Abstract
Posttranslational modification of histones through acetylation, methylation, and phosphorylation is a common mode of regulating chromatin structure and, therefore, diverse nuclear processes. One such modification, methylated histone H3 at lysine-4 (H3-meK4), colocalizes with hyperacetylated histones H3 and H4 in mammalian chromatin. Whereas activators directly recruit acetyltransferases, the process whereby H3-meK4 is established is unknown. We tested whether the hematopoietic-specific activators NF-E2 and GATA-1, which mediate transactivation of the β-globin genes, induce both histone acetylation and H3-meK4. Through the use of NF-E2- and GATA-1-null cell lines, we show that both activators induce H3 acetylation at the promoter upon transcriptional activation. However, analysis of H3-mek4 revealed that NF-E2 and GATA-1 differentially regulate chromatin modifications at the βmajor promoter. NF-E2, but not GATA-1, induces H3-meK4 at the promoter. Thus, under conditions in which NF-E2 and GATA-1 activate the transcription of an endogenous gene at least 570-fold, these activators differ in their capacity to induce H3-meK4. Despite strong H3-meK4 at hypersensitive site 2 of the upstream locus control region, neither factor was required to establish H3-meK4 at this site. These results support a model in which multiple tissue-specific activators collectively function to assemble a composite histone modification pattern, consisting of overlapping histone acetylation and methylation. As GATA-1 induced H3 acetylation, but not H3-meK4, at the promoter, H3 acetylation and H3-meK4 components of a composite histone modification pattern can be established independently.
Keywords: globin, chromosome, erythroid, epigenetic
Chromatin structure at the promoters and over broad chromosomal segments is a critical determinant of gene expression, and therefore, chromatin modification is an essential step in transcriptional control. The acetylation of core histones in nucleosomes represents a major mode of chromatin modification (1–3). Histone acetylation increases the accessibility of nucleosomal DNA through structural transitions at the level of the nucleosome (4, 5) and higher-order chromatin structure (6). In addition to directly modifying chromatin structure, the acetyllysine residue can be recognized by regulatory factors, e.g., bromodomain-containing coactivators (7–9). Such coactivators catalyze additional chromatin modification and interact with components of the transcriptional machinery, thereby stimulating transcription (10).
Besides acetylation, histones are subjected to methylation, phosphorylation, and ubiquitination (11). Given the combinatorial complexity of histone modifications, a “histone code” hypothesis has been proposed, which assumes that distinct patterns of modifications confer unique functional consequences (12). For example, distinct methyltransferases methylate histone H3 at either lysine-4 (H3-meK4) (13–15) or lysine-9 (H3-meK9) (16), and the distribution of H3-meK4 and H3-meK9 delineates functionally unique chromosome segments. Analysis of histone modifications within the chicken β-globin locus (17), heterochromatic regions of fission yeast (18), and the inactive human X chromosome (19) revealed that H3-meK9 was enriched in hypoacetylated, transcriptionally inactive chromatin, whereas H3-meK4 occurred predominantly at hyperacetylated, transcriptionally active chromatin. H3-meK9 is recognized by the chromodomain of heterochromatin protein 1 (HP1) (20–22), which participates in heterochromatin formation and gene silencing (23). H3-meK4 inhibits binding of the nucleosome remodeling deacetylase (NURD) repressor complex to the amino-terminal tail of histone H3, which may favor the transcriptionally active state (21). Thus, analogous to the acetylated H4–bromodomain interaction, methylated species of H3 engage in functionally important protein–protein interactions.
The ultimate test of the histone code hypothesis, especially in the context of mammalian transcriptional control, will require detailed knowledge of how complex histone modification patterns are established and maintained within native chromatin domains. Localized acetylation can be established by means of direct recruitment of histone acetyltransferases (HATs) by DNA-bound activators (2). However, there is still much to be learned about how broad histone modification patterns arise across entire chromosomal segments (24). Chromatin immunoprecipitation (ChIP) analysis has enabled the definition of locus-specific, broad histone modification patterns (25, 26). The β-globin locus, containing the erythroid-specific and developmentally regulated β-globin genes, has been a particularly informative system for investigating the structure/function of histone modification patterns (25, 26). H3 and H4 acetylation exists throughout the entire ≈60-kb chicken β-globin locus in erythroid cells (27, 28). Rather than having uniform acetylation, the murine β-globin locus in adult erythroid cells is characterized by hyperacetylation at the locus control region (LCR), which confers high-level transcription to the β-globin genes (29–31), and at the adult β-globin genes (βmajor and βminor) residing at the 3′ side of the locus (32, 33) (Fig. 1). In contrast, the embryonic/fetal β-globin genes (Εy and βH1), located centrally within the locus, reside within an ≈30-kb hypoacetylated subdomain. Thus, tissue-specific acetylation patterns within mammalian chromatin domains can be segregated into structurally unique subdomains. However, histone acetylation can be enriched within entire mammalian domains, as histone H4 acetylated at lysine-5 exists throughout the human α-globin locus in erythroid cells (34).
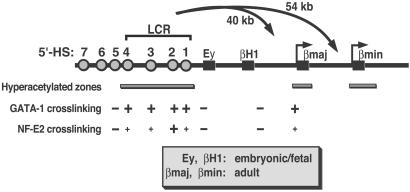
Fig 1.
Organization of the murine β-globin locus. The locus contains two embryonic/fetal (Ey and βH1) and two adult (βmajor and βminor) β-globin genes. Hypersensitive site 2 (HS2) resides 40 and 54 kb upstream of the βmajor and βminor promoters, respectively. The β-globin genes are depicted as boxes, and HSs are depicted as circles. Bars below the locus depict hyperacetylated zones defined by ChIP analysis with anti-acetylated H3 and H4 antibodies in MEL cells and in 14.5 days postcoitum (dpc) fetal liver (32). GATA-1 and NF-E2 crosslinking data, derived from ChIP analysis in MEL and G1E-ER-GATA-1 cells, are also depicted (40). Large +, strong crosslinking; small +, weak crosslinking; −, no or very weak crosslinking.
How are composite histone modification patterns consisting of acetylation and methylation established? Histone acetyltransferases and methyltransferases might be recruited to a chromatin template by a single activator. Alternatively, multiple activators might be required to recruit both enzymes. We have used the endogenous murine β-globin locus to investigate this issue. Two hematopoietic-specific activators required for β-globin expression, GATA-1 (35–37) and NF-E2 (38, 39), cooperatively recruit RNA polymerase II (pol II) to the murine adult β-globin promoters (40). GATA-1 is the founding member of the GATA family of zinc-finger activators, which control diverse aspects of development (41). Erythroid progenitors in GATA-1-null mice fail to survive and mature, and the embryos die of anemia (42). Whereas GATA-1 sites are distributed throughout the β-globin locus, GATA-1 binds to hypersensitive site (HS)1, HS2, HS3, and HS4, and to the βmajor promoter in vivo (40) (Fig. 1). NF-E2 is a heterodimeric transcription factor consisting of a hematopoietic-specific basic leucine zipper subunit, p45/NF-E2 (38, 39), and a broadly expressed subunit, a member of the Maf family of basic leucine zipper proteins (43). NF-E2 is required for megakaryopoiesis (44) and has been implicated in the control of β-globin expression (45–48). Conserved, consensus NF-E2 binding sites reside within HS2 of the LCR (49), whereas additional nonconserved, imperfect sites exist at other HSs of the LCR and at the βmajor promoter (40). NF-E2 binds to HS2, and, to a lesser extent HS1, HS3, and HS4, and to the βmajor promoter in vivo (40, 50) (Fig. 1).
Given the GATA-1-NF-E2 cooperativity to recruit pol II, it is instructive to ask whether these factors induce identical or distinct histone modifications at the promoter and neighboring chromatin. We showed that NF-E2 induced H3 and, to a lesser extent, H4 acetylation at the adult β-globin promoters (47). The potential role of NF-E2 in regulating histone methylation and of GATA-1 in regulating histone acetylation and methylation have not been described. ChIP analysis revealed that NF-E2 strongly induced H3-meK4 at and near the adult β-globin promoters, whereas both NF-E2 and GATA-1 induced hyperacetylation. These results are discussed vis-à-vis prospective mechanisms for how composite histone modification patterns are established in mammalian chromatin.
Materials and Methods
Cell Culture and Transfection.
Mouse erythroleukemia (MEL) and CB3 cells were maintained in Dulbecco's modified Eagle's medium (Biofluids, Rockville, MD) containing 1% antibiotic/antimycotic (GIBCO/BRL) and 10% FBS. The p45/NF-E2 expression vector has been described (51, 52). Stably transfected clones of CB3 cells expressing p45/NF-E2 (CB3–6 and CB3–9) were selected and maintained in 1 mg/ml G418 sulfate (Calbiochem). G1E cells (53) were maintained in Iscove's modified Dulbecco's medium (GIBCO/BRL) containing 2% penicillin/streptomycin (GIBCO/BRL), 2 units/ml erythropoietin, 120 nM monothioglycerol (Sigma), 15% FBS, and 0.6% conditioned medium from a Kit-ligand-producing CHO cell line. G1E-ER-GATA-1 cells, which stably express GATA-1 as a fusion to the human estrogen receptor ligand binding domain (54) were maintained identical to G1E cells except that media contained 1 μg/ml puromycin.
Quantitative ChIP Assay.
ChIP analysis was performed as described (25, 47). MEL, CB3, CB3-6, and CB3-9 cells were incubated for 4 days with 1.5% DMSO (Sigma). G1E and G1E-ER-GATA-1 cells were incubated for 48 h with 1 μM tamoxifen (Sigma). Immunoprecipitated DNA was analyzed by real-time PCR (Applied Biosystems Prism 7000). Primers were designed by PRIMER EXPRESS 1.0 software (PE Applied Biosystems) to amplify 50- to 150-bp subregions within the endpoints of the corresponding standard PCR primers (32). Primers were based on Hbbd haplotype sequences (GenBank accession nos. Z13985, X14061, AF128269, and AF133300). Samples from at least two independent immunoprecipitations were analyzed. Product was measured by SYBR green fluorescence in 25-μl reactions, and the amount of product was determined relative to a standard curve generated from a titration of input chromatin. Dissociation curves after amplification showed that primer pairs generated single products.
Forward and Reverse Primers for Quantitative ChIP Assay.
(5′–3′): HS2, AGTCAATTCTCTACTCCCCACCCT and ACTGCTGTGCTCAAGCCTGAT; IVR3, TGTGCTAGCCTCAAGCTCACA and TCCCAGCACTCAGAAGAAGGA; IVR5, GTATGCTCAATTCAAATGTACCTTATTTTAA and TTACCTCTTTATTTCACTTTTACACATAGCTAA; βmajor promoter, CAGGGAGAAATATGCTTGTCATCA and GTGAGCAGATTGGCCCTTACC; βmajor-3′, GCCCTGGCTCACAAGTACCA and TTCACAGGCAAGAGCAGGAA; IVR6, ATAGGAAAGAAAATGCACACATAGATTC and CCCACGCCTCATTTATACTTTCAG; βminor promoter, CCTCACCCTGCAAGGTAACAC and TGCTCAGCTTTATATACCCAATGC; IVR16, TGGCCATTTTTACTATGTTAATTTTGC and TAGACTTGTCATGGTTATGGATTGG.
Antibodies.
Rabbit anti-pol II (N-20, sc-899) was obtained from Santa Cruz Biotechnology. Anti-acetylated histone H3 (06-599), anti-tetraacetylated H4 (06-866), and anti-H3-meK4 (07-030) antibodies were obtained from Upstate Biotechnology (Lake Placid, NY). Preimmune serum served as controls for each of the antibodies. For Western analysis, protein A–peroxidase (Sigma) and goat anti-rat IgG conjugated with horseradish peroxidase (Santa Cruz Biotechnology) were used to detect p45/NF-E2 and GATA-1, respectively.
RNA Isolation and Analysis by Quantitative RT-PCR.
RNA and protein were prepared from the same cell cultures used for ChIP. Total RNA was purified with Trizol (GIBCO/BRL). RNA (1 μg) was used to prepare cDNA by annealing with 250 ng of a 5:1 mixture of random and oligo(dT) primers heated at 68°C for 10 min. This procedure was followed by incubation with reverse transcriptase (Superscript II, GIBCO/BRL) combined with 10 mM DTT, RNasin (Promega), and 0.5 mM dNTPs at 42°C for 1 h. The reaction mixture was diluted to a final volume of 150 μl and heat inactivated at 95–99°C for 5 min. RT-PCRs (25 μl) contained 2.5 μl of cDNA, 12.5 μl of SYBR green (Applied Biosystems), and the appropriate primers. Product accumulation was monitored by SYBR green fluorescence. Relative expression levels were determined from a standard curve of serial dilutions of MEL cDNA samples. Forward and reverse primers for real-time RT-PCRs (5′-3′): β-globin, CAGCCTCAGTGAGCTCCACTG and GATCATATTGCCCAGGAGCC; GAPDH, GAAGGTACGGAGTCAACGGATTT and GAATTTGACCATGGGTGGAAT.
Protein Analysis.
Total protein was prepared by boiling cells for 5 min in SDS sample buffer [50 mM Tris (pH 6.8)/100 mM DTT/2% SDS/0.1% bromophenol blue/10% glycerol]. Extracts from 1 × 105 cells were resolved on SDS/polyacrylamide gels, transferred to Immobilon P membranes (Millipore), and analyzed by Western blotting. Proteins were visualized by using ECL-Plus (Amersham Pharmacia).
Results and Discussion
NF-E2 Requirement for Establishing an Overlapping Pattern of H3-meK4 and H3 and H4 Acetylation.
A quantitative real-time PCR-based ChIP assay was developed and used to determine whether the endogenous murine β-globin locus was enriched in H3-meK4 in erythroid cells and, if so, whether the hematopoietic activator NF-E2 regulates H3-meK4. Previously, we showed that NF-E2 induced H3, and to a lesser extent, H4 acetylation at and near the adult β-globin promoters (47). H3-meK4 was measured at the βmajor promoter in erythroleukemia cells expressing NF-E2 (MEL), in NF-E2-null erythroleukemia cells (CB3) (45), and in two clonal lines of CB3 cells (CB3-6 and CB3-9) that stably express NF-E2 at physiological levels (Fig. 2A). NF-E2 exists as two isoforms that result from alternative translational start sites, although no functional distinctions have been ascribed to the isoforms (38, 39). NF-E2 expression in CB3 cells reactivated the silent βmajor gene (Fig. 2B), as has been described (45, 46), demonstrating the NF-E2 requirement for β-globin transcription. Real-time PCR was used to quantitate βmajor promoter DNA immunoprecipitated by anti-H3-meK4 or preimmune (PI) antibodies. Assays were done under conditions in which product accumulated linearly with respect to input DNA (Fig. 2C). Representative amplification plots are depicted in Fig. 2D. Strong methylation was apparent at the βmajor promoter in MEL cells, whereas considerably less was evident in CB3 cells. SYBR green fluorescence reflected the expected homogenous PCR product, as shown by dissociation analysis (Fig. 2E).
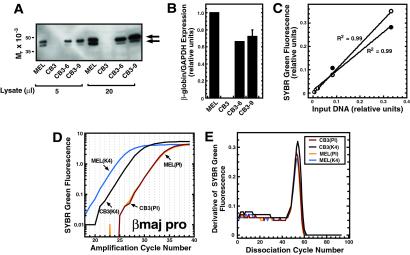
Fig 2.
Quantitative ChIP analysis reveals differential H3-meK4 at the βmajor promoter in MEL and CB3 cells. Cells were incubated for 4 days with 1.5% DMSO. (A) p45/NF-E2 expression in DMSO-induced CB3, MEL, CB3-6, and CB3-9 cells. The arrows depict p45/NF-E2 isoforms. (B) Quantitative real-time RT-PCR analysis of β-globin expression in CB3, MEL, CB3-6, and CB3-9 cells. Relative levels of β-globin expression were normalized to the expression of GAPDH. The graph depicts data from three (MEL, CB3, and CB3-9) and two (CB3-6) independent experiments. (C) SYBR green fluorescence (relative units) was plotted vs. the initial MEL and CB3 cell input DNA concentration. The plot illustrates the linearity and range of signals used to measure the relative amounts of target DNA in samples. (○) CB3 cells. (•) MEL cells. (D) A representative amplification plot for quantitative analysis of H3-meK4 at the βmajor promoter in MEL and CB3 cells. PI, preimmune. (E) Dissociation curves of the amplicons illustrated in D. The homogeneity of the curves reflects the generation of a single amplicon.
We compared H3-meK4 and H3 and H4 acetylation among MEL, CB3, CB3-6, and CB3-9 cells (Fig. 3). Analysis of H3-meK4 at functionally distinct sites of the β-globin locus in MEL cells yielded a variable distribution at these sites (Fig. 3A). H3-meK4 was high at the βmajor and βminor promoters and at the 3′ end of the βmajor gene. By contrast, H3-meK4 was low at an intergenic site (IVR-6) between the βmajor and βminor genes, and at both upstream (IVR-3, IVR-5) and downstream (IVR-16) intergenic sites. This pattern resembled the acetylation pattern mapped previously by using a semiquantitative ChIP assay (32). The quantitative ChIP assay was also used to measure histone H3 and H4 acetylation with antibodies recognizing multiple acetylated forms of these histones (Fig. 3 B and C, respectively). This analysis confirmed that H3-meK4, and H3 and H4 acetylation shared a similar pattern. The three modifications were not uniformly enriched throughout the β-globin domain, but rather, had a highly variable distribution at functionally distinct sites. This result differs from the chicken β-globin locus, in which high-level modifications were broadly distributed throughout the domain (17).
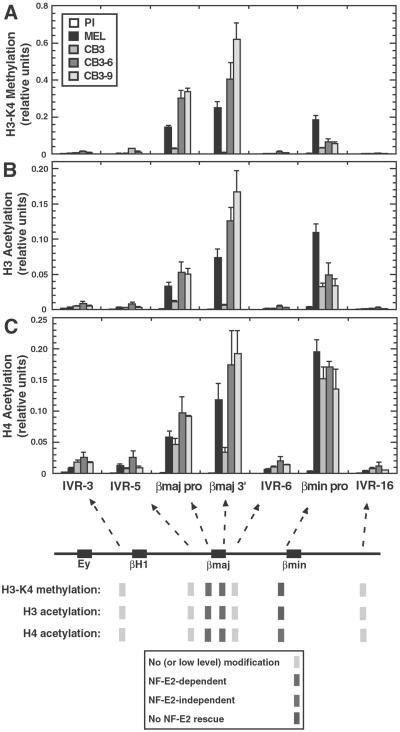
Fig 3.
NF-E2-dependent H3-meK4 and H3 and H4 acetylation patterns of the endogenous murine β-globin locus. Cells were incubated for 4 days with 1.5% DMSO. (A) H3-meK4 pattern of the murine β-globin locus in DMSO-induced MEL, CB3, CB3-6, and CB3-9. The relative level of H3-meK4 was determined quantitatively and plotted as a function of the position within the locus. (B and C) H3 and H4 acetylation patterns of the β-globin locus in DMSO-induced MEL, CB3, CB3-6, and CB3-9 cells. The relative levels of H3 and H4 acetylation were determined quantitatively and plotted as a function of the position within the locus. Number of independent ChIP samples analyzed: MEL, n = 5; CB3, n = 5; CB3-6, n = 3; and CB3-9, n = 2 (IVR-3, IVR-5, βmajor promoter, IVR-6, and IVR-16); and n = 3 (βmajor 3′ and βminor promoter).
CB3 cells had strongly reduced H3-meK4 at all sites that were shown to be methylated in MEL cells. NF-E2 expression rescued defective H3-meK4 at the βmajor promoter and βmajor 3′ (a 10-fold and 69-fold increase, respectively), but not at the βminor promoter, in CB3-6 and CB3-9 cells. The failure of NF-E2 to rescue H3-meK4 at the βminor promoter is consistent with quantitative RT-PCR analysis with βmajor- and βminor-specific primers showing that NF-E2 reactivates only βmajor expression in CB3 cells (unpublished data); differential regulation of βmajor and βminor expression has been observed during normal murine erythropoiesis (55) and upon the differentiation of MEL cells (50, 56). Similarly, CB3 cells had reduced levels of acetylated H3, consistent with our previous semiquantitative analysis (47). NF-E2 expression increased H3 acetylation at the βmajor promoter and βmajor 3′ 4.6-fold and 23-fold, respectively. H3 acetylation at the βminor promoter was not rescued by NF-E2 expression. H4 acetylation at the promoters was very similar between MEL and CB3 cells. NF-E2 expression increased H4 acetylation at the βmajor promoter and βmajor 3′ 2.1-fold and 5.0-fold, respectively. H4 acetylation at the βminor promoter was NF-E2-independent. Thus, H3-meK4 had a distribution similar to H3 and H4 acetylation. Furthermore, the establishment of both H3-meK4 and H3 acetylation at the βmajor promoter and βmajor 3′ required NF-E2, providing an example of a mammalian activator inducing H3-meK4 in vivo.
Because NF-E2 can be crosslinked to the βmajor promoter (40, 50), NF-E2 resides in close proximity to the acetylated and methylated chromatin of the promoter. Although it is unclear as to whether these observations result from LCR-bound NF-E2 contacting the promoter, or NF-E2 bound directly to nonconsensus sites on the promoter, the result is consistent with a direct action of NF-E2 to induce acetylation and H3-meK4. Further support for a direct function of NF-E2 at the promoter comes from the use of a combined ChIP-in vivo footprinting assay (57). Chromatin was immunoprecipitated with an NF-E2 antibody, and the resulting DNA was subjected to footprinting analysis with dimethyl sulfate to detect sequence-specific protein–DNA interactions. A footprint was detected over an imperfect NF-E2 site on the βmajor promoter in MEL cells, consistent with NF-E2 binding directly to this site.
GATA-1 Induces Histone H3 Hyperacetylation, but Not H3-meK4, at the βmajor Promoter.
As NF-E2 cooperates with GATA-1 to recruit pol II to the adult βmajor promoter (40), we asked whether both activators are able to induce H3-meK4 and acetylation or whether they have distinct activities. H3-meK4 and H3 acetylation were analyzed in physiologically relevant G1E cells, proerythroblast-like cells derived from GATA-1-null murine embryonic stem cells (53), and G1E cells stably expressing an estrogen receptor-GATA-1 fusion protein (ER-GATA-1) (54) (Fig. 4A). In contrast to a previous report indicating that ER-GATA-2 and GATA-2 have opposing effects on erythroid differentiation (58), ER-GATA-1 and GATA-1 both induce erythroid differentiation of G1E cells (54, 59, 60). Whereas ER-GATA-1 activation in G1E-ER-GATA-1 cells strongly induced β-globin expression (574-fold) (Fig. 4B), both G1E and G1E-ER-GATA-1 cells had similar levels of H3-meK4 at the βmajor promoter (Fig. 4C). In contrast, GATA-1 induced H3 acetylation, analogous to NF-E2 in CB3 cells (Fig. 3B) and strongly induced pol II loading on the promoter (Fig. 4C). GATA-1 can be crosslinked to the βmajor promoter (39), which contains consensus GATA-1 sites, consistent with the direct action of GATA-1 to induce acetylation and pol II recruitment. A direct action is further supported by the result that treatment of G1E-ER-GATA-1 cells for 3 h with tamoxifen induces H3 acetylation at the βmajor promoter (Gerd Blobel, personal communication), similar to results described herein with a 48-h treatment. However, one cannot rule out the possibility that ER-GATA-1 regulates histone acetylation through an indirect mechanism involving changes in expression of other factors that regulate the β-globin genes.
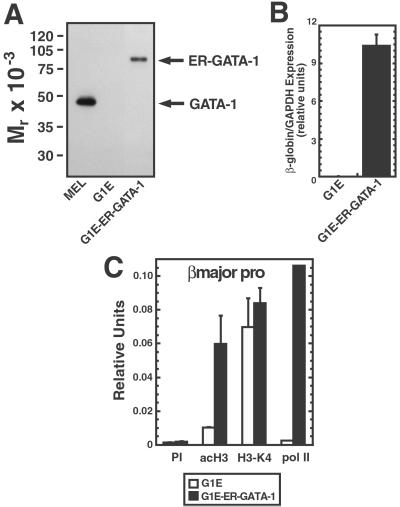
Fig 4.
In contrast to NF-E2, GATA-1 does not induce H3-meK4 at the βmajor promoter. G1E-GATA-1 and G1E cells were incubated for 48 h with 1 μM tamoxifen. (A) Western blot analysis of GATA-1 and ER-GATA-1 expression in total cell lysates from MEL, G1E, and G1E-ER-GATA cell lines. (B) Real-time RT-PCR analysis of β-globin RNA expression in G1E and G1E-ER-GATA cells. Relative expression of β-globin RNA levels normalized to GAPDH. The graph depicts data from four (G1E) and five (G1E-ER-GATA-1) independent RNA isolations. (C) Real-time PCR ChIP analysis of H3 acetylation, H3-meK4, and pol II binding at the βmajor promoter. Samples from three independent ChIP experiments were analyzed quantitatively.
ER-GATA-1 activation in G1E cells increased erythroid Krüppel-like factor (EKLF) transcripts (59). In addition, other studies have shown that the EKLF promoter is induced by GATA-1 in transient transfection assays and in transgenic mice (61, 62). EKLF is a critical regulator of definitive erythropoiesis (63) and functions through HS3 of the LCR (64). As shown in Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org, however, EKLF protein levels were nearly identical in our G1E and G1E-ER-GATA-1 lines. This result is inconsistent with a mechanism in which GATA-1 increases EKLF levels as a critical step in inducing acetylation at the βmajor promoter in this system. ER-GATA-1 activation also increased Friend of GATA-1 (FOG-1) transcripts (59). GATA-1 utilizes FOG-1 to both activate and repress target genes (65, 66). By contrast, ER-GATA-1 activation had no effect on p45/NF-E2 protein levels (40) and decreased GATA-2 transcripts (59). Importantly, as GATA-2 expression is high in G1E cells and chromatin at the adult β-globin genes is hypoacetylated in these cells, GATA-2 lacks the activity of GATA-1 to establish hyperacetylation at this site. As GATA-2 interacts with HDAC3, this finding might explain its inability to substitute for GATA-1 to establish hyperacetylation (67).
Based on existing knowledge of how GATA-1 and NF-E2 function, one could not predict whether these factors would have similar or different influences on histone acetylation and methylation. Both factors interact with CBP/p300 (68), which could mediate changes in histone acetylation. However, NF-E2, but not GATA-1, increased the histone acetyltransferase activity of CBP toward nucleosomal substrates in vitro (69), suggesting that NF-E2 might uniquely regulate histone acetylation. Neither factor, nor related proteins, has been shown to interact with histone methyltransferases. Because FOG-1 interacts with GATA-1, but not NF-E2, GATA-1 might exert a mechanistically unique activity to regulate transcription.
The results of Figs. 3 and 4 show that NF-E2 has an activity shared with GATA-1 to induce H3 hyperacetylation and a unique activity to induce H3-meK4. Hypoacetylation in GATA-1-null cells cannot be attributed to decreased levels of NF-E2 and vice versa (40), indicating that both activators are required to induce hyperacetylation. The collective actions of the two factors thereby establish a composite histone modification pattern. Alternatively, because GATA-1 is required only to establish hyperacetylation, it is possible that GATA-1 is critical for activating transcription, and hyperacetylation is transcription-dependent. No qualitative differences in the activities of GATA-1 and NF-E2 in transcription assays have been described previously, to our knowledge. We have shown that pol II recruitment to HS1, HS2, and HS3 of the LCR is more dependent on GATA-1 than NF-E2 (40). However, this differential activity is likely unrelated to the differential chromatin modifying activity. Both GATA-1 and NF-E2 are required for pol II recruitment to the βmajor promoter, which may reflect the cooperative actions of these factors to establish the composite histone modification pattern.
H3-meK4 at Hypersensitive Site 2 Is NF-E2- and GATA-1-Independent.
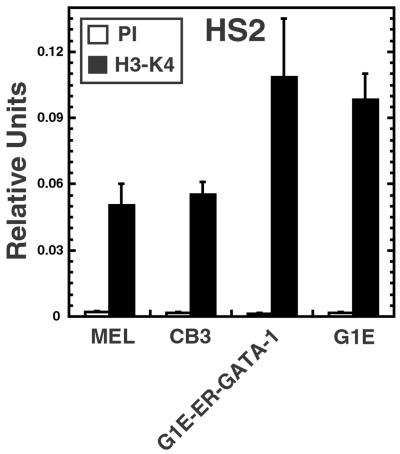
Because all sites of H3-meK4 in the vicinity of the adult β-globin genes were dependent on NF-E2, it was important to ask whether NF-E2 is required to establish methylation at other sites within the β-globin domain. ChIP analysis revealed strong H3-meK4 at HS2 in MEL cells (Fig. 5). Interestingly, CB3 cells lacking NF-E2 had an equivalent level of H3-meK4. H3-meK4 was similarly high at HS2 in GATA-1-null G1E cells, and the expression of ER-GATA-1 did not change the methylation state. This result supports a model in which NF-E2, but not GATA-1, establishes H3-meK4 within a subregion of the locus containing the adult β-globin genes, whereas the establishment of methylation far upstream at HS2 requires factors distinct from NF-E2 and GATA-1.
Fig 5.
H3-meK4 at hypersensitive site 2 is NF-E2- and GATA-1-independent. MEL and CB3 cells were incubated for 4 days with 1.5% DMSO. G1E-GATA-1 and G1E cells were incubated for 48 h with 1 μM tamoxifen. Quantitative PCR ChIP analysis of H3-meK4 at HS2 in MEL, CB3, G1E-ER-GATA-1, and G1E cells was done. Samples from five (MEL and CB3) and three (G1E-ER-GATA-1 and G1E) independent ChIP experiments were analyzed quantitatively, relative to a standard curve generated from input chromatin. The plot depicts relative levels of H3-meK4 at HS2 in the cell lines.
Establishment of Composite Histone Modification Patterns in Mammalian Chromatin Domains: Mechanistic Considerations.
Two lines of evidence presented in this study support distinct activator requirements for establishing acetylation and methylation components of a composite histone modification pattern. First, analysis of histone modifications in G1E cells revealed a dissociation of events involved in establishing H3-meK4 and H3 hyperacetylation (Fig. 4C). As H3-meK4 was enriched in hypoacetylated chromatin at the βmajor promoter in G1E cells, H3 hyperacetylation is not required for the establishment of H3-meK4. Inversely, H3-meK4 is insufficient to induce H3 acetylation. Thus, the establishment of H3-meK4 precedes hyperacetylation. The result is consistent with a model in which the establishment of H3-meK4 and H3 acetylation requires multiple activators, potentially with unique activities. H3 hyperacetylation requires GATA-1, whereas H3-meK4 does not at the βmajor promoter. Second, we have shown that two tissue-specific activators, GATA-1 and NF-E2, which strongly activate transcription of an endogenous gene, have distinct activities to induce H3-meK4 and acetylation at the βmajor promoter. GATA-1 and NF-E2 share the ability to induce H3 acetylation, whereas only NF-E2 induces H3-meK4. Although the possibility cannot be excluded that GATA-1 would induce H3-meK4 in other contexts, GATA-1 differs from NF-E2 in lacking this ability at the βmajor promoter. Intriguingly, neither GATA-1 nor NF-E2 is required for the establishment of H3-meK4 at HS2, indicating that additional factors function with GATA-1 and NF-E2 to establish the composite histone modification pattern. Although these conclusions are based on the analysis of the endogenous β-globin locus in living cells, MEL, CB3, and G1E cells represent cell lines derived in different ways. Additional mechanisms might contribute to the establishment of the composite histone modification pattern during normal erythropoiesis.
The differential chromatin-modifying activities of GATA-1 and NF-E2 associated with the transcriptional activation of the βmajor promoter strongly support the hypothesis that the establishment of a composite histone modification pattern requires activators with qualitatively distinct activities. A test of this hypothesis will require comparative analysis of the activities of multiple activators that regulate chromatin structure in complex mammalian systems, such as the β-globin domain. Such analyses should reveal whether activators can be categorized into a finite number of groups, based on their intrinsic activities to induce unique histone modifications. Establishment of physiological chromatin structure would require the coordinated actions of representative members of each group. This finding would have broad mechanistic implications for understanding genetic networks that control cell and organismal function.
Supplementary Material
Acknowledgments
We thank Mitchell Weiss, Yaakov Ben-David, and Jim Bieker for kindly providing G1E cells, CB3 cells, and the 4B9 anti-EKLF antibody, respectively. We also thank Hogune Im and David A. Wassarman for a critical review of the manuscript. This work was funded by National Institutes of Health Grants DK50107 and DK55700 (to E.H.B.). E.H.B. is a Scholar of the Leukemia Society of America and a Shaw Scientist.
Abbreviations
ChIP, chromatin immunoprecipitation
LCR, locus control region
pol II, RNA polymerase II
HS, hypersensitive site
MEL, mouse erythroleukemia
EKLF, erythroid Krüppel-like factor
PI, preimmune
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Workman J. L. & Kingston, R. E. (1998) Annu. Rev. Biochem. 67, 545-579. [DOI] [PubMed] [Google Scholar]
- 2.Roth S. Y., Denu, J. M. & Allis, C. D. (2001) Annu. Rev. Biochem. 70, 81-120. [DOI] [PubMed] [Google Scholar]
- 3.Eberharter A. & Becker, P. B. (2002) EMBO Rep. 3, 224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee D. Y., Hayes, J. J., Pruss, D. & Wolffe, A. P. (1993) Cell 72, 73-84. [DOI] [PubMed] [Google Scholar]
- 5.Vettese-Dadey M., Grant, P. A., Hebbes, T. R., Crane-Robinson, C., Allis, C. D. & Workman, J. L. (1996) EMBO J. 15, 2508-2518. [PMC free article] [PubMed] [Google Scholar]
- 6.Tse C., Sera, T., Wolffe, A. P. & Hansen, J. C. (1998) Mol. Cell. Biol. 18, 4629-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhalluin C., Carlson, J. E., Zeng, L., He, C., Aggarwal, A. K. & Zhou, M. M. (1999) Nature 399, 491-496. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson R. H., Ladurner, A. G., King, D. S. & Tjian, R. (2000) Science 288, 1422-1425. [DOI] [PubMed] [Google Scholar]
- 9.Mamorstein R. & Berger, S. L. (2001) Gene 272, 1-9. [DOI] [PubMed] [Google Scholar]
- 10.Kadonaga J. T. (1998) Cell 92, 307-313. [DOI] [PubMed] [Google Scholar]
- 11.Jenuwein T. & Allis, C. D. (2001) Science 293, 1074-1080. [DOI] [PubMed] [Google Scholar]
- 12.Strahl B. D. & Allis, C. D. (2000) Nature 403, 41-45. [DOI] [PubMed] [Google Scholar]
- 13.Wang H., Cao, R., Xia, L., Erdjument-Bromage, H., Borchers, C., Tempst, P. & Zhang, Y. (2001) Mol. Cell 8, 1207-1217. [DOI] [PubMed] [Google Scholar]
- 14.Briggs S. D., Bryk, M., Strahl, B. D., Cheung, W. L., Davie, J. K., Dent, S. Y., Winston, F. & Allis, C. D. (2001) Genes Dev. 15, 3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishioka K., Chuikov, S., Sarma, K., Erdjument-Bromage, H., Allis, C. D., Tempst, P. & Reinberg, D. (2002) Genes Dev. 16, 479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rea S., Eisenhaber, F., O'Carroll, D., Strahl, B. D., Sun, Z. W., Schmid, M., Opravil, S., Mechtler, K., Ponting, C. P., Allis, C. D. & Jenuwein, T. (2000) Nature 406, 593-599. [DOI] [PubMed] [Google Scholar]
- 17.Litt M. D., Simpson, M., Gaszner, M., Allis, C. D. & Felsenfeld, G. (2001) Science 293, 2453-2455. [DOI] [PubMed] [Google Scholar]
- 18.Nakayama J., Rice, J. C., Strahl, B. D., Allis, C. D. & Grewal, S. I. (2001) Science 292, 110-113. [DOI] [PubMed] [Google Scholar]
- 19.Boggs B. A., Cheung, P., Heard, E., Spector, D. L., Chinault, A. C. & Allis, C. D. (2002) Nat. Genet. 30, 73-76. [DOI] [PubMed] [Google Scholar]
- 20.Lachner M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. (2001) Nature 410, 116-120. [DOI] [PubMed] [Google Scholar]
- 21.Bannister A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C. & Kouzarides, T. (2001) Nature 410, 120-124. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs S. A., Taverna, S. D., Zhang, Y., Briggs, S. D., Li, J., Eissenberg, J. C., Allis, C. D. & Khorasanizadeh, S. (2001) EMBO J. 20, 5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eissenberg J. C. & Elgin, S. C. (2000) Curr. Opin. Genet. Dev. 10, 204-210. [DOI] [PubMed] [Google Scholar]
- 24.Forsberg E. C. & Bresnick, E. H. (2001) BioEssays 23, 820-830. [DOI] [PubMed] [Google Scholar]
- 25.Johnson K. D. & Bresnick, E. H. (2002) Methods 26, 27-36. [DOI] [PubMed] [Google Scholar]
- 26.Bulger M., Sawado, T., Schubeler, D. & Groudine, M. (2002) Curr. Opin. Genet. Dev. 12, 170-177. [DOI] [PubMed] [Google Scholar]
- 27.Hebbes T. R., Clayton, A. L., Thorne, A. W. & Crane-Robinson, C. (1994) EMBO J. 13, 1823-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litt M. D., Simpson, M., Recillas-Targa, F., Prioleau, M. N. & Felsenfeld, G. (2001) EMBO J. 20, 2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grosveld F., van Assendelft, G. B., Greaves, D. R. & Kollias, G. (1987) Cell 51, 975-985. [DOI] [PubMed] [Google Scholar]
- 30.Epner E., Reik, A., Cimbora, D., Telling, A., Bender, M. A., Fiering, S., Enver, T., Martin, D. I., Kennedy, M., Keller, G. & Groudine, M. (1998) Mol. Cell 2, 447-455. [DOI] [PubMed] [Google Scholar]
- 31.Navas P. A., Li, Q., Peterson, K. R., Swank, R. A., Rohde, A., Roy, J. & Stamatoyannopoulos, G. (2002) Hum. Mol. Genet. 11, 893-903. [DOI] [PubMed] [Google Scholar]
- 32.Forsberg E. C., Downs, K. M., Christensen, H. M., Im, H., Nuzzi, P. A. & Bresnick, E. H. (2000) Proc. Natl. Acad. Sci. USA 97, 14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schubeler D., Groudine, M. & Bender, M. A. (2001) Proc. Natl. Acad. Sci. USA 98, 11432-11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anguita E., Johnson, C. A., Wood, W. G., Turner, B. M. & Higgs, D. R. (2001) Proc. Natl. Acad. Sci. USA 98, 12114-12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai S. F., Martin, D. I., Zon, L. I., D'Andrea, A. D., Wong, G. G. & Orkin, S. H. (1989) Nature 339, 446-451. [DOI] [PubMed] [Google Scholar]
- 36.Evans T. & Felsenfeld, G. (1989) Cell 58, 877-885. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto M., Ko, L. J., Leonard, M. W., Beug, H., Orkin, S. H. & Engel, J. D. (1990) Genes Dev. 4, 1650-1662. [DOI] [PubMed] [Google Scholar]
- 38.Andrews N. C., Erdjument-Bromage, H., Davidson, M. B., Tempst, P. & Orkin, S. H. (1993) Nature 362, 722-728. [DOI] [PubMed] [Google Scholar]
- 39.Ney P. A., Andrews, N. C., Jane, S. M., Safer, B., Purucker, M. E., Weremowicz, S., Morton, C. C., Goff, S. C., Orkin, S. H. & Nienhuis, A. W. (1993) Mol. Cell. Biol. 13, 5604-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson K. D., Grass, J. D., Boyer, M. E., Kiekhaefer, C. M., Blobel, G. A., Weiss, M. J. & Bresnick, E. H. (2002) Proc. Natl. Acad. Sci. USA 99, 11760-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cantor A. B. & Orkin, S. H. (2002) Oncogene 21, 3368-3376. [DOI] [PubMed] [Google Scholar]
- 42.Fujiwara Y., Browne, C. P., Cunniff, K., Goff, S. C. & Orkin, S. H. (1996) Proc. Natl. Acad. Sci. USA 93, 12355-12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrews N. C., Kotkow, K. J., Ney, P. A., Erdjument-Bromage, H., Tempst, P. & Orkin, S. H. (1993) Proc. Natl. Acad. Sci. USA 90, 11488-11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shivdasani R. A., Rosenblatt, M. F., Zucker-Franklin, D., Jackson, C. W., Hunt, P., Saris, C. J. & Orkin, S. H. (1995) Cell 81, 695-704. [DOI] [PubMed] [Google Scholar]
- 45.Lu S. J., Rowan, S., Bani, M. R. & Ben-David, Y. (1994) Proc. Natl. Acad. Sci. USA 91, 8398-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotkow K. J. & Orkin, S. H. (1995) Mol. Cell. Biol. 15, 4640-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson K. D., Christensen, H. M., Zhao, B. & Bresnick, E. H. (2001) Mol. Cell 8, 465-471. [DOI] [PubMed] [Google Scholar]
- 48.Forsberg E. C., Downs, K. M. & Bresnick, E. H. (2000) Blood 96, 334-339. [PubMed] [Google Scholar]
- 49.Talbot D. & Grosveld, F. (1991) EMBO J. 10, 1391-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sawado T., Igarashi, K. & Groudine, M. (2001) Proc. Natl. Acad. Sci. USA 98, 10226-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bean T. L. & Ney, P. A. (1997) Nucleic Acids Res. 25, 2509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mosser E. A., Kasanov, J. D., Forsberg, E. C., Kay, B. K., Ney, P. A. & Bresnick, E. H. (1998) Biochemistry 37, 13686-13695. [DOI] [PubMed] [Google Scholar]
- 53.Weiss M. J., Yu, C. & Orkin, S. H. (1997) Mol. Cell. Biol. 17, 1642-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gregory T., Yu, C., Ma, A., Orkin, S. H., Blobel, G. A. & Weiss, M. J. (1999) Blood 94, 87-96. [PubMed] [Google Scholar]
- 55.Whitney J. B. (1977) Cell 12, 863-871. [DOI] [PubMed] [Google Scholar]
- 56.Ganguly S. & Skoultchi, A. I. (1985) J. Biol. Chem. 260, 167-173. [PubMed] [Google Scholar]
- 57.Lee Kang S. H., Vieira, K. & Bungert, J. (2002) Nucleic Acids Res. 30, e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kitajima K., Masuhara, M., Era, T., Enver, T. & Nakano, T. (2002) EMBO J. 17, 3060-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crispino J. D., Lodish, M. B., MacKay, J. P. & Orkin, S. H. (1999) Mol. Cell 3, 219-228. [DOI] [PubMed] [Google Scholar]
- 60.Shirihai O. S., Gregory, T., Yu, C., Orkin, S. H. & Weiss, M. J. (2000) EMBO J. 19, 2492-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crossley M., Tsang, A. P., Bieker, J. J. & Orkin, S. H. (1994) J. Biol. Chem. 269, 15440-15444. [PubMed] [Google Scholar]
- 62.Anderson K. P., Crable, S. C. & Lingrel, J. B. (2000) Blood 95, 1652-1655. [PubMed] [Google Scholar]
- 63.Perkins A. C., Sharpe, A. H. & Orkin, S. H. (1995) Nature 375, 318-322. [DOI] [PubMed] [Google Scholar]
- 64.Gillemans N., Tewari, R., Lindeboom, F., Rottier, R., de Wit, T., Wijgerde, M., Grosveld, F. & Philipsen, S. (1998) Genes Dev. 12, 2863-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsang A. P., Visvader, J. E., Turner, C. A., Fujuwara, Y., Yu, C., Weiss, M. J., Crossley, M. & Orkin, S. H. (1997) Cell 90, 109-119. [DOI] [PubMed] [Google Scholar]
- 66.Cantor A. B., Katz, S. G. & Orkin, S. H. (2002) Mol. Cell. Biol. 22, 4268-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ozawa Y., Towatari, M., Tsuzuki, S., Hayakawa, F., Maeda, T., Miyata, Y., Tanimoto, M. & Saito, H. (2001) Blood 98, 2116-2123. [DOI] [PubMed] [Google Scholar]
- 68.Blobel G. A. (2000) Blood 95, 745-755. [PubMed] [Google Scholar]
- 69.Chen C. J., Deng, Z., Kim, A. Y., Blobel, G. A. & Lieberman, P. M. (2001) Mol. Cell. Biol. 21, 476-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.