Abstract
Extracellular signals are transduced into cells through mitogen-activated protein kinases (MAPKs), which are activated by their upstream kinases. Recently, families of scaffolding proteins have been identified to tether specific combinations of these kinases along specific signaling pathways. Here we describe a protein, JLP (c-Jun NH2-terminal kinase-associated leucine zipper protein), which acts as a scaffolding protein to bring together Max and c-Myc along with JNK (c-Jun NH2-terminal kinase) and p38MAPK, as well as their upstream kinases MKK4 (MAPK kinase 4) and MEKK3 (MAPK kinase kinase 3). Thus, JLP defines a family of scaffolding proteins that bring MAPKs and their target transcription factors together for the execution of specific signaling pathways.
The mitogen-activated protein kinase (MAPK) family consists of a group of kinases responsive to a variety of environmental stimuli. MAPKs can be subdivided into three groups: extracellular signal-regulated kinase (ERK), c-Jun NH2-terminal kinase (JNK), and p38MAPK (1–8). Although ERK has been shown to be activated primarily by proliferative signals, JNK and p38MAPK are activated by genotoxic as well as cytotoxic stress signals (7–10). The structural organization of these kinases into specific signaling modules appears to be facilitated by scaffolding proteins such as STE5 in yeast (11–14) and JNK/stress-activated protein kinase-associated protein (JSAP) and JNK interacting proteins 1–3 in mammalian cells (15–20). These scaffolding proteins tether different MAPK kinase kinases (MEKKs), MAPK kinases (MKKs), and MAPKs into close proximity so that the successive phosphorylation events can occur efficiently, thus conferring specificity to a particular combination of kinases for activation. Although it is well documented that these phosphorylation cascades lead to the activation of transcription factors such as Fos, Jun, and Myc, the precise mechanisms through which the specific transcription factors are recruited is not known (1, 4). In our search for proteins that associate with transcription factors such as Max and Myc, we identified a scaffolding protein, which we termed JLP for JNK-associated leucine zipper protein. Here we show that JLP brings together Max and c-Myc along with JNK and p38MAPK, as well as their upstream kinases MKK4 and MEKK3. Thus, JLP defines a family of scaffolding proteins that bring MAPKs and their target transcription factors together for the execution of specific signaling pathways.
Materials and Methods
Cloning of JLP.
Human Max was expressed as bacterial recombinant protein, which was 32P-labeled in vitro with heart muscle kinase (Sigma) by using [γ-32P]ATP. The Max probe was added in the hybridization buffer Hyb75 (20 mM Hepes-KOH, pH 7.7/75 mM KCl/0.1 mM EDTA/2.5 mM MgCl2/0.05% Nonidet P-40/1% nonfat milk/10 mM DTT) and used to screen the λgt11 expression library derived from 32Dcl3 cells (21) as described (22). Four overlapping cDNAs were used to generate a cDNA sequence encoding for the full-length protein of JLP.
S Tagging and Mutation of JLP.
PCR was used to generate a fragment of JLP sequence (3,250–4,083 bp) whose 3′ end contained the coding sequence of S tag (KETAAAKFERQHMDS) followed by a stop codon. Substitution of all leucine residues [amino acids 117, 124, 131, 145, 152, and 159 in leucine zipper I (LZI) and amino acids 413 and 420 in leucine zipper II (LZII)] of JLP with alanine residues was carried out by site-directed mutagenesis and fusion PCR by using M2 cDNA as the template as described (23). All mutations and deletion constructs were verified by sequence analysis. JLP-S 3′ deletion mutants were created by digestion of the WT JLP-S cDNA EcoRI/MluI fragment with SmlI, MslI, PvuII, BssSI, SpeI, and Bsu36I. JLP-S domain constructs were generated by PCR. The regions spanned by domain II, II/LZII, and IIΔ are 640–1,356 bp, 640–1,551 bp, and 792–1,356 bp, respectively. JLP-I-S and JLP-I/LZI–S are the two shortest 3′ deletion mutants. Dominant negative mutant ΔJBD was created by deletion of the amino acids of JLP from 1 to 107 and from 197 to 209 by using fusion PCR. All of these mutants and domains have the S tag at their C terminal.
Bacterial Recombinant Proteins and Gel-Shift Assays.
The cDNA inserts of M2 and M2LZI corresponding to nucleotides 114-1471 of JLP cDNA were blunt-ended and subcloned into the XmaI-digested and blunt-ended pDS56-derived vector (Qiagen, Chatsworth, CA) to place the 6-histidine tag at the N terminus. MC15 host cells were transformed with plasmid DNAs, and the recombinant proteins were induced in the presence of 1 mM isopropyl-β-d-thiogalactopyranoside and purified by Ni2+-NTA agarose resin (Qiagen) under denaturing conditions by using the manufacturer's protocol. The recombinant proteins were renatured by stepwise dialysis to remove guanidine-HCl. Recombinant Max protein was produced similarly as described (24). Gel-shift assays were performed as described (24).
Western Blot Analysis.
Western blot analysis was performed similarly as described (25). The specific antibody against FLAG tag (M2) was purchased from Sigma, and the specific antibodies against S tag, GST tag, hemagglutinin (HA) tag, ERK2, Max (C17), and c-Myc (N262) were from Santa Cruz Biotechnology. Rabbit antibodies against recombinant JLP were produced in-house.
In Vivo Pull-Down Assay.
The assay was performed as described (26) by using buffer S (250 mM Tris·HCl, pH 7.5/137 mM NaCl/1% Nonidet P-40/0.1% SDS/0.5% sodium deoxycholate/1 mM PMSF/2 μg/ml pepstatin/2 μg/ml leupeptin/1.9 μg/ml aprotinin/1 mM Na3VO4) for lysis. Cell lysate (400–500 μg) was mixed with the S-protein agarose beads (Novagen) at 4°C for 1 h. The beads of each sample was washed four times with buffer S, and the precipitates were incubated with sample buffer at 37°C for 30 min before they were boiled for 7 min. They were then subjected to SDS/PAGE, followed by Western blot analysis.
Indirect Immunofluorescence Staining of JLP.
These studies were conducted similarly as described (27) by using JLP C-terminal-specific antibody. The cells were mounted and analyzed under a fluorescence microscope (Olympus IX70/Fluoview).
Results and Discussion
Identification of a Max-Binding Protein JLP.
In our search for proteins that associate with the Myc/Max family of proteins, we screened a λgt11 expression library of the mouse myeloid cell line, 32Dcl3, with 32P-labeled Max probe, which yielded 13 positive clones (22). These clones were found to be derived from mRNAs that encode two different classes of proteins. One class was the basic helix–loop–helix/leucine zipper proteins such as c-Myc and Mad, which have been reported to associate with Max (28–30). The second class consisted of proline-rich proteins containing many charged amino acids. The nucleotide sequence of one set of the overlapping clones revealed the presence of a single large ORF of 3.9 kb encoding a polypeptide of 1,307 aa (Fig. 1A). This protein contains LZI, LZII, and a C-terminal domain that shows extensive homology with Caenorhabditis elegans protein ZK1098.10. In addition, this protein shares 69% homology with JSAP1 and JNK interacting protein 3 that act as scaffolding proteins for the JNK signaling pathway (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). This protein has also been found to share sequence similarity with other partially cloned cDNAs and EST sequences such as KIAA0516. This protein, described in Fig. 1A, was named JLP.
Fig 1.
Sequence and expression of murine JLP. (A) The deduced protein sequence of JLP is given. The conserved domains A, B, and C are boxed. The heptad repeat of leucine and homologous hydrophobic amino acids in LZI and LZII are circled. The putative SH2 and SH3 binding sites are shaded and underlined, respectively. (B) A full-length cDNA of JLP was subcloned into a modified form of pSG5 mammalian expression vector (Stratagene) and used for transient transfection into COS7 cells. The endogenous JLP of 32Dcl3 cells was immunoprecipitated from cell lysates with preimmune antibody (PI) and the JLP N-terminal-specific antibody (I). The immunoprecipitates were resolved by SDS/PAGE together with the lysates from COS7 cells transfected with vector alone (V) or expression plasmid for JLP. The Western blot analysis was performed with the same JLP antibody. (C) Subcellular localization of JLP. The subcellular localization of endogenous JLP in Swiss3T3 cells was analyzed by indirect immunofluorescence studies. Unstimulated cells (US) were permeablized with Triton X-100, followed by RNase A treatment and immunostaining with preimmune antibody (PI/US) or the JLP C-terminal-specific antibody (JLP/US), followed by the fluorescein-conjugated anti-rabbit IgG secondary antibody (green). The cell nuclei were stained with propidium iodide (red). Cells had also been exposed to UV (254 nm, 5 min), followed by 30-min incubation before they were stained with the JLP-specific antibody (JLP/UV).
Northern blot analysis using the JLP cDNA probe shows a ubiquitous pattern of expression (data not shown). Ectopic expression of JLP in COS7 cells followed by Western blot analysis revealed that the 180-kDa protein encoded by this cDNA was identical to the endogenous JLP that could be immunoprecipitated from 32Dcl3 cells (Fig. 1B). Because the ORF contains 1,307 codons, which should encode for a polypeptide of 143 kDa, it is possible that JLP undergoes one or more forms of posttranslational modification. Immunofluorescence studies using Swiss3T3 cells revealed that JLP was a primarily cytoplasmic protein with a punctate distribution (Fig. 1C). However, JLP attained a perinuclear distribution on exposure to UV (Fig. 1C), suggesting that the cytoplasmic distribution of JLP changes dynamically in response to stress signals such as UV radiation.
JLP/Max Interactions Are Mediated by Their Leucine Zipper Domains.
Because JLP was isolated as a Max-binding protein, we next proceeded to demonstrate its binding to Max in vivo and in vitro by coimmunoprecipitation. For this, recombinant proteins (produced in Escherichia coli) of Max and the N-terminal region of JLP (M2), which contained LZI and LZII, were prepared. The purified recombinant proteins were mixed and immunoprecipitated either with the preimmune serum or Max-specific antibody. Western blot analysis shown in Fig. 2A demonstrates that M2 (N-terminal domain of JLP) could be readily immunoprecipitated from the mixture by Max-specific antibody, but not by the preimmune serum, suggesting that the N-terminal domain of JLP containing the two leucine zippers associates with Max. The precipitation of M2 was not caused by the nonspecific binding of the Max antibody to M2 because these antibodies could not precipitate M2 in the absence of Max (Fig. 2A).
Fig 2.
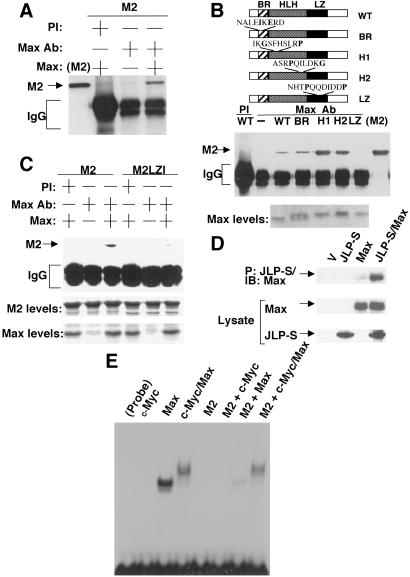
Association of JLP with Max. (A) Bacterial recombinant Max protein was mixed with the recombinant protein M2 (the N-terminal region of JLP containing LZI and LZII) in the binding buffer (22) and immunoprecipitated with preimmune serum (PI) or Max-specific antibody (Max Ab). The immunocomplexes were subjected to Western blot analysis with the JLP N terminal-specific antibody. (B) A series of bacterial recombinant WT Max and its mutants were used (24). (Top) The mutations were introduced in the basic region (BR), the first helix (H1), the second helix (H2), and the leucine zipper (LZ). The binding studies were carried out by using the method described in A. (Middle) The mapping results from the coimmunoprecipitation experiments. The lane labeled M2 contains the recombinant M2, loaded directly onto the gel. (Bottom) The Coomassie blue-stained gels for the WT and mutants of Max (2 μg each). (C) Mapping of the domain of JLP involved in association with Max. Recombinant WT JLP (M2) or a mutant where the leucines of the first leucine zipper were replaced by alanines (M2LZI) was used in a coimmunoprecipitation experiment with WT Max protein as described in A. The lower two panels show the Coomassie blue-stained gels for the levels of M2, M2LZI, and Max recombinant proteins (5 μg each). (D) In vivo association of JLP with Max. COS7 cells were transiently transfected to express Max with or without S-tagged JLP (JLP-S). The cell lysates were subjected to a pull-down assay using the S-protein agarose. The precipitates as well as the total cell lysates were analyzed with the specific antibodies against the S tag or Max. (E) Effect of JLP on Max homodimerization and c-Myc/Max heterodimerization. Electrophoretic mobility-shift assay was performed by using 32P-labeled double-stranded oligonucleotide, CM1 containing an E-box (CACGTG). Recombinant c-Myc and Max, as described (24), were mixed with the labeled probe in the presence or absence of recombinant M2. The DNA/protein complexes were resolved in a native nondenaturing polyacrylamide gel (4%).
To determine the domains responsible for JLP–Max association, we carried out the coimmunoprecipitation experiments described above with several mutants of Max in which the basic helix–loop–helix and leucine zipper domains were mutated (ref. 24, Fig. 2B). The recombinant WT and mutant forms of Max were incubated with M2, followed by immunoprecipitation with the Max antibody, and analyzed by SDS/PAGE. Results of these experiments show that although the WT Max and the basic helix–loop–helix mutants could be coimmunoprecipitated with M2 (Fig. 2B), the mutant containing alterations in the leucine zipper could not. These results demonstrate that the leucine zipper domain of Max is essential for its interaction with M2. It is interesting to note that the amount of M2 coimmunoprecipitated with helix–loop–helix mutant proteins of Max was greater than that seen with the WT Max. This increase is probably because mutations in the helix–loop–helix domain abolish Max homodimer formation, resulting in greater availability of Max monomers for association with M2.
To determine the domain of M2 responsible for its association with Max, a mutant form of JLP (M2LZI) was prepared, in which all of the leucine residues of LZI were mutated to alanine. This mutant contained no mutations in LZII. Immunoprecipitation experiments carried out with this recombinant protein showed that it failed to associate with Max, whereas its WT counterpart readily did so (Fig. 2C). Similar experiments with a mutant form of M2 in which the leucines of LZII were mutated to alanine did not affect JLP interactions with Max (data not shown). These studies demonstrate that LZI of M2 is important for interaction with Max, and LZII does not participate in this interaction. To demonstrate association between Max and JLP in vivo, a mammalian expression vector encoding the full-length JLP, which was tagged with S peptide (S tag) and WT Max were coexpressed in COS7 cells. After transient transfection, the S-tagged JLP protein (JLP-S) was precipitated by using S-protein agarose (Novagen), and the precipitate was examined for the presence of Max by Western blot analysis. These results demonstrate that Max can associate readily with JLP in vivo (Fig. 2D).
Effect of JLP on Max Homodimer and Myc–Max Heterodimer Formation.
To gain an understanding of the role of JLP in Max-mediated functions, we next investigated its relative affinities toward Max–Max homodimerization and c-Myc–Max heterodimerization reactions. For this study, we carried out electrophorectic mobility-shift assays that can distinguish between Max–Max homodimers and Myc–Max heterodimers by using recombinant c-Myc, Max, and M2 proteins. A 32P-labeled double-stranded oligonucleotide containing the Max-binding site E-box (CACGTG) was used in the assay. Results obtained from this experiment showed that this probe readily bound to both Max–Max homodimers and c-Myc–Max heterodimers as evidenced by the appearance of shifted bands of different mobilities (Fig. 2E). When recombinant c-Myc alone or M2 alone was used in this assay, there was no detectable binding of the probe to either of these two proteins. Interestingly, addition of the M2 protein to the reaction mixture containing recombinant Max, and the labeled probe was found to result in a disruption of Max–Max homodimer formation. However, when recombinant M2 was added to the reaction mixture containing recombinant c-Myc and Max, little or no effect was observed on the formation of c-Myc–Max heterodimers. These results suggest that JLP has a high affinity for Max–Max homodimers and its interaction with Max results in the dissociation of these homodimers. However, JLP's affinity for Max appears to be less than that of c-Myc as evidenced by its inability to dissociate c-Myc–Max heterodimers.
Interaction of JLP with JNK and p38MAPK Signaling Modules.
Because JLP shares homology with the JNK-scaffolding proteins JSAP1 and JNK interacting protein 3, it was of interest to determine whether JLP could also associate with JNK in a similar manner. To test, HA-tagged JNK1 (HA-JNK1) was coexpressed along with JLP-S in COS7 cells and a pull-down assay was performed by using the S-protein agarose. The precipitates were analyzed for the presence of HA-JNK1 by Western blot analysis with a HA-specific antibody. Results of this experiment presented in Fig. 3A show that JNK1 could associate with JLP. When the pull-down assays were performed with cell lysates expressing JLP or JNK1 alone, such coprecipitation was not seen, suggesting that this is a specific interaction between JNK1 and JLP. Such an interaction with endogenous JLP and JNK1 could also be demonstrated (Fig. 3B). Similar pull-down experiments with coexpressed FLAG-p38MAPKα or endogenous ERK2 indicated that JLP-S could also associate with p38MAPKα, but not ERK2 (Fig. 3A).
Fig 3.
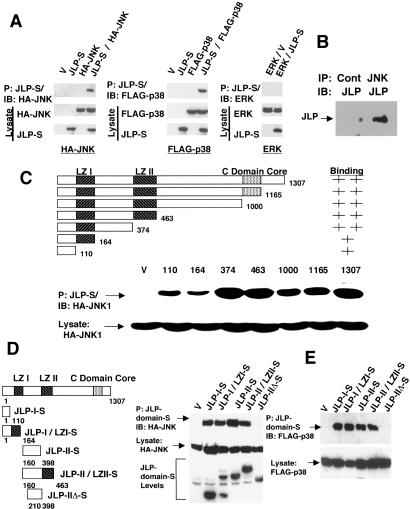
Association of JLP with JNK1 and p38MAPKα. (A) The lysates from COS7 cells expressing HA-JNK1 (HA-JNK), FLAG-p38MAPKα (Flag-p38), or endogenous ERK2 with or without JLP-S were precipitated with the S-protein agarose. Cell lysates were prepared as described (26). The precipitates and the lysates were analyzed with the specific antibodies against the S tag, HA tag, FLAG tag, and ERK2. (B) In vivo association of JLP and JNK1. COS7 cells were lysed and immounoprecipitated with JNK1-specific antibody (JNK) or a control rabbit antibody (Cont). The immunoprecipitates were subjected to Western blot analysis with the JLP C terminal-specific antibody (JLP). (C) Deletion mapping of JNK1-interacting domain of JLP. HA-JNK1 was coexpressed with the WT or 3′ deletion mutants of JLP in COS7 cells. All proteins were S-tagged. The lysates were subjected to the pull-down assay with the S-protein agarose. The precipitates and total cell lysates were analyzed with anti-HA antibody. (D) Five mutants of JLP that were C-terminally S-tagged were expressed in COS7 cells along with HA-JNK1. These mutants were JLP-I-S (amino acids 1–110), JLP-I/LZI-S (amino acids 1–164), JLP-II-S (amino acids 160–398), JLP-II/LZII-S (amino acids 160–463), and JLP-IIΔ-S (amino acids 210–398). The cell lysates were precipitated with S-protein agarose, and the precipitates and the total cell lysates were analyzed with the anti-HA antibody on a Western blot. The lower panel shows the expression levels of the JLP mutants detected by using the anti-S-tag antibody. (E) To map the domains of JLP associating with p38MAPKα, pull-down assays using the same set of C-terminally S-tagged proteins of different regions of JLP described in D were used. The proteins were expressed together with FLAG-p38MAPKα in COS7 cells, and the lysates were precipitated with S-protein agarose. The precipitates and the total cell lysates were analyzed with the anti-FLAG tag antibody on a Western blot.
To determine the domain involved in the association between JLP and JNK1, a series of 3′ deletion mutants of JLP, all of which were S-tagged at the 3′ end, were prepared. After transfection of expression plasmids into COS7 cells, pull-down assays were carried out by using the S-protein agarose beads, which showed that all of the deletion mutants bound to JNK1 (Fig. 3C). However, mutants spanning amino acids 1–110 and 1–164 showed reduced levels of association, suggesting that the region spanning 1–374 may contain multiple binding sites for JNK1. To further determine the sequences of JLP involved in this association, three additional S-tagged JLP-deletion mutants encoding amino acids 160–398, 160–463, or 210–398 were constructed and coexpressed along with HA-JNK1 in COS7 cell. S-protein agarose pull-down experiments using these transfectants indicated that the protein fragments containing the region spanning positions 1–110 and 160–398 could coprecipitate JNK1, whereas a shorter polypeptide lacking amino acids 160–209 but containing amino acids 210–398 failed to do so (Fig. 3D). These results suggest that the extreme N terminus (1–110) and the region containing amino acids 160–209, which are located between the two leucine zipper domains are involved in the association of JLP with JNK1. These results also suggest that either one of these regions is sufficient for JLP–JNK association.
To map the domain of JLP that interacts with p38MAPKα, we performed similar experiments using the above expression vectors together with the p38MAPKα expression vector in COS7 cells. Results of this experiment presented in Fig. 3E show that the JLP-S protein fragments containing the region spanning positions 1–110 and 160–398 could also bring down p38MAPKα. As in the case of JNK, the region of JLP spanning amino acids 210–398 failed to coprecipitate p38MAPKα. Because the domain spanning amino acids 1–110 or 160–209 is sufficient to interact with either JNK or p38MAPK, it is possible that at a given moment a single JLP molecule can tether both JNK and p38MAPK together by using one site for each of the molecules. Such a versatile arrangement could allow JNK and/or p38MAPK to phosphorylate factors binding to LZI, LZII, or regions nearby. JLP was also found to form oligomers (data not shown), which further facilitates the tethering of JNK and p38MAPK. It appears that JLP is a scaffolding protein that is designed uniquely to bring JNK and p38MAPK signaling modules together.
Interaction of JLP with MEKK3 and MKK4.
Both JNK and p38MAPK are activated by an upstream MKK. JNK is activated by MKK4 and MKK7 whereas p38MAPK is activated by MKK3, MKK4, and MKK6 (4). It is significant to note here that MKK4 can activate both JNK and p38MAPK. It was therefore of interest to determine whether JLP is involved in bringing together these kinases to MKK4. To determine the ability of JLP to associate with these upstream kinases that activate JNK and p38MAPK, HA-tagged MKK3 or GST-tagged MKK4 were coexpressed along with S-tagged JLP, and the pull-down assays were performed with the S-protein agarose beads. These studies indicated that JLP could associate with MKK4, but not with MKK3 (Fig. 4A). A similar study was performed to identify the MEKK isoform upstream of MKK4. Coexpression of HA epitope-tagged MEKK3 along with S-tagged JLP in COS7 cells followed by pull-down assays indicated that JLP specifically interacted with MEKK3 (Fig. 4A). Taken together, these results strongly suggest that JLP is a scaffolding protein that brings JNK1 and p38MAPKα in close proximity to the upstream kinases MKK4 and MEKK3 to form a functional MEKK3-MKK4-JNK/p38MAPK signaling module.
Fig 4.
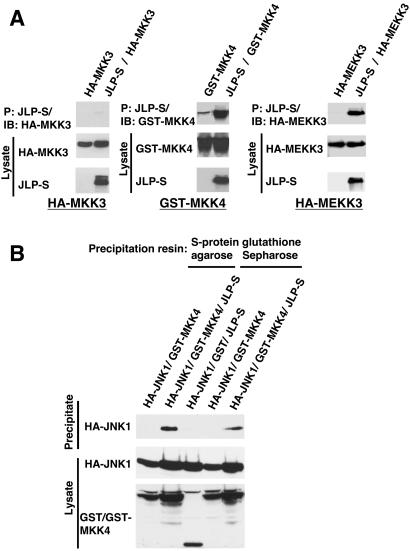
In vivo association of JLP with the upstream kinases. (A) COS7 cells were transiently transfected to express the upstream kinases (GST-tagged MKK4, HA-tagged MEKK3, and MKK3) with or without JLP-S. Cell lysates from the transfected cells were subjected to a pull-down assay using the S-protein agarose (26). The precipitates and lysates were analyzed with the corresponding specific antibodies against the S tag, HA tag, or GST tag. (B) To demonstrate ternary complex formation between JLP, JNK1, and MKK4, COS7 cells were transiently transfected to express HA-JNK1 and GST or GST-tagged MKK4 with or without JLP-S as indicated. Cell lysates from the transfected cells were subjected to pull-down assays using the S-protein agarose or glutathione Sepharose. The precipitates and lysates were analyzed with the corresponding specific antibodies against the HA tag or GST tag.
To show that JLP could function as a scaffolding protein that can tether JNK and its upstream kinase, MKK4 simultaneously, we transiently transfected COS7 cells to express HA-JNK1 and GST-MKK4 with or without JLP-S and carried out pull-down assays with glutathione Sepharose or S-protein agarose. The precipitates and cell lysates were subjected to Western blots using HA antibody or GST antibody. Results presented in Fig. 4B show that GST-MKK4 could pull down HA-JNK1 only when cotransfected with JLP, suggesting that JLP can function as a scaffolding protein to tether multiple factors in the signaling module simultaneously.
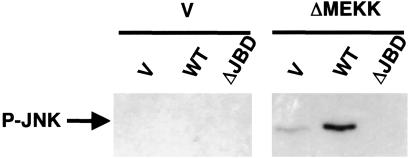
To demonstrate the role of JLP in JNK activation, NIH 3T3 cells were transiently transfected with expression plasmids for JNK1-S, p38MAPK, MKK4, and MEKK3 along with WT JLP or a dominant negative mutant of JLP (Fig. 5) and a dominant positive mutant of MEKK1 (ΔMEKK). Previous studies have shown that ΔMEKK activates JNK and p38MAPK pathways in a constitutive manner (19). JNK1-S was precipitated by S-protein agarose and examined by phospho-JNK antibody. Results show that activation of JNK was enhanced by coexpression of JLP and abolished by the dominant mutant of JLP. These studies further suggest that an important function of JLP could be the assembly of components of JNK pathway for further activation.
Fig 5.
Modulation of JNK phosphorylation by JLP. NIH 3T3 cells were transiently transfected with expression plasmids for JNK1-S, FLAG-p38 MAPK, HA-MKK4, and HA-MEKK3 together with JLP-HA WT, its dominant negative mutant containing the deletion of the JNK binding domains (ΔJBD) or an empty vector (V) in the presence or absence of the dominant positive mutant of MEKK1 (ΔMEKK) as indicated. Dominant negative mutant ΔJBD was created by deletion of the amino acids of JLP from 1–107 and 197–209. One day after transfection, cells were lysed and the cell lysates from the transfected cells were subjected to pull-down assays using the S-protein agarose. The precipitates were analyzed with the phospho-specific antibody against JNK1.
Interaction of JLP with c-Myc.
Because the function of Max is intimately associated with that of Myc, we next examined whether JLP can interact with Myc. For this, we transiently expressed JLP-S and WT c-Myc in COS7 cells, and JLP-S/Myc interaction was studied by using S-protein agarose pull-down assays. Our results presented in Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org, show that c-Myc could readily associate with JLP. Studies with deletions mutants of JLP suggest that the very N-terminal region (amino acids 1–110) and the region (amino acids 160–209) between the two leucine zipper domains are necessary for the association of JLP with c-Myc and each region is sufficient to associate with c-Myc.
Biochemical Function of JLP.
Analysis of the structural features and binding partners of JLP provides interesting clues to its function. JLP is a scaffolding protein that can bring together MEKK3, MKK4, and JNK as well as p38MAPK with a set of transcription factors such as c-Myc and Max. Although JLP exhibits sequence homology to JSAP1, it differs from the latter in its ability to tether two distinct downstream MAPKs, namely JNK and p38MAPK. Because JLP also associates with Max through LZI it is possible that other transcription factors containing leucine zipper motifs can associate with LZII. In addition to the leucine zipper domains, JLP has three putative Src homology (SH)2 and SH3 binding sites, which can potentially tether SH2- and SH3-containing kinases to the multiprotein complexes. By doing so, JLP may provide a link for other signaling pathways mediated by those SH2/SH3-containing kinases to the transcription factors. Because JLP has a ubiquitous expression pattern, JLP can function as a scaffolding protein in many cell types, especially for the ubiquitously expressed MAPKs, such as JNK1, JNK2, p38MAPKα, and p38MAPKβ.
Our findings that JLP can interact with both the kinase signaling modules and transcription factors have important implications for cell signaling mechanisms. Although the crucial role of JNK as well as p38MAPK signaling modules in different signaling pathways has been fully recognized, the mechanism(s) through which these kinase modules activate different sets of transcription factors under different physiological conditions have largely remained unknown. Our findings that JLP can tether MEKK3, MKK4, and JNK/p38MAPK along with Myc and Max suggests a mechanism through which JLP can function as a signaling conduit to transmit extracellular signals to transcription factors. Such a scaffolding function of JLP can accelerate the transmission of signals to the target proteins in addition to insulating the signals from other signaling events in the neighboring milieu. Thus, JLP can function as a positive regulator for the JNK/p38MAPK signaling module. Furthermore, JLP is likely to be involved in shuttling the signaling proteins to different intracellular sites, depending on the signal strength or origin. In fact, such differential localization of signaling kinases and other proteins in specific regions of the cell has been observed during different aspects of cell signaling. It has been observed that the scaffolding proteins, in association with transport proteins such as kinesins, facilitate subcellular protein trafficking. In this context, it is noteworthy that JNK interacting protein/JSAP associate with kinesin-1 (27, 31, 32), which is known to be involved in the active transportation of different signaling proteins along microtubules. Our recent studies show a similar association between JLP and kinesin-1 (Q. Nguyen and E.P.R., unpublished work). On the basis of these observations, it can be predicted that JLP, in association with other transport proteins, is involved in shuttling the preorganized signaling complex to different sites, such as cytoplasmic to pernuclear regions. In fact, the dynamic cytoplasmic to perinuclear localization of JLP in response to UV irradiation suggests such a role for JLP.
Supplementary Material
Acknowledgments
We are grateful to Dr. Roger Davis for the human cDNAs of JNK1 and p38MAPKα and Dr. Richard R. Vaillancourt for the cDNAs of MEKK3 and MKK3. We thank Dr. John Jenkins and Ms. Christine Addison for their help with expression screening, Mr. Richard V. Mettus for sequence analysis, and Mr. Akhileshwar Khanal for cloning the 5′ end of the human JLP cDNA. C.M.L. was supported by the Daniel Swan Fellowship and National Institutes of Health Postdoctoral Training Grant CA09214. This work was supported by grants from the National Cancer Institute (CA79086), National Institutes of Health (GM 49897), and Fels Foundation.
Abbreviations
MAPK, mitogen-activated protein kinase
MKK, MAPK kinase
MEKK, MAPK kinase kinase
ERK, extracellular signal-regulated kinase
JNK, c-Jun NH2-terminal kinase
JLP, JNK-associated leucine zipper protein
JSAP, JNK/stress-activated protein kinase-associated protein
HA, hemagglutinin
LZI, leucine zipper I
LZII, leucine zipper II
SH, Src homology
References
- 1.Brunet A. & Pouyssegur, J. (1997) Essays Biochem. 32, 1-16. [PubMed] [Google Scholar]
- 2.Cano E. & Mahadevan, L. C. (1995) Trends Biochem. Sci. 20, 117-122. [DOI] [PubMed] [Google Scholar]
- 3.Davis R. J. (1995) Mol. Reprod. Dev. 42, 459-467. [DOI] [PubMed] [Google Scholar]
- 4.Dhanasekaran N. & Reddy, E. P. (1998) Oncogene 17, 1447-1455. [DOI] [PubMed] [Google Scholar]
- 5.English J., Pearson, G., Wilsbacher, J., Swantek, J., Karandikar, M., Xu, S. & Cobb, M. H. (1999) Exp. Cell Res. 253, 255-270. [DOI] [PubMed] [Google Scholar]
- 6.Minden A. & Karin, M. (1997) Biochim. Biophys. Acta 1333, F85-F104. [DOI] [PubMed] [Google Scholar]
- 7.Kyriakis J. M. & Avruch, J. (1996) BioEssays 18, 567-577. [DOI] [PubMed] [Google Scholar]
- 8.Nebreda A. R. & Porras, A. (2000) Trends Biochem. Sci. 25, 257-260. [DOI] [PubMed] [Google Scholar]
- 9.Obata T., Brown, G. E. & Yaffe, M. B. (2000) Crit. Care Med. 28, N67-N77. [DOI] [PubMed] [Google Scholar]
- 10.Raingeaud J., Gupta, S., Rogers, J. S., Dickens, M., Han, J., Ulevitch, R. J. & Davis, R. J. (1995) J. Biol. Chem. 270, 7420-7426. [DOI] [PubMed] [Google Scholar]
- 11.Choi K. Y., Satterberg, B., Lyons, D. M. & Elion, E. A. (1994) Cell 78, 499-512. [DOI] [PubMed] [Google Scholar]
- 12.Marcus S., Polverino, A., Barr, M. & Wigler, M. (1994) Proc. Natl. Acad. Sci. USA 91, 7762-7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Posas F. & Saito, H. (1997) Science 276, 1702-1705. [DOI] [PubMed] [Google Scholar]
- 14.Yablonski D., Marbach, I. & Levitzki, A. (1996) Proc. Natl. Acad. Sci. USA 93, 13864-13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elion E. A. (1998) Science 281, 1625-1626. [DOI] [PubMed] [Google Scholar]
- 16.Kelkar N., Gupta, S., Dickens, M. & Davis, R. J. (2000) Mol. Cell. Biol. 20, 1030-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaeffer H. J., Catling, A. D., Eblen, S. T., Collier, L. S., Krauss, A. & Weber, M. J. (1998) Science 281, 1668-1671. [DOI] [PubMed] [Google Scholar]
- 18.Yasuda J., Whitmarsh, A. J., Cavanagh, J., Sharma, M. & Davis, R. J. (1999) Mol. Cell. Biol. 19, 7245-7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito M., Yoshioka, K., Akechi, M., Yamashita, S., Takamatsu, N., Sugiyama, K., Hibi, M., Nakabeppu, Y., Shiba, T. & Yamamoto, K. I. (1999) Mol. Cell. Biol. 19, 7539-7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito M., Akechi, M., Hirose, R., Ichimura, M., Takamatsu, N., Xu, P., Nakabeppu, Y., Tadayoshi, S., Yamamoto, K. & Yoshioka, K. (2000) Gene 255, 229-234. [DOI] [PubMed] [Google Scholar]
- 21.Valtieri M., Tweardy, D. J., Caracciolo, D., Johnson, K., Mavilio, F., Altmann, S., Santoli, D. & Rovera, G. (1987) J. Immunol. 138, 3829-3835. [PubMed] [Google Scholar]
- 22.Blanar M. A. & Rutter, W. J. (1992) Science 256, 1014-1018. [DOI] [PubMed] [Google Scholar]
- 23.Ho S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. (1989) Gene 77, 51-59. [DOI] [PubMed] [Google Scholar]
- 24.Reddy C. D., Dasgupta, P., Saikumar, P., Dudek, H., Rauscher, F. D. & Reddy, E. P. (1992) Oncogene 7, 2085-2092. [PubMed] [Google Scholar]
- 25.Oh I. H. & Reddy, E. P. (1998) Mol. Cell. Biol. 18, 499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noguchi K., Kitanaka, C., Yamana, H., Kokubu, A., Mochizuki, T. & Kuchino, Y. (1999) J. Biol. Chem. 274, 32580-32587. [DOI] [PubMed] [Google Scholar]
- 27.Whitmarsh A. J., Kuan, C. Y., Kennedy, N. J., Kelkar, N., Haydar, T. F., Mordes, J. P., Appel, M., Rossini, A. A., Jones, S. N., Flavell, R. A., et al. (2001) Genes Dev. 15, 2421-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blackwood E. M. & Eisenman, R. N. (1991) Science 251, 1211-1217. [DOI] [PubMed] [Google Scholar]
- 29.Blackwood E. M., Luscher, B. & Eisenman, R. N. (1992) Genes Dev. 6, 71-80. [DOI] [PubMed] [Google Scholar]
- 30.Ayer D. E., Kretzner, L. & Eisenman, R. N. (1993) Cell 72, 211-222. [DOI] [PubMed] [Google Scholar]
- 31.Bowman A. B., Kamal, A., Ritchings, B. W., Philp, A. V., McGrail, M., Gindhart, J. G. & Goldstein, L. S. (2000) Cell 103, 583-594. [DOI] [PubMed] [Google Scholar]
- 32.Verhey K. J., Meyer, D., Deehan, R., Blenis, J., Schnapp, B. J., Rapoport, T. A. & Margolis, B. (2001) J. Cell Biol. 152, 959-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.