. 88, 353-361. [DOI] [PubMed] [Google Scholar]
- 20.Farver O., Jeuken, L. J., Canters, G. W. & Pecht, I. (2000) Eur. J. Biochem. 267, 3123-3129. [DOI] [PubMed] [Google Scholar]
- 21.Vogelstein B., Lane, D. & Levine, A. J. (2000) Nature 408, 307-310. [DOI] [PubMed] [Google Scholar]
- 22.Schuler M. & Green, D. R. (2001) Biochem. Soc. Trans. 29, 684-688. [DOI] [PubMed] [Google Scholar]
- 23.Schuler M., Bossy-Wetzel, E., Goldstein, J. C., Fitzgerald, P. & Green, D. R. (2000) J. Biol. Chem. 275, 7337-7342. [DOI] [PubMed] [Google Scholar]
- 24.Marchenko N. D., Zaika, A. & Moll, U. M. (2000) J. Biol. Chem. 275, 16202-16212. [DOI] [PubMed] [Google Scholar]
- 25.Ding H. F., McGill, G., Rowan, S., Schmaltz, C., Shimamura, A. & Fisher, D. E. (1998) J. Biol. Chem. 273, 28378-28383. [DOI] [PubMed] [Google Scholar]
- 26.Miyashita T. & Reed, J. C. (1995) Cell 80, 293-299. [DOI] [PubMed] [Google Scholar]
- 27.Gottlieb T. M. & Oren, M. (1998) Semin. Cancer Biol. 8, 359-368. [DOI] [PubMed] [Google Scholar]
- 28.Gross A., Jockel, J., Wei, M. C. & Korsmeyer, S. J. (1998) EMBO J. 17, 3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desagher S., Osen-Sand, A., Nichols, A., Eskes, R., Montessuit, S., Lauper, S., Maundrell, K., Antonsson, B. & Martinou, J. C. (1999) J. Cell Biol. 144, 891-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maki C. G., Huibregtse, J. M. & Howley, P. M. (1996) Cancer Res. 56, 2649-2654. [PubMed] [Google Scholar]
- 31.Haupt Y., Maya, R., Kazaz, A. & Oren, M. (1997) Nature 387, 296-299. [DOI] [PubMed] [Google Scholar]
- 32.Asher G., Lotem, J., Kama, R., Sachs, L. & Shaul, Y. (2002) Proc. Natl. Acad. Sci. USA 99, 3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang N.-S., Pratt, N., Heath, J., Schultz, L., Sleve, D., Carey, G. B. & Zevotek, N. (2001) J. Biol. Chem. 276, 3361-3370. [DOI] [PubMed] [Google Scholar]
- 34.Xie Z.-D., Hershberger, C. D., Shankar, S., Ye, R. W. & Chakrabarty, A. M. (1996) J. Bacteriol. 178, 4990-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukhopadhyay S., Shankar, S., Walden, W. & Chakrabarty, A. M. (1997) J. Biol. Chem. 272, 17815-17820. [DOI] [PubMed] [Google Scholar]
- 36.Chopade B. A., Shankar, S., Sundin, G. W., Mukhopadhyay, S. & Chakrabarty, A. M. (1997) J. Bacteriol. 179, 2181-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sundin G. W., Shankar, S., Chugani, S. A., Chopade, B. A., Kavanaugh-Black, A. & Chakrabarty, A. M. (1996) Mol. Microbiol. 20, 965-979. [DOI] [PubMed] [Google Scholar]
- 38.Kanovsky M., Raffo, A., Drew, L., Rosal, R., Do, T., Friedman, F. K., Rubinstein, P., Visser, J., Robinson, R., Brandt-Rauf, P. W., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 12438-12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maki C. G. (1999) J. Biol. Chem. 274, 16531-16535. [DOI] [PubMed] [Google Scholar]
- 40.Kirn D. H. (2000) J. Clin. Invest. 105, 837-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed J. C. (1999) J. Clin. Oncol. 17, 2941-2953. [DOI] [PubMed] [Google Scholar]
- 42.Thornborrow E. C., Patel, S., Mastropietro, A. E., Schwartzfarb, E. M. & Manfredi, J. J. (2002) Oncogene 21, 990-999. [DOI] [PubMed] [Google Scholar]
- 43.Ronan S. G., Han, M. C. & Das Gupta, T. K. (1988) Semin. Oncol. 15, 558-565. [PubMed] [Google Scholar]
- 44.Ronan S. G., Farolan, M. J., McDonald, A., Manaligod, J. R. & Das Gupta, T. K. (1994) J. Cutan. Pathol. 21, 494-499. [DOI] [PubMed] [Google Scholar]
- 45.Das Gupta T. K., Ronan, S. G., Beattie, C. W., Shilkaitis, A. L. & Amoss, M. S., Jr. (1989) Pediatr. Dermatol. 6, 289-299. [DOI] [PubMed] [Google Scholar]
- 46.Hieken T. J., Ronan, S. G., Farolen, M., Shilkaitis, A. L. & Das Gupta, T. K. (1999) Cancer 85, 375-382. [DOI] [PubMed] [Google Scholar]