Abstract
Helicobacter pylori is a causative agent of gastritis and peptic ulcer. cagA+ H. pylori strains are more virulent than cagA− strains and are associated with gastric carcinoma. The cagA gene product, CagA, is injected by the bacterium into gastric epithelial cells and subsequently undergoes tyrosine phosphorylation. The phosphorylated CagA specifically binds SHP-2 phosphatase, activates the phosphatase activity, and thereby induces morphological transformation of cells. CagA proteins of most Western H. pylori isolates have a 34-amino acid sequence that variably repeats among different strains. Here, we show that the repeat sequence contains a tyrosine phosphorylation site. CagA proteins having more repeats were found to undergo greater tyrosine phosphorylation, to exhibit increased SHP-2 binding, and to induce greater morphological changes. In contrast, predominant CagA proteins specified by H. pylori strains isolated in East Asia, where gastric carcinoma is prevalent, had a distinct tyrosine phosphorylation sequence at the region corresponding to the repeat sequence of Western CagA. This East Asian-specific sequence conferred stronger SHP-2 binding and morphologically transforming activities to Western CagA. Finally, a critical amino acid residue that determines SHP-2 binding activity among different CagA proteins was identified. Our results indicate that the potential of individual CagA to perturb host-cell functions is determined by the degree of SHP-2 binding activity, which depends in turn on the number and sequences of tyrosine phosphorylation sites. The presence of distinctly structured CagA proteins in Western and East Asian H. pylori isolates may underlie the strikingly different incidences of gastric carcinoma in these two geographic areas.
A spiral Gram-negative microaerophilic bacterium, Helicobacter pylori, is present in the stomachs of at least half of the world's populations. It causes chronic inflammation that is frequently associated with atrophic gastritis and peptic ulcer disease. Epidemiological studies have suggested, and animal studies have confirmed, that the chronic infection of H. pylori is an important risk factor for the development of gastric carcinomas (1–5). Accordingly, the World Health Organization has classified H. pylori as a class I carcinogen. The presence of a functional cag pathogenicity island (cagPAI), a 40-kb DNA segment integrated in the H. pylori chromosome (6, 7), is associated with enhanced virulence as measured by the mucosal inflammation (8, 9). The cagPAI DNA segment contains genes constituting a type IV secretion apparatus, as well as the cagA gene that encodes an ≈145-kDa CagA protein (6, 7, 10). Compared with cagA− H. pylori strains, cagA+ strains significantly increase the risk of developing severe gastritis and gastric carcinoma (11–13).
H. pylori elicits its pathological activity at least partly through direct contact with the target gastric epithelial cells. Of note is an induction of a growth factor-like cellular morphological change termed the hummingbird phenotype, which is characterized by dramatic cell elongation that occurs by means of the attachment of cagA+ but not cagA− H. pylori strains to the cells (14, 15). During the bacterium–gastric epithelial cell interaction, H. pylori injects CagA directly into the attached cells by means of the bacterial type IV secretion apparatus (16–19). The translocated CagA protein localizes to the inner surface of the plasma membrane and subsequently undergoes tyrosine phosphorylation in the host cells by the Src family protein tyrosine kinase (20, 21). We have recently shown that CagA binds an Src homology 2 (SH2)-containing tyrosine phosphatase SHP-2 in a tyrosine phosphorylation-dependent manner and stimulates the phosphatase activity of SHP-2 (22). Membrane tethering and activation of SHP-2 by the tyrosine phosphorylated CagA are necessary and sufficient for the induction of the hummingbird phenotype. Because SHP-2 plays an important role in both cell growth and cell motility, deregulation of SHP-2 by CagA may be involved in the induction of abnormal proliferation and movement of gastric epithelial cells, a cellular condition eventually leading to gastritis and gastric carcinoma.
Although CagA is an H. pylori virulence factor, the presence of CagA is not sufficient for the prediction of disease outcome in H. pylori infection. Structural analyses have shown that the CagA protein varies in size in different strains (10, 23–25). The size variation correlates with the presence of a variable number of repeat sequences located in the C-terminal region of CagA and also involves amino acid sequence diversity both within and outside the repeat region. These sequence variations raise an intriguing possibility that the biological activity of CagA can vary from one strain to the next, which may influence the pathogenicities of different cagA+ H. pylori strains.
Here, we show that the potential of individual CagA to perturb host-cell functions is determined by the degree of SHP-2 binding activity, which depends in turn on the number and sequences of tyrosine phosphorylation sites. We also provide evidence that prevalent CagA proteins in East Asian countries are significantly more potent in binding SHP-2 and in inducing cellular morphological changes than are CagA proteins of Western isolates. Differences in the biological activities of Western and East Asian CagA proteins may underlie the striking difference in gastric cancer incidence between these two geographic areas.
Materials and Methods
Expression Vector Constructs.
The cagA gene was isolated from H. pylori standard strain NCTC11637 and was C-terminal hemagglutinin (HA)- or Flag-epitope-tagged. CagA mutants were generated from the HA-tagged, 11637-CagA by using a Chameleon site-directed mutagenesis kit (Stratagene). Human SHP-2 cDNA was tagged with Myc-epitope as described (22). To disable the N-terminal SH2 domain of SHP-2 (SHP-2-SH2ΔN-Myc), Arg-32 (R32) and His-53 (H53) were changed to alanine residues. Similarly, R138 and H169 were replaced with alanines to disable C-terminal SH2 domain function (SHP-2-SH2ΔC-Myc). SHP-2-SH2ΔNC-Myc was generated by substituting R32, H53, R138, and H169 with alanine residues. The DNAs were inserted into the pSP65SRα mammalian expression vector. All CagA constructs were HA-epitope-tagged unless otherwise noted.
Cell Culture and Transfection.
Human AGS gastric epithelial cells and monkey COS-7 cells were cultured in RPMI medium 1640 and DMEM, respectively, with 10% (vol/vol) FBS. CagA and SHP-2 expression vectors were transfected into the cells as described (22). Thirty μg of plasmids was transfected into AGS cells (1.8 × 106 cells) by using 30 μl of Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. COS-7 cells (1.5 × 106 cells) were transfected with 20 μg of plasmids by using the calcium phosphate method. The morphology of the AGS cells was observed at 17 h after transfection. AGS cells were transiently transfected with an expression vector for WT or mutant CagA (n = 3). Cells showing the hummingbird phenotype were counted in 10 different fields in each of three dishes (the area of one field = 0.25 mm2).
Antibodies.
HA epitope-specific polyclonal antibody Y-11 (Santa Cruz) and anti-HA monoclonal antibody 12CA5 were used as primary antibodies for immunoblotting and immunoprecipitation of HA-tagged CagA, respectively. Anti-Myc monoclonal antibody 9E10 and anti-Flag monoclonal antibody M2 (Sigma) were used for immunoblotting and immunoprecipitation. Anti-phosphotyrosine monoclonal antibody 4G10 was purchased from Upstate Biotechnology (Lake Placid, NY) and used for immunoblotting.
Immunoprecipitation and Immunoblotting.
AGS or COS-7 cells were harvested at 36 h after transfection and lysed in lysis buffer (50 mM Tris⋅HCl, pH 7.5/100 mM NaCl/5 mM EDTA/1% Brij-35) containing 2 mM Na3VO4, 2 mM PMSF, 10 μg/ml leupeptin, 10 μg/ml trypsin inhibitor, and 10 μg/ml aprotinin (22). The cell lysates were treated with the appropriate antibody, and then immune complexes were trapped on protein A- or protein G-Sepharose beads. The immunoprecipitates were subjected to SDS/PAGE. Proteins transferred to poly(vinylidene difluoride) (PVDF) membrane filters were incubated in primary antibodies at 0.3 μg/ml (anti-HA, anti-SHP-2, anti-Myc, or anti-phosphotyrosine antibody) or 6 μg/ml (anti-Flag antibody) in PBS containing 0.6% Tween-20, 1% skim milk, and 0.1% BSA and then visualized by using Western Blot Chemiluminescence Reagent (NEN). Intensities of chemiluminescence on the immunoblotted filter were quantitated by using a luminescent image analyzer (LAS-1000, Fuji).
Results
Identification of in Vivo Tyrosine Phosphorylation Sites in the EPIYA-Containing Repeat Sequence of CagA.
The CagA protein from H. pylori standard strain NCTC11637 (11637-CagA, see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org) possesses five glutamic acid–proline–isoleucine-tyrosine-alanine (EPIYA/D1/R1) motifs that are potential targets of tyrosine phosphorylation (10, 23). Phosphorylation of 11637-CagA was observed with WT but not mutant in which all of the tyrosine residues present in all of the EPIYA motifs were replaced by alanine (22). This result indicates that at least one of the EPIYA motifs was phosphorylated in vivo. Furthermore, the tyrosine phosphorylation was essential for the interaction of CagA with SHP-2.
First, we wished to know which EPIYA motif(s) was phosphorylated and was involved in SHP-2 binding in vivo. The first and second EPIYA motifs (designated EPIYA-A and EPIYA-B, respectively) are present in the CagA proteins specified by almost all isolates of H. pylori, whereas the remaining three EPIYA motifs (EPIYA-C) were made by duplication of an EPIYA-containing 34-amino acid sequence stretch (10). Because the sequence exists in various numbers ranging from 0 to 3 in most CagA proteins from H. pylori isolated in Western countries such as those of Europe, America, and Australia (Fig. 5 and Table 1, which are published as supporting information on the PNAS web site; refs. 23–25), we designated it the “Western CagA-specific sequence” (WSS; Fig. 1a). The major CagA proteins carried by Western isolates have a single WSS and, thus, are classified as EPIYA-A-B-C types (23, 24), whereas 11637-CagA is classified EPIYA-A-B-C-C-C type because it has three repeats of WSS.
Fig 1.
Identification of tyrosine phosphorylation sites of CagA and their roles in SHP-2 binding and morphological transformation. (a) The EPIYA motifs present in NCTC11637 CagA (Upper) and schematic view of the EPIYA mutants (Lower). Three repeats of the Western CagA-specific sequence (WSS) are each boxed. All CagA constructs were C-terminal HA-tagged. WT: WT 11637-CagA; PR: phosphorylation-resistant CagA. (b and c) AGS cells were transfected with WT-CagA, each of EPIYA mutant, or a control vector. CagA proteins then were immunoprecipitated from cell lysates with anti-HA. The immunoprecipitates (IP) and total cell lysates (TCL) were immunoblotted (IB) with antibodies as described. (d) AGS cells were transiently transfected with an expression vector for WT or EPIYA mutant of CagA. Cells showing the hummingbird phenotype were counted.
To determine the in vivo tyrosine phosphorylation sites of Western CagA, we generated EPIYA mutants, abCCC and ABccc, from 11637-CagA (Fig. 1a). In the abCCC mutant, EPIYA-A and EPIYA-B were converted into a phosphorylation-resistant EPIAA sequence. Similarly, three EPIYA-C motifs were replaced by EPIAA to make the ABccc mutant. In AGS cells, abCCC was efficiently phosphorylated, whereas ABccc was not (Fig. 1b), indicating that the tyrosine residues in EPIYA-C motifs of WSS are the major, if not exclusive, in vivo phosphorylation sites of Western CagA in AGS cells. Recently, Backert et al. and Stein et al. reported similar conclusions (20, 26). Next, we generated a series of 11637-CagA mutants in which three EPIYA-C motifs had been converted into EPIAA in all possible combinations (Fig. 1a). Upon expression in AGS human gastric epithelial cells, the levels of CagA phosphorylation were proportional to the number of EPIYA-C but were independent of the EPIYA-C positions (Fig. 1c), indicating that each of the three EPIYA-C motifs in 11637-CagA was equally phosphorylated in AGS cells.
Correlation Among Number of Tyrosine Phosphorylation Sites, SHP-2-Binding Activity, and Hummingbird Phenotype Induction of CagA.
CagA binds SHP-2 in a tyrosine phosphorylation-dependent manner (22). Upon binding, CagA activates SHP-2 and induces in gastric epithelial cells a highly elongated cell morphology termed the hummingbird phenotype (22). To determine the relationship between the degree of CagA phosphorylation and biological activity of CagA, we examined SHP-2 complex formation and the hummingbird phenotype induction by each of the EPIYA mutants. As expected, ABccc, which did not undergo tyrosine phosphorylation, failed to bind SHP-2. In contrast, CagA proteins having a single EPIYA-C, while receiving tyrosine phosphorylation, were capable of binding SHP-2. Elevated CagA phosphorylation, as a result of an increased number of EPIYA-C, significantly increased CagA-SHP-2 complex formation and potentiated the ability of CagA to induce the hummingbird phenotype (Fig. 1 c and d). Thus, among the Western CagA proteins, the number of EPIYA-C, which increases as a result of WSS duplication, is a critical determinant of CagA to disturb cell signaling as a virulence factor.
Involvement of the SH2 Domains of SHP-2 in CagA Binding.
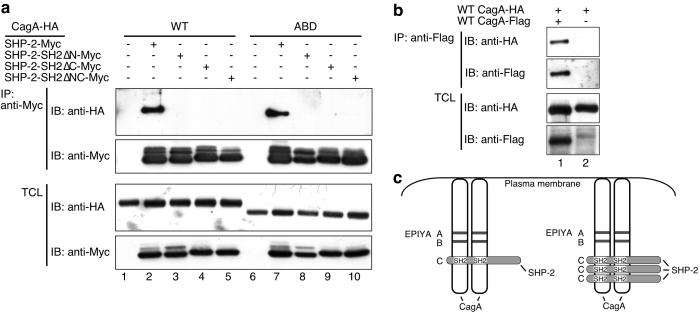
SHP-2 possesses two SH2 domains, the N-SH2 and C-SH2, each of which can independently bind a phosphotyrosine-containing peptide (27). The observation that CagA with a single EPIYA-C was still capable of binding SHP-2 suggested that occupation of one of the two SH2 domains of SHP-2 may be sufficient for the complex formation between CagA and SHP-2. To our surprise, however, inactivation of either of the SH2 domains by introducing point mutations abolished CagA-binding activity of SHP-2 (Fig. 2a). This result indicates that both of the SH2 domains are required for the stable complex formation with CagA. With this notion in mind, we wondered if CagA proteins are present as multimers such as dimers in the host cells. To address this, we performed cotransfection experiments and found that HA-tagged CagA and Flag-tagged CagA were capable of forming a physical complex within the cell (Fig. 2b). The result indicates that a single SHP-2 protein may interact simultaneously with a tyrosine-phosphorylated CagA dimer via its two SH2 domains to form a stable complex (Fig. 2c). Because ectopic coexpression of SHP-2 did not increase the CagA-CagA complex formation, CagA oligomerization did not seem to require SHP-2 (data not shown).
Fig 2.
Role of the SHP-2 SH2 domains in the interaction between SHP-2 and CagA. (a) COS-7 cells were cotransfected with expression vectors for HA-tagged CagA and Myc-tagged, WT or mutant SHP-2 lacking one or both of the functional SH2 domains. Cell lysates were subjected to immunoprecipitation (IP) with anti-Myc. Immunoprecipitates and total cell lysates (TCL) were immunoblotted (IB) with the indicated antibodies. N-SH2 domain, C-SH2 domain, and both of the SH2 domains were inactivated in SH2ΔN-Myc, SH2ΔC-Myc, and SH2ΔNC-Myc, respectively. WT: WT 11637-CagA. ABD variant was generated from WT 11637-CagA by replacing the entire EPIYA region with that of F32-CagA. (b) COS-7 cells were cotransfected with expression vectors for HA-tagged CagA and Flag-tagged CagA. Cell lysates were subjected to immunoprecipitation (IP) with anti-Flag. Immunoprecipitates and total cell lysates (TCL) were immunoblotted (IB) with the indicated antibodies. (c) A model of CagA–SHP-2 interaction.
Identification and Characterization of Tyrosine Phosphorylation Sequence Unique to East Asian CagA.
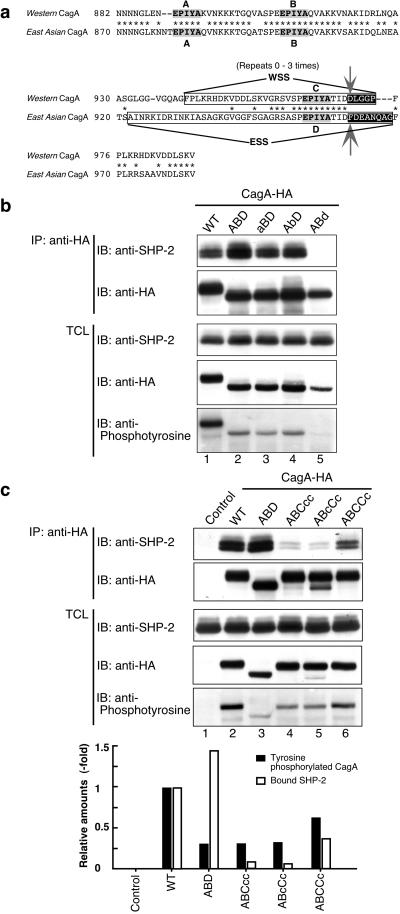
The amino acid sequence of the CagA proteins specified by H. pylori strains isolated in East Asian countries, where the incidence of gastric carcinoma is among the highest in the world, is significantly different from that of Western CagA (23–25, 28–30). Most notably, predominant East Asian CagA proteins do not have the WSS but, instead, possess a distinct sequence that we designated “East Asian CagA-specific sequence (ESS)” in the corresponding region (Fig. 3a, and see also Fig. 5; refs. 23 and 24). This observation raises an intriguing possibility that East Asian CagA is not functionally equal to Western CagA and the difference may influence the pathogenicities as well as clinical outcomes of different cagA+ H. pylori strains.
Fig 3.
Identification and functional characterization of East Asian CagA-specific sequence for tyrosine phosphorylation. (a) Sequence alignment of the EPIYA regions for Western (11637 strain) and East Asian CagA proteins. The Western CagA-specific sequence (WSS) and the East Asian (F32 strain) CagA-specific sequence (ESS) are boxed. Nonconserved residues at the C-terminal regions of EPIYA-C and EPIYA-D are black-boxed. Arrows indicate residues replaced for generation of ABC970DFcc and ABD961FD mutants of CagA. (b and c) AGS cells were transfected with WT-CagA, EPIYA mutant, or a control empty vector. CagA proteins were immunoprecipitated from the cell lysates with anti-HA. The immunoprecipitates (IP) and total cell lysates (TCL) were immunoblotted (IB) with antibodies as described. Protein expression levels of each CagA variants were normalized with band intensities obtained from anti-HA immunoblotting. The relative amounts of tyrosine phosphorylated CagA (black bars) and CagA-bound SHP-2 (white bars) were calculated with the values for WT 11637-CagA taken as 1. The aBD, AbD, and ABd variants were generated from ABD by replacing tyrosine residues constituting EPIYA-A, -B, and -D with alanine residues, respectively.
To address the above possibility, we wished to identify the tyrosine phosphorylation site of East Asian CagA proteins because they do not have a WSS. Intriguingly, ESS possesses an EPIYA motif, denoted EPIYA-D, and the sequence surrounding EPIYA-D of ESS is highly homologous to that surrounding EPIYA-C of WSS (Fig. 3a). This sequence information suggests that EPIYA-D undergoes tyrosine phosphorylation and subsequently constitutes a SHP-2-binding site that is specific to East Asian CagA. To determine whether EPIYA-D is in fact such a site, we generated a 11637-CagA mutant, ABD, in which the entire EPIYA region (from residue 868 to residue 1,086, containing EPIYA-A, EPIYA-B, and three EPIYA-C motifs) was replaced with that of F32-CagA (from residue 856 to residue 1,012, containing EPIYA-A, -B, and -D motifs; see Fig. 6, which is published as supporting information on the PNAS web site) isolated from a Japanese gastric cancer patient. We also generated a series of ABD-derivatives, aBD, AbD, and ABd, in which each of the EPIYA was replaced by EPIAA. Upon expression in AGS cells, ABD became phosphorylated at the tyrosine residue within EPIYA-D (Fig. 3b). This tyrosine phosphorylation was an essential prerequisite for the interaction of ABD with SHP-2 (Fig. 3b), and, again, the binding required the two SH2 domains of SHP-2 (Fig. 2a). To compare directly the SHP-2 binding activities of ESS and WSS, complex formations between SHP-2 and ABD (1xESS), ABCcc (1xWSS), ABcCc (1xWSS), ABCCc (2xWSS), or WT 11637-CagA (3xWSS) were examined (Fig. 3c). In AGS cells, the tyrosine phosphorylation levels of ABD were significantly lower than those of WT 11637-CagA or ABCCc, but were comparable with those of ABCcc or AbcCc, indicating that ESS and WSS are equally capable of phosphorylation in AGS cells. On the other hand, ABD showed substantially greater ability to form a complex with SHP-2 than did any of the tested types of Western CagA, including 11637-CagA having 3× WSS (Fig. 3c). This result indicates that the SHP-2 binding affinity of ESS is significantly higher than that of WSS.
Delineation of Amino Acid Residues That Determine SHP-2-Binding Affinity Among Distinctly Structured CagA Proteins.
SH2 domains recognize phosphopeptide motifs composed of phosphotyrosine (pY) followed by several C-terminal residues. Given this, we generated ABCDcc from ABCcc, which represents the major Western CagA proteins with a single WSS, by replacing the five residues immediately following pY (i.e., positions pY + 1 to pY + 5) with an eight-amino acid sequence from the corresponding region of F32-CagA (see Fig. 3a, black-boxed sequences). We also generated a point mutant of ABCcc, ABC970DFcc, in which the aspartic acid residue at the pY + 5 position, the only amino acid different between WSS and ESS within the C-terminal five residues from pY, was replaced with phenylalanine to mimic the ESS sequence (Fig. 3a arrows). Reciprocally, ABD961FD CagA mutant was made from ABD by substituting the phenylalanine residue at the pY + 5 position with aspartic acid to mimic WSS. When expressed in AGS cells, ABCDcc, ABC970DFcc, and ABD961FD underwent tyrosine phosphorylation at levels similar to those of ABCcc or ABD having a single phosphorylation site, but the levels were significantly less than those of 11637-CagA having three phosphorylation sites (Fig. 4a). Despite this, ABCDcc and ABC970DFcc exhibited much a stronger ability to form a complex with SHP-2 compared with the complex-forming activity of ABCcc (Fig. 4a). Indeed, their SHP-2-binding activities were even greater than that of 11637-CagA. On the other hand, binding of ABD961FD to SHP-2 was substantially reduced when compared with that of ABD. Hence, differential SHP-2-binding activities between Western CagA and East Asian CagA are attributable to the amino acid difference at the pY + 5 position. Finally, we compared induction of the hummingbird phenotype by these CagA derivatives. As expected, those having stronger SHP-2-binding activities (ABD, ABC970DFcc, and ABCDcc) exhibited substantially stronger activities to induce the hummingbird phenotype than those with weaker SHP-2-binding activities (ABCcc and ABD961FD) did (Fig. 4b). This finding indicates that prevalent East Asian CagA proteins are more active biologically than are most Western CagA proteins in disturbing host-cell signaling pathways as a virulence factor.
Fig 4.
Delineation of CagA amino acid sequences involved in SHP-2 interaction. (a) AGS cells were transfected with the indicated CagA expression vectors. WT or mutant CagA proteins then were immunoprecipitated from the cell lysates with anti-HA. The immunoprecipitates (IP) and total cell lysates (TCL) were immunoblotted (IB) with antibodies as described. ABCDcc mutant was made from ABCcc by replacing a 5-aa sequence that immediately follows the phosphotyrosine (pY) with an 8-aa sequence from the corresponding region of F32-CagA (see Fig. 3a, black-boxed sequences). A point mutant of ABCcc, ABC970DFcc, was made by replacing aspartic acid at the position pY + 5 with phenylalanine to mimic the ESS sequence (Fig. 3a arrows). Reciprocally, ABD961FD was made from ABD by substituting phenylalanine at the position pY + 5 with aspartic acid to mimic WSS. (b) AGS cells used in a then were examined for the induction of the hummingbird phenotype.
Discussion
We demonstrate in this work that CagA is heterogeneous with respect to its potential for undergoing tyrosine phosphorylation, SHP-2 binding, and induction of cellular morphological changes. Such biological diversities among different CagA proteins are caused by the variations in the number and sequences of tyrosine phosphorylation sites of the molecule. CagA proteins specified by H. pylori strains isolated in Western countries (such as those of Europe, America, and Australia) are characterized by the presence of a SHP-2-binding sequence termed WSS. Whereas predominant CagA proteins in Western isolates possess a single WSS, some have two or three WSS repeats created by sequence duplication at the C-terminal repeat region (10, 23–25). Our results indicate that the tyrosine residue constituting the EPIYA motif in WSS undergoes phosphorylation and becomes a docking site for the SH2 domains of SHP-2. Hence, CagA proteins with a large number of WSS are expected to be more active biologically than those with a small number of WSS because they interact more effectively with SHP-2 phosphatases and more severely perturb SHP-2-dependent signaling pathways. This notion is strongly supported by the observation that the ability of CagA to induce the hummingbird phenotype, a hallmark of the CagA biological activity, is proportional to the number of WSS. This proportionality in turn suggests the presence of cagA+ H. pylori strains with various levels of virulence, depending on the biological activity of CagA. Our conclusion is highly consistent with results of recent epidemiological studies showing that H. pylori strains carrying CagA proteins with multiple repeat sequences are associated with severe gastric mucosal atrophy, intestinal metaplasia, and gastric carcinoma (23, 24).
In contrast to the Western CagA proteins characterized by the presence of WSS, prevalent CagA proteins specified by H. pylori strains isolated in East Asian countries such as Japan and Korea have a distinct sequence termed ESS at the region corresponding to WSS. Replacement of WSS with ESS confers stronger SHP-2-binding activity and elevated morphology-transforming activity to Western CagA proteins. Thus, our results indicate that WSS is a low-affinity SHP-2-binding site, whereas ESS is a high-affinity site for SHP-2 binding. A majority of Western CagA proteins are less active because they have a single WSS and only weakly bind SHP-2, whereas some CagA proteins that have multiple WSS exhibit increased SHP-2-binding activity and, hence, are more active than those with a single WSS. In contrast, the predominant CagA proteins circulating in East Asia possess a high-affinity SHP-2-binding site within ESS and are more active biologically than most if not all of the Western CagA proteins. The CagA–SHP-2 interaction requires both the SH2 domains of SHP-2. De Souza and coworkers (31) recently reported that the two SH2 domains from SHP-2 bind to highly related sequences, and the consensus ligand-binding motif for the N- and C-SH2 domains of SHP-2 is pY-(S/T/A/V/I)-X-(V/I/L)-X-(W/F). Intriguingly, the consensus motif perfectly matches the SHP-2-binding site of East Asian CagA, pY-A-T-I-D-F. Furthermore, replacement of the pY + 5 position from W/F with any other amino acids, such as aspartic acid in the case of WSS in Western CagA, reduces the binding affinity to SHP-2. Hence, differential SHP-2-binding activities observed between WSS and ESS of CagA proteins are caused by the difference of a single amino acid at the pY + 5 position. Identification of the critical amino acid residues involved in SHP-2 binding, as presented in this work, may have important clinical value for H. pylori infection, as it enables us to predict the virulence of individual cagA+ strains based on the CagA sequences.
We propose a previously uncharacterized model for the CagA-SHP-2 interaction: a single SHP-2 is capable of binding two tyrosine-phosphorylated CagA proteins. This model is based on the observation that the interaction of SHP-2 with CagA having a single tyrosine phosphorylation site requires both the N- and C-SH2 domains of SHP-2 and is supported by the recent finding that both of the SH2 domains recognize very similar tyrosine phosphorylated sequences as noted above (31). Obviously, the model does not exclude the possibility that a SHP-2 molecule can also bind a single CagA protein having two or more WSS once they are tyrosine-phosphorylated. In either case, simultaneous occupancy of the two SH2 domains of SHP-2 may stabilize the CagA-SHP-2 complexes and potently stimulate the SHP-2 phosphatase activity, as has been reported (22, 32–35).
Our results indicate that the structural differences are substantially associated with functional differences between East Asian and Western CagA proteins. Endemic circulation of H. pylori populations carrying biologically more active CagA proteins in East Asian countries such as Japan and Korea, where the incidence of gastric carcinoma is among the highest in the world, may be involved in increasing the risk of gastric carcinoma in these geographic areas.
Supplementary Material
Acknowledgments
We thank Dr. Robert A. Weinberg for critical review of the manuscript. This work was supported by grants-in-aid for science research from the Ministry of Education, Science, Sports, and Culture of Japan, by a Research Grant from the Human Frontier Science Program Organization, by a grant of the Virtual Research Institute of Aging of Nippon Boehringer Ingelheim, and by a grant from the Akiyama Foundation.
Abbreviations
HA, hemagglutinin
WSS, Western CagA-specific sequence
ESS, East Asian CagA-specific sequence
SH2, Src homology 2
pY, phosphotyrosine
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AB090073–AB090075, AB090079–AB090081, AB090085, AB090087–AB090089, AB090091–AB090098, AB090100, AB090101, AB090103–AB090105, AB090107–AB090113, AB090115, AB090117–AB090127, AB090129, AB090132–AB090139, and AB090141–AB090154).
References
- 1.Huang J. Q., Sridhar, S., Chen, Y. & Hunt, R. H. (1998) Gastroenterology 114, 1169-1179. [DOI] [PubMed] [Google Scholar]
- 2.Danesh J. (1999) Aliment. Pharmacol. Ther. 13, 851-856. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe T., Tada, M., Nagai, H., Sasaki, S. & Nakao, M. (1998) Gastroenterology 115, 642-648. [DOI] [PubMed] [Google Scholar]
- 4.Honda S., Fujioka, T., Tokieda, M., Satoh, R., Nishizono, A. & Nasu, M. (1998) Cancer Res. 58, 4255-4259. [PubMed] [Google Scholar]
- 5.Uemura N., Okamoto, S., Yamamoto, S., Matsumura, N., Yamaguchi, S., Yamakido, M., Taniyama, K., Sasaki, N. & Schlemper, R. J. (2001) N. Engl. J. Med. 345, 784-789. [DOI] [PubMed] [Google Scholar]
- 6.Censini S., Lange, C., Xiang, Z., Crabtree, J. E., Ghiara, P., Borodovsky, M., Rappuoli, R. & Covacci, A. (1996) Proc. Natl. Acad. Sci. USA 93, 14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akopyants N. S., Clifton, S. W., Kersulyte, D., Crabtree, J. E., Youree, B. E., Reece, C. A., Bukanov, N. O., Drazek, E. S., Roe, B. A. & Berg, D. E. (1998) Mol. Microbiol. 28, 37-53. [DOI] [PubMed] [Google Scholar]
- 8.Kuipers E. J., Perez-Perez, G. I., Meuwissen, S. G. & Blaser, M. J. (1995) J. Natl. Cancer Inst. 87, 1777-1780. [DOI] [PubMed] [Google Scholar]
- 9.Peek R. M., Miller, G. G., Tham, K. T., Perez-Perez, G. I., Zhao, X., Atherton, J. C. & Blaser, M. J. (1995) Lab. Invest. 73, 760-770. [PubMed] [Google Scholar]
- 10.Covacci A., Censini, S., Bugnoli, M., Petracca, R., Burroni, D., Macchia, G., Massone, A., Papini, E., Xiang, Z., Figura, N., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 5791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaser M. J., Perez-Perez, G. I., Kleanthous, H., Cover, T. L., Peek, R. M., Chyou, P. H., Stemmermann, G. N. & Nomura, A. (1995) Cancer Res. 55, 2111-2115. [PubMed] [Google Scholar]
- 12.Parsonnet J., Friedman, G. D., Orentreich, N. & Vogelman, H. (1997) Gut 40, 297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rugge M., Busatto, G., Cassaro, M., Shiao, Y. H., Russo, V., Leandro, G., Avellini, C., Fabiano, A., Sidoni, A. & Covacci, A. (1999) Cancer 85, 2506-2511. [PubMed] [Google Scholar]
- 14.Segal E. D., Lange, C., Covacci, A., Tompkins, L. S. & Falkow, S. (1997) Proc. Natl. Acad. Sci. USA 94, 7595-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segal E. D., Cha, J., Lo, J., Falkow, S. & Tompkins, L. S. (1999) Proc. Natl. Acad. Sci. USA 96, 14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asahi M., Azuma, T., Ito, S., Ito, Y., Suto, H., Nagai, Y., Tsubokawa, M., Tohyama, Y., Maeda, S., Omata, M., et al. (2000) J. Exp. Med. 191, 593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein M., Rappuoli, R. & Covacci, A. (2000) Proc. Natl. Acad. Sci. USA 97, 1263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Odenbreit S., Puls, J., Sedlmaier, B., Gerland, E., Fischer, W. & Haas, R. (2000) Science 287, 1497-1500. [DOI] [PubMed] [Google Scholar]
- 19.Backert S., Ziska, E., Brinkmann, V., Zimny-Arndt, U., Fauconnier, A., Jungblut, P. R., Naumann, M. & Meyer, T. F. (2000) Cell. Microbiol. 2, 155-164. [DOI] [PubMed] [Google Scholar]
- 20.Stein M., Bagnoli, F., Halenbeck, R., Rappuoli, R., Fantl, W. J. & Covacci, A. (2002) Mol. Microbiol. 43, 971-980. [DOI] [PubMed] [Google Scholar]
- 21.Selbach M., Moese, S., Hauck, C. R., Meyer, T. F. & Backert, S. (2002) J. Biol. Chem. 277, 6775-6778. [DOI] [PubMed] [Google Scholar]
- 22.Higashi H., Tsutsumi, R., Muto, S., Sugiyama, T., Azuma, T., Asaka, M. & Hatakeyama, M. (2002) Science 295, 683-686. [DOI] [PubMed] [Google Scholar]
- 23.Yamaoka Y., Kodama, T., Kashima, K., Graham, D. Y. & Sepulveda, A. R. (1998) J. Clin. Microbiol. 36, 2258-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaoka Y., El-Zimaity, H. M., Gutierrez, O., Figura, N., Kim, J. G., Kodama, T., Kashima, K., Graham, D. Y. & Kim, J. K. (1999) Gastroenterology 117, 342-349. [DOI] [PubMed] [Google Scholar]
- 25.Yamaoka Y., Osato, M. S., Sepulveda, A. R., Gutierrez, O., Figura, N., Kim, J. G., Kodama, T., Kashima, K. & Graham, D. Y. (2000) Epidemiol. Infect. 124, 91-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Backert S., Moese, S., Selbach, M., Brinkmann, V. & Meyer, T. F. (2001) Mol. Microbiol. 42, 631-644. [DOI] [PubMed] [Google Scholar]
- 27.Hof P., Pluskey, S., Dhe-Paganon, S., Eck, M. J. & Shoelson, S. E. (1998) Cell 92, 441-450. [DOI] [PubMed] [Google Scholar]
- 28.Miehlke S., Kibler, K., Kim, J. G., Figura, N., Small, S. M., Graham, D. Y. & Go, M. F. (1996) Am. J. Gastroenterol. 91, 1322-1325. [PubMed] [Google Scholar]
- 29.Alm R. A., Ling, L. S., Moir, D. T., King, B. L., Brown, E. D., Doig, P. C., Smith, D. R., Noonan, B., Guild, B. C., deJonge, B. L., et al. (1999) Nature 397, 176-180. [DOI] [PubMed] [Google Scholar]
- 30.Kersulyte D., Mukhopadhyay, A. K., Velapatino, B., Su, W., Pan, Z., Garcia, C., Hernandez, V., Valdez, Y., Mistry, R. S., Gilman, R. H., et al. (2000) J. Bacteriol. 182, 3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Souza D., Fabri, L. J., Nash, A., Hilton, D. J., Nicola, N. A. & Baca, M. (2002) Biochemistry 41, 9229-9236. [DOI] [PubMed] [Google Scholar]
- 32.Lechleider R. J., Sugimoto, S., Bennett, A. M., Kashishian, A. S., Cooper, J. A., Shoelson, S. E., Walsh, C. T. & Neel, B. G. (1993) J. Biol. Chem. 268, 21478-21481. [PubMed] [Google Scholar]
- 33.Sugimoto S., Wandless, T. J., Shoelson, S. E., Neel, B. G. & Walsh, C. T. (1994) J. Biol. Chem. 269, 13614-13622. [PubMed] [Google Scholar]
- 34.Pluskey S., Wandless, T. J., Walsh, C. T. & Shoelson, S. E. (1995) J. Biol. Chem. 270, 2897-2990. [DOI] [PubMed] [Google Scholar]
- 35.Eck M. J., Pluskey, S., Trub, T., Harrison, S. C. & Shoelson, S. E. (1996) Nature 379, 277-280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.