Abstract
Although nuclear factor (NF)-κB plays a central role in mediating cytokine-stimulated human inducible nitric-oxide synthase (hiNOS) gene transcription, very little is known about the factors involved in silencing of the hiNOS promoter. NF-κB-repressing factor (NRF) interacts with a specific negative regulatory element (NRE) to mediate transcriptional repression of certain NF-κB responsive genes. By sequence comparison with the IFN-β and IL-8 promoters, we identified an NRE in the hiNOS promoter located at −6.7 kb upstream. In A549 and HeLa human cells, constitutive NRF mRNA expression is detected by RT-PCR. Gel shift assay showed constitutive NRF binding to the hiNOS NRE. Mutation of the −6.7-kb NRE site in the hiNOS promoter resulted in loss of NRF binding and increased basal but not cytokine-stimulated hiNOS transcription in promoter transfection experiments. Interestingly, overexpression of NRF suppressed both basal and cytokine-induced hiNOS promoter activity that depended on an intact cis-acting NRE motif. By using stably transformed HeLa cells with the tetracycline on/off expression system, reduction of cellular NRF by expressing antisense NRF increased basal iNOS promoter activity and resulted in constitutive iNOS mRNA expression. These data demonstrate that the transacting NRF protein is involved in constitutive silencing of the hiNOS gene by binding to a cis-acting NRE upstream in the hiNOS promoter.
The inducible isoform of nitric-oxide synthase (iNOS) catalyzes the production of nitric oxide (NO), which participates in the physiologic and pathophysiologic conditions of every organ system (1–7). The NO generated by iNOS from its substrate l-arginine has beneficial effects (e.g., antimicrobial, antiatherogenic, antiapoptotic; refs. 8–10), whereas aberrant iNOS expression and excessive NO production are observed in a large variety of diseases (e.g., septic shock, multiple sclerosis, rheumatoid arthritis, ulcerative colitis, and certain cancers; refs. 6 and 11–13).
Although, iNOS activity can be regulated by posttranscriptional mechanisms (14–17), the rodent and human iNOS (hiNOS) genes are regulated predominantly at the transcriptional level (18–21). Our laboratory, as well as others, has shown that nuclear factor (NF)-κB and signal transducer and activator of transcription (Stat)-1 transcription factors play a key role in mediating the induction of the rodent and human iNOS genes by lipopolysaccharide (LPS) and/or tumor necrosis factor α (TNF-α), IL-1β, and IFN-γ (22–27). DNA sequence analysis of the hiNOS promoter from −3.8 to −7.2 kb identified four putative NF-κB cis-regulatory transcription factor-binding sites upstream of −4.7 kb. Site-directed mutagenesis of these sites revealed that NF-κB motif at −5.8 kb was required for cytokine-induced hiNOS promoter activity (25, 27).
Although NF-κB plays a central role in mediating cytokine-stimulated human iNOS gene transcription (28), very little is known about the factors involved in silencing of the hiNOS promoter, especially at its basal level. Previously, we reported that overexpression of an IκBα superrepressor inhibited NF-κB DNA-binding activity and suppressed iNOS expression (29). The tumor suppressor gene product p53 also inhibited hiNOS promoter activity (30). Genes that promote malignant transformation, such as Ras and RhoA, were recently reported to inhibit iNOS transcription, whereas genes with tumor suppressor activity, such as RhoB, enhanced iNOS expression (31). Thus, repressive regulation of hiNOS transcription is complex and warrants further elucidation.
NF-κB-repressing factor (NRF) was identified as a constitutively expressed silencer protein that binds to the negative regulatory element (NRE) in the IFN-β promoter and represses the basal transcription of this gene (32–34). Reduction of NRF protein level through expression of NRF antisense RNA resulted in basal activation of IFN-β gene transcription (33). In vitro, NF-κB proteins bind to purified NRF by a direct protein–protein interaction, and NRF can inhibit NF-κB-enhancing activity. Recently, NRF was found to have a dual role in IL-8 transcription. In the absence of stimulation, NRF is involved in transcriptional silencing, but, in the presence of IL-1β, it is required for full induction of the IL-8 promoter (35).
Considering the critical importance of NF-κB in regulating hiNOS transcription, we hypothesized that NRF might have a modulatory role for transcriptional repression of hiNOS gene expression. By comparing the DNA sequence of hiNOS, IL-8, and IFN-β promoters, we identified a potential NRE binding site for NRF located at −6.7 kb upstream in the hiNOS promoter. Using gel shift assays and promoter transfection experiments, we showed constitutive silencing of the hiNOS gene by NRF binding to the cis-acting NRE.
Methods
Cell Lines and Reagents.
The A549 human lung epithelial cell line was obtained from American Type Culture Collection (ATCC) and cultured in F-12K medium (GIBCO/BRL) supplemented with 10% heat inactivated, low endotoxin FBS (GIBCO/BRL), 100 units/ml penicillin, 100 μg/ml streptomycin, and 15 mM Hepes (pH 7.4). The human cervix epithelial cell line HeLa was also obtained from ATCC and cultured in Dulbecco's modified Eagle's medium complemented with 10% FCS, low endotoxin FBS (GIBCO/BRL), 100 units/ml penicillin, 100 μg/ml streptomycin, and 15 mM Hepes (pH 7.4). Unless indicated, cells were stimulated with a cytokine mixture consisting of 1,000 units/ml human TNF-α (R & D Systems), 100 units/ml IL-1β (provided by C. Reynolds, National Cancer Institute), and human 250 units/ml IFN-γ (R & D Systems or Roche Pharmaceuticals), which were purified-recombinant proteins
Plasmid Constructs.
The hiNOS promoter reporter plasmid piNOS(7.2)Luc contains −7.2 kb of upstream 5′-flanking DNA linked to the luciferase reporter gene and has been described (24, 25). Mutation of the −6.7-kb NRE element was generated from the piNOS(7.2)Luc reporter plasmid by using the QuickChange mutagenesis kit according to manufacturer recommendations (Stratagene). Mutations were confirmed by DNA sequence analysis by the University of Pittsburgh DNA Sequencing Facility and are shown in Table 1. Expression plasmids encoding the human NRF containing the NRE DNA-binding domain (DBD) and the NF-κB-binding domain (pMBC-NRF or VP16NRF) have been reported (33, 35).
Table 1.
Sequence comparison of the NRE sites in IFN-β, IL-8, and hiNOS promoters
| Construct | NRE sequence |
|---|---|
| Gene | |
| IFN-β promoter | (−60) AATTCCTCTGA (−50) |
| IL-8 promoter | (−1415) AATTCCTCTGA (−1405) |
| hiNOS promoter | (−6749) AATTCCTCAGC (−6739) |
| Oligonucleotide | |
| Core hiNOS NRE (WT −6.7) | aattcgAATTCCTCAGCcgaaca |
| Core hiNOS NRE (Mut. −6.7) | aattcgAATTCCCCCGCcgaaca |
Underlined nucleotides are the point mutants.
Transient Transfections and Activity Assays.
DNA transfections of cells were carried out in six-well plates (Corning) by using Lipofectamine (GIBCO) as described (24, 25). Briefly, cells were exposed to serum-free medium containing 1 μg of DNA and 20 μg of liposomes for 4 h, washed, and replenished with medium supplemented with 5% calf serum. Preliminary transfection experiments showed optimal transfection efficiency and low toxicity with a DNA:liposome ratio of 1:20. To control for transfection efficiency between groups, 0.25 μg of a plasmid containing a cytomegalovirus promoter-driven β-galactosidase gene (pIEP-Lacz) was added to each well. As a positive control, cells were transfected with PRSV-Luc while transfection of the promoterless plasmid pXP2 served as negative control. Cells were lysed with Reporter lysis buffer (Promega) or buffer containing 1% Triton X-100, 5 mM DTT, 50% glycerol, 10 mM EDTA, and 125 mM Tris phosphate (pH 7.8). Luciferase activity was assayed with 40 μl of lysate in a Berthold (Nashua, NH) AutoLumat LB 953 luminometer by using a commercially available kit (Promega). β-Galactosidase activity was determined as recommended (Promega) by using a 96-well multiplate reader with softmax software (Molecular Devices). Luciferase activity was normalized to β-galactosidase activity or protein. Cotransfection experiments with NRF expression vectors included an additional 1.0 μg of the indicated expression plasmid.
Preparation of Nuclear Protein.
Briefly, the cytokine-stimulated cells were washed and scraped into phosphate-buffered solution, and centrifuged at 4,500 rpm for 5 min in a Microfuge (Beckman). The pelleted cells were suspended in buffer A (10 mM Tris, pH 7.5/1.5 mM MgCl2/10 mM KCl/0.5% Nonidet P-40) at approximately 10 times the packed cell volume and lysed by gentle pipetting. Nuclei were recovered by microcentrifugation at 7,000 rpm for 5 min. The nuclear proteins were extracted at 4°C by gentle resuspension of the nuclei (at approximately 2 times the packed nuclear volume) of buffer containing the following: 20 mM Tris (pH 7.5), 10% glycerol, 1.5 mM MgC12, 420 mM NaCl, and 0.2 mM EDTA, followed by 30 min of platform rotation. The nuclear protein suspension was cleared by microcentrifugation at 13,000 rpm for 15 min. The supernatants were collected and were frozen at −80°C or directly used in gel shift assays. All buffers contained the following additions: 1–2 μg/ml each of aprotinin, chymostatin, leupeptin, and pepstatin; 0.2 mM PMSF; 0.5 mM DTT; and 0.1 mM sodium-vanadate. All steps were carried out on ice or at 4°C. Protein concentrations were measured by using the Bio-Rad protein assay using BSA as a standard.
Electrophoretic Mobility-Shift Assays.
All oligonucleotides were ordered from Life Technologies (Grand Island, NY); their sequences are listed in Table 1. DNA probes were prepared by end-labeling with [γ-32P]dATP (DuPont/NEN) and T4 polynucleotide kinase (Boehringer Mannheim) and purified in TEN (Tris-EDTA-sodium chloride) by using G-50 resin columns (Whatman). Typically, 5 μl (5–10 μg) of nuclear proteins were incubated with ≈100,000 cpm of 32P-labeled oligonucleotides (≈0.5 ng) for 30 min at room temperature. The nuclear proteins and various oligonucleotide probes were incubated in a buffer containing 10 mM Tris (pH 7.5), 10% glycerol, and 0.2% Nonidet P-40. Additionally, 2–4 μg of poly(dI-dC) (Boehringer Mannheim) was included as a nonspecific competitor DNA. Protein–DNA complexes were resolved on 4% nondenaturing polyacrylamide gels in 0.4× TBE running buffer (450 mM Tris/borate/1 μM EDTA, pH 8.0). After electrophoresis, gels were dried and subjected to autoradiography. Antibody supershift experiments included the addition of 2 μl of various antibodies (34).
RT-PCR Analysis.
First-strand cDNA synthesis was performed with 5 μg of total RNA isolated from the aorta by using Superscript II (GIBCO/BRL) according to the manufacturer's instructions. The resulting cDNA was amplified by RT-PCR using the following: human beta-actin primers (sense, 5′-ATGGA TGATG ATATC GCCGC GCT-3′, and antisense, 5′-GACTC CATGC CCAGG AAGG A-3′); human NRF primers (sense, 5′-CCAAA TTCCA TGCGA GACCT CG-3′, and antisense, 5′-TATTT TTGGG GATGT CGGCA GG-3′); and human iNOS primers (sense, 5′-ACAAG CTGGC CTCGC TCTGG AAAGA-3′, and antisense, 5′-TCCAT GCAGA CAACC TTGGG GTTGA AG-3′). The protocol consisted of 25–30 cycles of incubation at 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, followed by extension for 10 min at 72°C. The amplified actin product (812 bp), NRF product (561 bp), and iNOS product (507 bp) were analyzed by 2% agarose gel electrophoresis and visualized by ethidium bromide staining under UV light.
Statistical Methods.
Data are presented as the mean ± SEM. Cultures were performed in duplicate or triplicate, and experiments were performed a minimum of three times. Data were analyzed by the Student t test or analysis of variance where appropriate, and P < 0.05 was considered statistically significant.
Results
An NRE cis-Element Is Identified in the hiNOS Promoter.
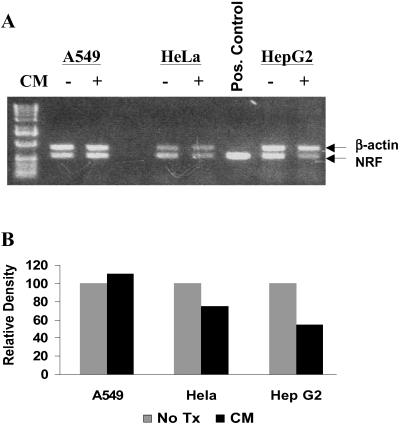
Sequence comparison of the hiNOS gene 5′-flanking region with the NRE binding motifs in the IFN-β and IL-8 promoters produced three potential NRE sites located between −8.0 kb and −6.5 kb upstream in the hiNOS promoter. The most promising match identified was an 11-nt NRE motif at −6.7 kb (Table 1). To determine a putative role for NRF in regulating hiNOS transcription, we initially examined for NRF mRNA expression in several human cell lines. Constitutive expression of NRF mRNA was seen in A549, HeLa, and HepG2 human cell lines by RT-PCR (Fig. 1A), consistent with a previous report of constitutive NRF protein expression (33). The addition of a cytokine mixture (CM) of TNF-α + IL-1β + IFN-γ known to induce iNOS expression did not significantly change the NRF mRNA levels, although there was a slight down-regulation of NRF mRNA expression in the HeLa and HepG2 cells (Fig. 1B).
Fig 1.
Endogenous NRF mRNA level in different human cell lines. (A) PCR gel for NRF and β-actin mRNA. Total RNA was extracted after treatment of cell with/without CM of TNF-α (1,000 units/ml), IL-1 (100 units/ml), and IFN-β (250 units/ml) for 8 h. RT-PCR was used to detect the mRNA level. NRF and β-actin bands are indicated by arrows. (B) Relative density was quantified by nih image software.
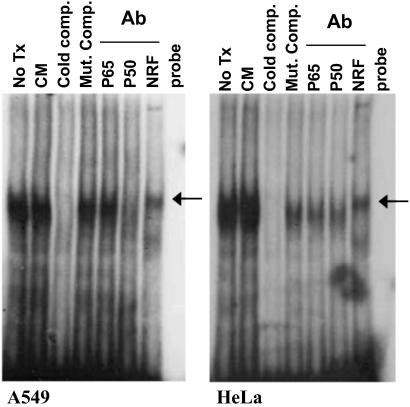
Constitutive Binding of NRF to the NRE DNA Element in the hiNOS Promoter.
The presence of an NRE-like DNA element does not indicate functional protein binding or transcriptional regulation. Therefore, we initially performed gel shift assays with the hiNOS-specific NRE located at −6.7 kb in the hiNOS promoter. Nuclear extracts of unstimulated A549 or HeLa cells showed strong protein binding to the −6.7-kb hiNOS NRE oligonucleotide (Fig. 2). This constitutive protein–DNA complex was not significantly altered by addition of cytokines (CM). Cold competition with excess unlabeled oligonucleotide, along with restoration of the protein–DNA complex by competition with a cold mutant NRE oligonucleotide, indicates specificity of protein binding to NRE. Supershift of the complex with anti-NRF antibody confirms identity of NRF in the protein–DNA complex. Antibody against p65 or p50 components of NF-κB failed to elicit a supershift.
Fig 2.
Constitutive binding of NRF to the NRE DNA element in the hiNOS promoter. Nuclear extracts from A549 cells and HeLa cells stimulated for 60 min with CM or left untreated were incubated with radiolabeled hiNOS promoter putative NRE oligonucleotide. Antibodies against p65 NF-κB (p65), p50 NF-κB (p50), or NRF (NRF) were added to the binding reactions where indicated. Competitions with cold unlabeled WT (Cold comp.) or mutant (Mut. Comp.) hiNOS NRE oligonucleotide were also indicated.
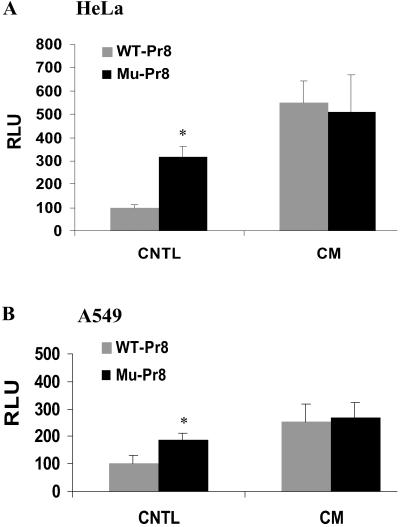
Basal Repression of hiNOS Promoter Activity Requires a Functional NRE at −6.7 kb Upstream.
To document a role for NRF-NRE interaction in regulating basal or CM-induced hiNOS transcription, we used a luciferase reporter gene under the control of a −7.2-kb fragment of hiNOS promoter. Parallel experiments were carried out with the wild-type (WT) hiNOS promoter containing an intact NRE, and a mutant −6.7 NRE contained within the −7.2-kb hiNOS plasmid. The −6.7-kb NRE mutant was generated by site-directed mutagenesis inserting a two-point mutation that was previously found to inactivate the NRE (33). After transient transfection, the WT −7.2-kb hiNOS promoter displayed low basal transcriptional activity in the HeLa or A549 cells (Fig. 3). As expected, cytokine stimulation induced a 3- to 4-fold increase in promoter activity, consistent with the fold-induction previously reported (24–27). Mutation of the −6.7 kb NRE resulted in significant activation of basal hiNOS promoter activity. This finding is indicative of basal repression of hiNOS gene transcription by NRF-NRE binding, and is consistent with the role of NRE in constitutive silencing of the IFN-β and IL-8 genes (33, 35). Noteworthy is that the increased basal promoter activity observed with the mutant NRE is nearly comparable to the level of CM inducibility. Interestingly, mutation of the NRE did not affect CM-induced hiNOS promoter activity, indicating that an intact NRE is not essential for cytokine activation of hiNOS transcription.
Fig 3.
Mutation analysis of NRE-site in hiNOS promoter. HeLa cells (A) and A549 cells (B) were respectively transfected with 1 μg of luciferase reporter plasmids containing the WT −7.2-kb hiNOS promoter (gray bars) or mutant −6.7-kb NRE (black bars). Luciferase activity was determined 6 h after stimulation with CM containing IL-1β, IFN-γ, and TNF-α. Values are expressed relative to the level of luciferase in control (untreated WT plasmid-transfected cells) and are the means of three independent transfection experiments. *, P < 0.05 vs. control.
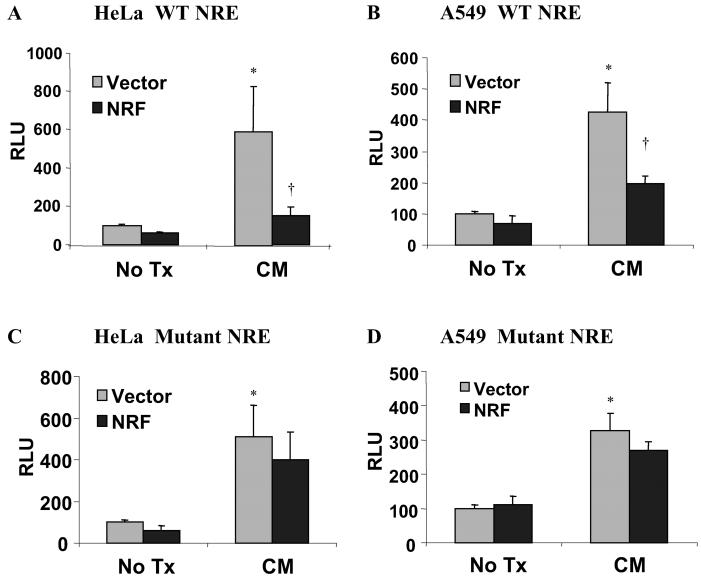
Overexpression of NRF Inhibits Basal and Cytokine-Induced hiNOS Promoter Activity.
To further implicate a role for NRF in repressing hiNOS transcription, we transiently cotransfected A549 and HeLa cells with −7.2-kb hiNOS promoter construct and the VP16NRF expression plasmid previously shown to be functional (33, 35). Overexpression of NRF suppressed both basal and CM induction of hiNOS promoter activity when an intact NRE was present (Fig. 4 A and B). Cotransfection with a control vector for NRF yielded reporter activity comparable to hiNOS promoter activity alone. The promoter activity of the unstimulated, vector-cotransfected cells was normalized to 100% relative light units (RLU) for both the WT and mutant NRE constructs, to allow for comparison to the NRF cotransfected cells. Similar results were seen when the pMBC-NRF expression vector (lacking the VP16 fusion protein) was used (data not shown). The repressive effect of NRF was lost when the −6.7-kb NRE was mutated (Fig. 4 C and D), supporting repression mediated by an intact NRF-NRE interaction.
Fig 4.
Overexpression of NRF suppresses both basal and CM-induced hiNOS transcription. HeLa cells (A) and A549 cells (B) were cotransfected with 1 μg of empty expression vector (gray bars) or cDNA encoding WT NRF (black bars) together with 0.5 μg of WT −7.2 hiNOS promoter construct and 0.1 μg of pSV-β-galactosidase. Luciferase activity was determined 6 h after stimulation with CM containing IL-1β, IFN-γ, and TNF-α. HeLa cells (C) and A549 cells (D) were cotransfected with 1 μg of empty expression vector (gray bars) or cDNA encoding WT NRF (black bars) together with 0.5 μg of NRE mutant −7.2 hiNOS promoter construct and 0.1 μg of pSV-β-galactosidase. Twenty-four hours later, cells were stimulated with CM containing IL-1β, IFN-γ, and TNF-α or left untreated. Six hours later, cells were lysed, and luciferase activity was determined. Values are expressed relative to the level of luciferase in control (untreated WT plasmid-transfected cells) and are the means of three independent transfection experiments. *, P < 0.05 vs. control, untreated empty vector cotransfected group; †, P < 0.05 vs. CM-treated empty vector cotransfected group.
NRF Is Required for Constitutive Repression of Endogenous hiNOS Gene Transcription.
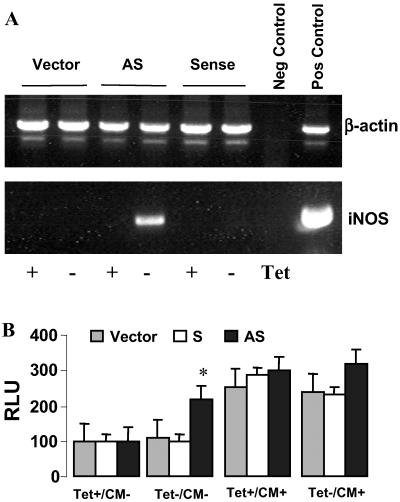
The experiments described above indicate an inhibitory role for NRF in regulating hiNOS transcription, but are performed in the somewhat artificial environment of transient promoter transfection experiments. To confirm the suppressive role of NRF on endogenous hiNOS transcription and subsequent hiNOS mRNA expression, we used stably transformed HeLa cells containing a plasmid expressing antisense NRF under the control of a tetracycline-sensitive promoter. The induction of antisense NRF mRNA with tetracycline withdrawal resulted in efficient reduction of nuclear NRF as previously determined by Western blot analysis of immunoprecipitated endogenous NRF (35). The expression of endogenous hiNOS mRNA was monitored by RT-PCR. No hiNOS mRNA was detected in the presence of tetracycline (Fig. 5A). Withdrawal of tetracycline in the cells stably transformed with anti-sense NRF led to a significant expression of endogenous hiNOS mRNA. In contrast, cells expressing full-length NRF sense mRNA, or cells transfected with empty vector, showed no endogenous hiNOS mRNA on withdrawal of tetracycline (Fig. 5A). We did not observe a significant increase in nitrite accumulation with antisense NRF activation, possibly due to the limits of detection in these cells (data not shown).
Fig 5.
NRF is required for basal silence of hiNOS promoter. HeLa cells were stably transfected with tetracycline-repressible plasmids pTBC containing the coding region of the NRF cDNA (NRF) or a 300-bp fragment of the NRF cDNA in antisense orientation or empty pTBC as described in ref. 35. Tetracycline (Tet) was removed from the culture medium for 48 h to induce NRF sense or antisense mRNA. (A) RT-PCR was used to detect the hiNOS mRNA level in above stably transfected cells with or without Tet withdrawal. (B) Above cell lines were respectively transfected with 1 μg of reporter plasmids containing the WT hiNOS promoter. Luciferase activity was determined 6 h after stimulation with CM containing IL-1β, IFN-γ, and TNF-α. *, P < 0.05 vs. vector or sense transfected cells.
Similar findings were observed when we examined the effect of antisense NRF on basal hiNOS promoter activity. Withdrawal of tetracycline in the HeLa cells stably transformed with antisense NRF (but not sense NRF or empty vector) resulted in more than doubling of basal hiNOS promoter activity in the absence of cytokines (Fig. 5B). Addition of cytokines produced a 3-fold increase in inducible promoter activity, but was not diminished by activation of antisense NRF with tetracycline removal, consistent with the lack of effect of mutant NRE on CM-induced hiNOS promoter activity (Fig. 3).
Discussion
Previously, we reported that NF-κB has a critical role in mediating cytokine-stimulated hiNOS gene transcription (25), and several functional cis-acting NF-κB response elements at −5 to −8 kb in the human iNOS promoter have been identified (25–27). In addition, inducible signal transducer and activator of transcription (Stat)-1 (27) and activator protein (AP)-1 (26) binding sites have also been reported. However, no information exists as to the transcriptional mechanisms that govern repression of hiNOS gene expression. This finding is particularly important given that hiNOS mRNA is usually absent in resting human cells, supporting a role for suppressive transcription factors. The major and novel findings of these experiments are the following: (i) identification of a specific cis-acting NRE DNA element at −6.7 kb in the hiNOS promoter that binds NRF and is involved in constitutive silencing of hiNOS transcription; (ii) determination that overexpressed NRF depressed both basal and cytokine-induced hiNOS transcription, which depends on an intact NRE site; and (iii) constitutive expression of hiNOS mRNA after cellular reduction of NRF by using antisense NRF mRNA.
NRE-related sequences have been identified in several promoters, including IFN-β, HIV-1, human T-lymphotrophic virus (HTLV)-1, IL-2Rα, and IL-8 (32, 36). The NRE DNA motifs constitute functionally active silencer elements that bind NRF and repress the enhancing activity of NF-κB/rel-binding sites within these promoters (33, 35, 36). Each of these promoters has one or two proximally located functional NRE binding sites within 1.5 kb upstream of the TATA box. The hiNOS promoter NRE at −6.7 kb is the first description of a far upstream NRE functioning as a transcriptional repressor motif. Supershift experiments in electrophoretic mobility-shift assays demonstrate that NRF binds to the promoter (Fig. 2). Using −7.2-kb hiNOS promoter construct, we studied the role of the potential NRE site located at −6.7 upstream of hiNOS promoter. A mutant NRE sequence leads to increased basal transcription from the hiNOS promoter (Fig. 3). This observation is consistent with a recent report where transfection of a nucleotide −6796 hiNOS promoter construct produced a 7.7-fold increase in luciferase activity after cytokine stimulation, whereas transfection of a nucleotide −6534 hiNOS promoter contract produced a 12.0-fold increase, suggesting the presence of a negative regulatory element from −6.8 to −6.5 kb in the hiNOS promoter (37). The hiNOS NRE site that we identified at −6.7 kb is located in this region.
The results presented in Fig. 1 show that NRF mRNAs are detected in all tested human cell lines. This finding indicates that NRF is abundant and available to participate in transcriptional regulation of target genes. Lowering the cellular amount of NRF by antisense mRNA expression resulted in detectable levels of iNOS mRNA, which was absent in unstimulated cells (Fig. 5). This derepression indicates the in vivo participation of NRF in the constitutive repression of hiNOS and highlights its homologous function in the regulation of basal promoter activity for the IL-8, IFN-β, and hiNOS genes (33, 35).
CM treatment of HeLa or A549 cells transfected with the WT hiNOS promoter resulted in more than 3- to 5-fold induction of promoter activity. The NRE mutagenesis studies (Fig. 3) showed that NRF functions to inhibit basal transcription, but was not required for cytokine-stimulated hiNOS promoter activity. Interestingly, overexpression of NRF decreased both basal and CM-induced hiNOS promoter activation dependent on the intact NRE site at −6.7 kb (Fig. 4). This result contrasts with the role of NRF in IL-8 gene expression where NRF repressed basal IL-8 promoter activity, but was required for strong activation of the IL-8 gene by IL-1 (35), suggesting that NRF exerts differential effects depending on cell type, environmental stimulation, and target gene cis-acting promoter elements. Interestingly, IFN-γ/IL-4 leads to maintenance of iNOS expression in primary human airway epithelium through production of soluble mediators (38); however, it is unknown whether NRF expression is altered in these cells.
It has been shown that NRF functions in promoter regulation through both DNA–protein interactions (NRF binding to NRE), as well as protein–protein interactions. NRF interacts directly with NF-κB proteins by an active repression mechanism (33, 35). There are multiple functional NF-κB elements in the hiNOS promoter (25–27). The NF-κB elements from −5.5 to −6.1 kb in the human iNOS promoter are spaced in approximate multiples of nucleosome units (200 bp), and this spacing may contribute to the 3D structure necessary for efficient hiNOS transcription. NRF contains a separable and position-independent repression domain (33). This repressing domain might contribute to its function in hiNOS via a suitable DNA-binding domain located at −6.7 and influence the 3D structure. Actually, we did observe that cotransfection of NRF expression plasmid decreased NF-κB reporter construct activation (data not shown). Unlike IFN-β and IL-8, the NRE site in hiNOS does not overlap the NF-κB sites in hiNOS promoter. Instead, there is a distance of ≈1 kb between the −6.7-kb NRE site and important NF-κB binding sites downstream at −5.8 kb and upstream at −8 kb in the hiNOS promoter (25–27). It is possible that the 3D DNA folding approximates NRF, with the NF-κB proteins allowing for direct protein–protein interaction. This result would be consistent with the ability of NRF to elicit an inhibitory effect on NF-κB elements at distances up to 2.5 kb from NRE (34).
Another noteworthy finding in this study is that reduction by antisense NRF mRNA derepressed hiNOS expression (Fig. 5), further confirming that the constitutive transcriptional silencing of hiNOS is mediated by the nuclear content of NRF in unstimulated cells. Because hiNOS expression has such profound physiologic effects, its basal regulation is strictly controlled. The expression of hiNOS and subsequent production of NO serves a protective role by increasing perfusion to the viscera and sites of inflammation. However, sustained overproduction can have detrimental effects, including refractory hypotension, cellular injury, and apoptosis (39). Thus, understanding the mechanism involved in basal repression of hiNOS gene might lead to new strategy for treatment of diseases.
Acknowledgments
This work was supported by National Institutes of Health Grant R01-GM52021 (to D.A.G.), and by the George H. A. Clowes, Jr., MD, FACS, Memorial Research Career Development Award of the American College of Surgeons (to D.A.G.).
Abbreviations
NO, nitric oxide
iNOS, inducible NO synthase
hiNOS, human iNOS
NF-κB, nuclear factor-κΒ
TNF-α, tumor necrosis factor α
NRF, NF-κB repressing factor
NRE, negative regulatory element
CM, cytokine mixture
RLU, relative light unit
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ignarro L. J., Buga, G. M., Wood, K. S., Byrns, R. E. & Chaudhuri, G. (1987) Proc. Natl. Acad. Sci. USA 84, 9265-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griscavage J. M., Rogers, N. E., Sherman, M. P. & Ignarro, L. J. (1993) J. Immunol. 151, 6329-6337. [PubMed] [Google Scholar]
- 3.Stuehr D. J. & Marletta, M. A. (1985) Proc. Natl. Acad. Sci. USA 82, 7738-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ignarro L. J., Napoli, C. & Loscalzo, J. (2002) Circ. Res. 90, 21-28. [DOI] [PubMed] [Google Scholar]
- 5.Guo F. H., De Raeve, H. R., Rice, T. W., Stuehr, D. J., Thunnissen, F. B. & Erzurum, S. C. (1995) Proc. Natl. Acad. Sci. USA 92, 7809-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moncada S. (1999) J. R. Soc. Med. 92, 164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murad F. (1997) Circulation 95, 1101-1103. [DOI] [PubMed] [Google Scholar]
- 8.Bogdan C. (2001) Nat. Immunol. 2, 907-916. [DOI] [PubMed] [Google Scholar]
- 9.Bogdan C., Rollinghoff, M. & Diefenbach, A. (2000) Immunol. Rev. 173, 17-26. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y. M., Bombeck, C. A. & Billiar, T. R. (1999) Circ. Res. 84, 253-256. [DOI] [PubMed] [Google Scholar]
- 11.Weinberg J. B., Granger, D. L., Pisetsky, D. S., Seldin, M. F., Misukonis, M. A., Mason, S. N., Pippen, A. M., Ruiz, P., Wood, E. R. & Gilkeson, G. S. (1994) J. Exp. Med. 179, 651-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroncke K. D., Fehsel, K. & Kolb-Bachofen, V. (1998) Clin. Exp. Immunol. 113, 147-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bogdan C. (2001) Trends Cell Biol. 11, 66-75. [DOI] [PubMed] [Google Scholar]
- 14.Tzeng E., Billiar, T. R., Robbins, P. D., Loftus, M. & Stuehr, D. J. (1995) Proc. Natl. Acad. Sci. USA 92, 11771-11775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y., Panda, K. & Stuehr, D. J. (2002) Biochemistry 41, 4618-4625. [DOI] [PubMed] [Google Scholar]
- 16.Eissa N. T., Yuan, J. W., Haggerty, C. M., Choo, E. K., Palmer, C. D. & Moss, J. (1998) Proc. Natl. Acad. Sci. USA 95, 7625-7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Pascual F., Hausding, M., Ihrig-Biedert, I., Furneaux, H., Levy, A. P., Forstermann, U. & Kleinert, H. (2000) J. Biol. Chem. 275, 26040-26049. [DOI] [PubMed] [Google Scholar]
- 18.Lowenstein C. J., Alley, E. W., Raval, P., Snowman, A. M., Snyder, S. H., Russell, S. W. & Murphy, W. J. (1993) Proc. Natl. Acad. Sci. USA 90, 9730-9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Q. W., Whisnant, R. & Nathan, C. (1993) J. Exp. Med. 177, 1779-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geller D. A., Nussler, A. K., Di Silvio, M., Lowenstein, C. J., Shapiro, R. A., Wang, S. C., Simmons, R. L. & Billiar, T. R. (1993) Proc. Natl. Acad. Sci. USA 90, 522-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geller D. A., Lowenstein, C. J., Shapiro, R. A., Nussler, A. K., Di Silvio, M., Wang, S. C., Nakayama, D. K., Simmons, R. L., Snyder, S. H. & Billiar, T. R. (1993) Proc. Natl. Acad. Sci. USA 90, 3491-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie Q. W., Kashiwabara, Y. & Nathan, C. (1994) J. Biol. Chem. 269, 4705-4708. [PubMed] [Google Scholar]
- 23.Gao J., Morrison, D. C., Parmely, T. J., Russell, S. W. & Murphy, W. J. (1997) J. Biol. Chem. 272, 1226-1230. [DOI] [PubMed] [Google Scholar]
- 24.de Vera M. E., Shapiro, R. A., Nussler, A. K., Mudgett, J. S., Simmons, R. L., Morris, S. M., Jr., Billiar, T. R. & Geller, D. A. (1996) Proc. Natl. Acad. Sci. USA 93, 1054-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor B. S., de Vera, M. E., Ganster, R. W., Wang, Q., Shapiro, R. A., Morris, S. M., Jr., Billiar, T. R. & Geller, D. A. (1998) J. Biol. Chem. 273, 15148-15156. [DOI] [PubMed] [Google Scholar]
- 26.Marks-Konczalik J., Chu, S. C. & Moss, J. (1998) J. Biol. Chem. 273, 22201-22208. [DOI] [PubMed] [Google Scholar]
- 27.Ganster R. W., Taylor, B. S., Shao, L. & Geller, D. A. (2001) Proc. Natl. Acad. Sci. USA 98, 8638-8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor B. S. & Geller, D. A. (2000) Shock 13, 413-424. [DOI] [PubMed] [Google Scholar]
- 29.Taylor B. S., Shao, L., Gambotto, A., Ganster, R. W. & Geller, D. A. (1999) Surgery 126, 142-147. [PubMed] [Google Scholar]
- 30.Forrester K., Ambs, S., Lupold, S. E., Kapust, R. B., Spillare, E. A., Weinberg, W. C., Felley-Bosco, E., Wang, X. W., Geller, D. A., Tzeng, E., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 2442-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delarue F. L., Taylor, B. S. & Sebti, S. M. (2001) Oncogene 20, 6531-6537. [DOI] [PubMed] [Google Scholar]
- 32.Nourbakhsh M. & Hauser, H. (1997) Immunobiology 198, 65-72. [DOI] [PubMed] [Google Scholar]
- 33.Nourbakhsh M. & Hauser, H. (1999) EMBO J. 18, 6415-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nourbakhsh M., Hoffmann, K. & Hauser, H. (1993) EMBO J. 12, 451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nourbakhsh M., Kalble, S., Dorrie, A., Hauser, H., Resch, K. & Kracht, M. (2001) J. Biol. Chem. 276, 4501-4508. [DOI] [PubMed] [Google Scholar]
- 36.Nourbakhsh M., Oumard, A., Schwarzer, M. & Hauser, H. (2000) Eur. Cytokine Network 11, 500-501. [PubMed] [Google Scholar]
- 37.Kristof A. S., Marks-Konczalik, J. & Moss, J. (2001) J. Biol. Chem. 276, 8445-8452. [DOI] [PubMed] [Google Scholar]
- 38.Guo F. H., Uetani, K., Haque, S. J., Williams, B. R., Dweik, R. A., Thunnissen, F. B., Calhoun, W. & Erzurum, S. C. (1997) J. Clin. Invest. 100, 829-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geller D. A. & Billiar, T. R. (1998) Cancer Metastasis Rev. 17, 7-23. [DOI] [PubMed] [Google Scholar]