Abstract
The cancer chemopreventive properties of selenium compounds are well documented, yet little is known of the mechanism(s) by which these agents inhibit carcinogenesis. We show that selenium in the form of selenomethionine (SeMet) can activate the p53 tumor suppressor protein by a redox mechanism that requires the redox factor Ref1. Assays to measure direct reduction/oxidation of p53 showed a SeMet-dependent response that was blocked by a dominant–negative Ref1. By using a peptide containing only p53 cysteine residues 275 and 277, we demonstrate the importance of these residues in the SeMet-induced response. SeMet induced sequence-specific DNA binding and transactivation by p53. Finally, cellular responses to SeMet were determined in mouse embryo fibroblasts wild-type or null for p53 genes. The evidence suggests that the DNA repair branch of the p53 pathway was activated. The central relevance of DNA repair to cancer prevention is discussed.
Keywords: cancer chemoprevention, selenium, APE/Ref1, p53 tumor suppressor gene, DNA repair
Selenium compounds in various forms are cancer-preventive. They have a 20-year history of mammary (1) and colon (2) cancer prevention in rodent models and are in phase II and III clinical trials for prostate cancer prevention (3). Several chemical forms of selenium have been used in laboratory studies. The prototype forms are sodium selenite, which causes single- and double-strand-break DNA damage, and selenomethionine (SeMet), which is relatively nontoxic and non-DNA-damaging (4–6). The mechanism(s) by which selenium compounds exert their anticancer properties is not known, although proposed mechanisms include cytosine methyltransferase inhibition (7), control of tumor angiogenesis (8), and inhibition of carcinogen bioactivation (9). Even less is known of the molecular genetic determinants of selenium action.
SeMet is the major component of dietary selenium, of which the recommended daily allowance (RDA) by the U.S. Food and Drug Administration is 50 μg per day. Cancer preventive use of selenium typically consists of 200 μg per day, exceeding the RDA by fourfold with no toxicity (3). Whether given in the diet or applied to tissue culture cells, SeMet undergoes a transsulfuration reaction through a selenohomocysteine intermediate to form selenocysteine, which is then incorporated specifically by a selenocystyl-tRNA into cellular selenoproteins, including thioredoxin reductase and glutathione peroxidase (10). Selenium increases the specific activity of these enzymes and promotes a cellular reducing environment (10). A number of cellular proteins are substrates for reduction by the thioredoxin reductase system, including Ref1 (redox factor-1) and p53 (11–13), which have been shown to interact under reducing conditions (11).
Activation of the p53 Tumor Suppressor.
In its normal role, p53 is a transcription factor that activates a number of downstream genes that function in cellular responses to DNA damage. Underscoring its importance in cancer, p53 is mutated and nonfunctional in most human cancers. We show that SeMet can activate p53 by a nongenotoxic mechanism that requires the redox factor Ref1 (13). The implication is that cells that retain functional p53, including perhaps p53+/− heterozygous cells, may be amenable to selenium chemoprevention. Indeed, we show that SeMet exerts differential effects on p53 wild-type vs. p53-null cells.
Materials and Methods
Cell Lines and Treatments.
We used cell lines carried over from previous use in other studies, whose p53 strategy is well documented (14–17). H1299 human lung cancer cells carrying deletions in both p53 alleles (ref. 14; hereafter referred to as p53-null) were transiently transfected with pCMV-p53 encoding wild-type human p53 48 h before SeMet treatments. Transient assays were chosen to minimize uncertainties because of genetic alterations intrinsic to the cell line. Other experiments used matched isogenic pairs of p53 wild-type and p53-null mouse embryo fibroblasts (MEFs; ref. 16) to demonstrate differential effects of SeMet dependent on p53 status. Additional matched pairs of p53 wild-type and p53-mutant (Val-143 → Ala) lines were used as indicated. Cells were treated with 20 μM SeMet (Sigma) for 15 h to induce the SeMet response (18). As a control, we used pyrrolidine dithiocarbamate (PDTC; Sigma), which is known to oxidize p53 cysteine residues (19). Additionally, we used a dominant–negative mutant redox-factor-1 (Ref-DN), which carried an alanine substitution for cysteine codon 65, predicted to block redox signaling to p53 and other Ref1-responsive proteins (20). The mutant Ref1 was driven by the cytomegalovirus (CMV) promoter of pcDNA3.1 to generate pcDNA-Ref65. A separate set of experiments confirmed the ability of the Ref65 mutant to act as a dominant–negative in overriding endogenous wild-type Ref1 (results not shown).
Direct p53 Reduction/Oxidation Assay (19).
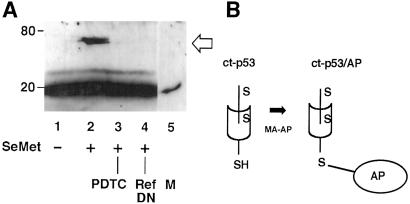
H1299 cells (p53-null) were transiently transfected with pCMV-p53 encoding wild-type human p53, 48 h before SeMet treatments. The fraction of reduced vs. oxidized p53 was determined by an assay that relies on the sulfhydryl-reactive reagent N-ethylmaleimide (NEM; Sigma). Cells were treated as indicated (Fig. 1) and then harvested by scraping in SEE buffer (0.1 M. sodium phosphate, pH 7.0/5 mM EDTA/5 mM EGTA) containing 20 mM NEM. Lysates were dialyzed overnight against two changes of PBS and then treated with 20 mM DTT for 30 min; they were dialyzed again and treated with 10 μg/ml 3-(maleimido-propioryl)-biocytin (MPB; Sigma) for 30 min. Samples were kept on ice throughout and dialysis was conducted at 4°C. The treatment protocol selectively labels the oxidized fraction of p53 cysteine residues by first blocking free sulfhydryl groups with NEM (19). Immune complexes were collected by using anti-p53 antibody Ab421, subjected to SDS/PAGE, and transferred to nitrocellulose. MPB-labeled p53 was detected by streptavidin-HRP conjugate. Controls consisted of bovine pancreatic chymotrypsinogen, which has five disulfide linkages but no free sulfhydryls, and rabbit muscle aldolase, which has eight free sulfhydryls but no disulfide linkages (results not shown). Alternatively, the reduced fraction of p53 cysteine residues was detected by a modified gel-shift assay (21). Cell lysates were collected in PBS containing 1% Triton X-100, then placed in reaction with EZ-link maleimide-activated alkaline phosphatase (Pierce). Lysates were subjected to immunoblotting by using Ab421 to detect p53 migrating as an alkaline-phosphatase-p53 complex. To identify the specific cysteine residue(s) that were redox-responsive to SeMet, we used a carboxyl-terminal peptide of p53 containing only p53 cysteines codons 275 and 277, beginning translation at internal methionine codon 246. Expression of the carboxyl-terminal peptide (ct-p53) was driven by pcDNA3.1 and migrated as a 20 kDa protein (Fig. 2). The covalently linked ct-p53/alkaline phosphatase protein migrated as an approximate 70-kDa band (Fig. 2). The carboxyl-terminal p53 fragment was generated by PCR using wild-type p53 cDNA as template. Primers 5′-TTTAAGCTTATGGGCGGCATGAACCGG-3′ and 5′-TTGAATTCTCAGTCTGAGTCAGGCCC-3′ were used in a 35-cycle standard procedure. After purification by PCR Magicprep (Promega), the 600-bp amplification product was ligated in pcDNA3.1 by using HindIII and EcoRI sites engineered in the primers.
Fig 1.
Reduction response of p53 protein to SeMet. (A) Reduction/oxidation of full-length p53. H1299 cells (p53-null) were transiently transfected with a wild-type p53 expression plasmid. Cells were treated with 20 μM SeMet and p53 protein reduction/oxidation assayed (19). Briefly, oxidized cysteine sulfhydryl residues were detected by lysis in the presence of NEM, which reacts with and blocks free sulfhydryls. Disulfide linkages then were reduced by DTT and labeled with the biotinylation reagent MPB; p53 was immunoprecipitated with Ab421, and biotinylated p53-detected by blotting with streptavidin HRP. Alternatively, p53 sulfhydryls were labeled directly with MPB to detect reduced p53 forms. The lanes reflect SeMet and other treatments. Lane 1, untreated cells; lane 2, cells treated with SeMet exhibit a p53 redox response; lane 3, abrogation of the p53 reduction response by dominant–negative Ref1 mutant (Ref-DN); lane 4, abrogation of the p53 reduction response by PDTC, known to oxidize p53 cysteines (positive control). Reduced and oxidized p53 cysteine residues were inversely correlated in SeMet-treated cells. Immune complexes were probed with Ab421 to ascertain equivalent gel loading. (B) Diagram of redox chemistry methodology used in A. Sulfhydryl groups were first covalently blocked by reaction with NEM (S-H to S-R modification), then disulfide-reduced by DTT and labeled with MPB for subsequent streptavidin detection (19). Alternatively, p53 sulfhydryls were labeled directly with MPB to detect reduced p53 forms.
Fig 2.
Redox regulation of p53 Cys-275 and/or Cys-277. (A) H1299 cells (p53-null) were transiently transfected with an expression plasmid encoding the carboxyl-terminal one-third of p53 protein. The carboxyl-terminal p53 peptide (ct-p53) migrates as a 20-kDa band. Reaction with maleimide-activated alkaline phosphatase (MA-AP) causes ct-p53 to migrate as a 70-kDa ct-p53/alkaline phosphatase (AP) conjugate, detected by Ab421 to p53. The lanes reflect SeMet and other treatments. Lane 1, untreated cells; lane 2, cells treated with SeMet exhibit a ct-p53 reduction response; lane 3, abrogation of the reduction response by PDTC; lane 4, abrogation of the reduction response by dominant–negative Ref1 mutant (Ref-DN); lane 5, peptide alone without modification by MA-AP, used to mark the position of unmodified peptide (M). (B) Diagram of redox chemistry used in A. A gel-shift assay was used to detect directly reduced forms of p53, specifically, of p53 residues 275 and/or 277. Free sulfhydryl groups were labeled with MA-AP, which forms a covalent linkage of sulfhydryl groups to AP. The resulting ctp53/AP conjugate migrates at approximately 70 kDa.
Ab1620 Epitope Assay.
H1299 cells (p53-null) were transiently transfected with pCMV-p53 encoding wild-type human p53 48 h before SeMet treatments. Human colon cancer RKO cells carrying endogenous wild-type p53 genes were used in some experiments. The presence of the 1620 epitope on p53 correlates with p53 activity (22). Cells were treated as indicated (Fig. 3), then lysates were subject to immunoprecipitation with Ab1620 (Oncogene Research Products, Cambridge, MA). After SDS/PAGE and transfer to nitrocellulose, p53 was detected by the DO1 antibody to p53 (Oncogene Research Products). To control for total p53 loading, lysates were probed directly with D01 without Ab1620 immunoprecipitation. Phosphorylated p53 was detected with phospho-p53 antibody sampler kit (Cell Signaling Technology, Beverly, MA).
Fig 3.
Promotion of p53 1620+ conformation in response to SeMet. H1299 cells transiently transfected with wild-type p53 were treated with 20 μM SeMet for 15 h, and immune complexes were collected by using Ab1620, a conformation-sensitive epitope present on active p53. The lanes reflect SeMet and other treatments. Lane 1, untreated cells; lane 2, cells treated with SeMet exhibit 1620+ p53; lane 3, abrogation of the 1620+ response by Ref-DN mutant; lane 4, abrogation of the 1620+ response by PDTC. (A) Immunoblots of p53 protein in 1620+ immune complexes. Total p53 was detected by blotting of lysates with antibody D01 without immunoprecipitation. (B) Quantification of three independent experiments, mean ± SD. P < 0.02, by Wilcoxon rank-sum test.
Electrophoretic Mobility Shift Assays (EMSA).
MCF7 and RKO cells carrying wild-type p53 genes or p53-null H1299 cells transfected with pCMV-p53 were used. Assays were conducted as described (15). Nuclear extracts were prepared from cells treated as indicated (Fig. 4A) and were incubated in the presence of 32P-labeled oligonucleotide corresponding to the p53-binding site of the human Gadd45 gene (Santa Cruz Biotechnology). Reactions contained anti-p53 antibody Ab421, known to generate supershifted p53 complexes (15). After electrophoresis in 6% native polyacrylamide gels (Invitrogen), shifted complexes were visualized by exposure to x-ray film. Negative controls consisted of a mutant oligonucleotide (Santa Cruz Biotechnology), omission of Ab421 antibody, and use of extracts from cells lacking p53. Extracts of UV-irradiated cells served as a positive control for p53 activation.
Fig 4.
Transcriptional activation of p53 by SeMet. (A) EMSA showing binding of p53 to a p53-responsive gene sequence. Extracts were prepared from H1299 cells transiently transfected with p53 expression plasmids. Lane 1, untreated cells; lane 2, cells treated with 20 μM SeMet for 15 h showed enhanced sequence-specific binding of p53; lane 3, cells treated with PDTC abrogates p53 activation by SeMet; lane 4, cells treated with UV radiation (positive control); lane 5, absence of binding to mutant oligonucleotide sequence (negative control; Santa Cruz Biotechnology). Arrow denotes the position of the Ab421-p53 supershifted complex. (B) Dependence of supershifted complex formation on Ab421. Lane 6, absence of Ab421; lane 7, addition of Ab421; lane 8, addition of Ab421 and DO1. Open arrow denotes the position of the doubly shifted complex. (C) Activation of a p53-responsive reporter gene by SeMet. H1299 cells were cotransfected with p53 expression vectors and pG13-CAT, a reporter construct that carries 13 p53 binding sites to measure p53 activation. Lane 1, untreated cells; lane 2, treatment with SeMet enhances p53-dependent transcription; lane 3, abrogation by PDTC; lane 4, abrogation by Ref-DN mutant; lane 5, lack of pG13-CAT activation by a p53-mutant (Val-143 → Ala; negative control). Quantification was by a CAT enzyme ELISA assay. Relative CAT enzyme units are shown; mean ± SD from five independent experiments (P < 0.01 by t test, using SIGMAPLOT software).
p53-Transactivation Assays (14).
The p53-responsive reporter plasmid pG13-CAT carries 13 p53-binding sites fused to a minimal promoter element 5′ of the bacterial chloramphenicol acetyltransferase (CAT) gene (14). The construct was introduced by stable transfection to MCF7 cells and selected for G418-resistance. Cells were treated as indicated (Fig. 4B). A derivative MCF7 line carrying a Ref1 mutant was similarly used. Some experiments used transient transfection of pG13-CAT together with the wild-type p53 vector pCMV-p53, in p53-null H1299 cells. Similar results were obtained irrespective of whether stable or transient transfectants were challenged with SeMet.
Cell Survival Assays (16).
To ascertain a role for p53 in the cellular response to selenium, MEFs wild-type or null for p53 genes were treated with 20 μM SeMet for 15 h and then treated with UV radiation. Untreated cells served as controls. Cell yield was determined after 7 days by a thiazolyl blue viability assay (Fig. 5A).
Fig 5.
Cellular responses to SeMet involving p53. (A) SeMet protected p53 wild-type but not p53-null cells from UV radiation. MEF cells were treated with 20 μM SeMet for 15 h, then with UV radiation at the indicated doses. Cell yield was determined after 7 days. SeMet protected wild-type MEFs, whereas p53-null cells remained UV sensitive. SeMet had no effect on survival of either cell line in the absence of DNA damage. Shown is the mean ± SD of three independent determinations (P < 0.02 by Wilcoxon rank-sum test). (B) DNA repair induction by SeMet in p53 wild-type but not p53 mutant cells. An isogenic matched pair of MCF7 cell lines were used (ref. 17). DNA repair was measured by HCR of a UV-damaged reporter gene. Cells carrying a p53 mutant (Val-143 → Ala) did not show induction of the HCR response (17); PDTC abrogated SeMet induction of the HCR response. Shown is the mean ± SD of five independent experiments (P < 0.01 by t test using SIGMAPLOT software).
Host-Cell Reactivation (HCR) Assays (15).
DNA repair was determined by HCR in a matched isogenic pair of p53 wild-type and p53-mutant cell lines, MCF7 and MCF7-mutp53 (Val-143 → Ala; ref. 17). The assay measures the capacity of a cell line to remove transcription-blocking UV lesions (15). UV-irradiated pSV2-CAT plasmid estimated to contain 20 UV photoproducts per molecule, was introduced by using Fugene reagent; cells were harvested for determination of CAT activity 60 h later. SeMet was added to the medium at a final concentration of 20 μM for the final 24 h of incubation.
Results
Biochemical Assays of p53 Activation by SeMet.
The mechanism of p53 activation by SeMet was explored. We used H1299 cells lacking p53 genes, wherein p53 expression vectors were transiently introduced. We examined redox regulation of full-length p53, as well as a truncated p53 allele (ct-p53 discussed below). Cells were treated with SeMet as described (18). By using a direct assay for p53 reduction/oxidation based on reacting free sulfhydryl groups with NEM (19), we found that SeMet promoted the reduction of p53 cysteine residues (Fig. 1). The introduction of a dominant–negative Ref1 (Ref-DN) mutant (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org) blocked p53 reduction, indicating a role for Ref1 in the SeMet response. Besides the Ref-DN mutant, an RNAi inhibitor also was used to deplete cells of wild-type Ref1. Inhibition of Ref1 by RNAi similarly blocked the p53 reduction response to SeMet (results not shown). Pyrrolidine dithiocarbamate (PDTC), a compound known to promote p53 oxidation (19), was used as a control. Like the Ref-DN mutant, PDTC blocked p53 reduction by SeMet. By using a carboxyl-terminal p53 peptide (ct-p53) containing only Cys-275 and -277, the peptide was found to be redox-responsive to SeMet (Fig. 2). Thus, p53 cysteine residue 275 or 277 is clearly identified as a substrate for SeMet-dependent redox regulation.
Epitope mapping showed that SeMet treatment revealed the antibody Ab1620 epitope on full-length p53. Epitope Ab1620 expression was promoted by SeMet and was blocked by the Ref-DN mutant or by PDTC (Fig. 3). Hence, Ab1620 expression correlated with the reduced form of p53 protein (Fig. 1). EMSA assays showed direct binding of p53 to a canonical p53-binding site, enhanced in extracts of SeMet-treated cells (Fig. 4A). An in vivo corollary to the EMSA result was obtained by the use of a p53-responsive reporter plasmid, pG13-CAT, consisting of thirteen p53-binding sites and a minimal promoter driving the CAT gene (14). Expression of the CAT reporter was induced by SeMet and was blocked by the Ref1-DN mutant or by PDTC (Fig. 4B). We conducted Western blots of Gadd45, an endogenous target gene that is transactivated by p53, and found that its induction followed the pattern of pG13-CAT induction (results not shown).
Taken together, the data suggest that SeMet activates p53 by a mechanism involving the redox factor Ref1. We asked whether p53 might be phosphorylated in response to SeMet, as is the case for the DNA-damage response (23). Using antibodies to phospho-p53 serine residues 6, 9, 15, 20, 37, 46, or 392, no evidence of p53 phosphorylation was observed (results not shown). Hence, the mechanism of p53 activation by SeMet differs from that caused by DNA damage (23, 24). Cells treated with SeMet alone showed no evidence of DNA damage (18) and, in fact, protected cells from exogenous DNA damage (see next section). Thioredoxin reductase (TR) activity was assayed as an indicator of the cellular reducing environment in SeMet-treated cells (25). TR-specific activity was enhanced three- to tenfold by SeMet (results not shown).
Cellular Effect of p53 Activation by SeMet.
By using transient expression assays, we showed the molecular events of p53 activation by SeMet in Figs. 1–4. Based on these data, we hypothesized that cells with normal p53 would respond differently to SeMet treatment than p53-null cells. MEF cells wild-type or null for p53 genes were treated with SeMet in concentrations ranging from 0 to 40 μM. At the SeMet concentrations used, we observed no growth inhibition nor apoptosis in either cell line (results not shown; and refs. 18 and 26). The rationale for investigating apoptosis was twofold: (i) one potential mechanism for cancer prevention is elimination of damaged cells by apoptosis; and (ii) p53 has been associated with apoptosis in some cell types. On the other hand, endogenous levels of DNA damage were perhaps insufficient to trigger apoptosis.
We used UV radiation to produce DNA damage, which then might uncover differences between p53 wild-type or p53-null cells treated with SeMet. Consistent with our recent findings in normal human fibroblasts (18), wild-type MEFs treated with SeMet were protected from UV radiation (Fig. 5A), whereas p53-null MEFs were unaffected by SeMet and remained UV sensitive (16, 27–29). We further tested a role for p53 by HCR experiments to measure DNA repair of a UV-damaged reporter gene in p53 wild-type or p53-mutant (Val-143 → Ala) cell lines. SeMet enhanced HCR in p53 wild-type but not the p53-mutant cells (Fig. 5B). Similar results were obtained by using an ELISA assay to detect 6–4 photoproducts (16). SeMet treatment enhanced the removal of 6–4 photoproducts from genomic DNA of UV-irradiated MEFs (results not shown).
It was important to validate that, before the use of DNA damage co-treatments as in Fig. 5A, SeMet treatment by itself did not cause DNA damage. The lack of DNA damage by SeMet alone was confirmed by alkaline electrophoresis assays (ref. 18, and results not shown). Indeed, SeMet alone in the 10–40 μM range apparently was not toxic to cells, as cells remained in cycle, as confirmed by flow cytometry (not shown) and by the demonstration of an unimpaired cell doubling time in the presence of SeMet (18). By necessity, cells were pretreated with SeMet in experiments employing UV radiation. Pretreatment was not required, however, as we conducted experiments in which cells were treated simultaneously with DNA-damaging agents 4-nitroquinoline-1-oxide (4NQO) or carboplatin in the presence or absence of SeMet (results not shown). Similar to UV radiation, SeMet protected p53 wild-type cells from 4NQO or carboplatin-induced DNA damage (Fig. 5B). Cells null for p53 genes remained sensitive to 4NQO or carboplatin (refs. 16 and 27–29; and results not shown).
Discussion
It is becoming clear that redox regulation is critical for a number of proteins. In the case of p53, an important implication is that p53 activity can be augmented by antioxidant mechanisms, thereby protecting cells from DNA damage. Inasmuch as DNA repair begins immediately after DNA damage, basal p53 activity and downstream effector gene expression are critical to the repair response (15, 16, 28, 29). Our study demonstrates that selenium concentration is a determinant of basal p53 activity, and that protection from DNA damage by SeMet is p53-dependent.
There are 10 cysteine residues in full-length human p53 that are potentially subject to redox regulation (24). We used a 20 kDa carboxyl-terminal fragment of p53 (ct-p53) to define further the p53 residue(s) responsive to selenium treatment. The ct-p53 construct initiates at Met-246, and contains only two cysteine residues at codons 275 and 277. It is clear (Fig. 2) that ct-p53 is strongly reduced in the selenium response, thereby implicating Cys-275 and/or Cys-277, although we cannot exclude a role for additional cysteines in full-length p53. Importantly, Cys-277 constitutes part of the active (sequence-specific DNA binding) site (24), consistent with our results showing enhanced sequence-specific binding in the presence of SeMet (Fig. 4A).
It is likely that selenium effects are pleiotropic. Most relevant in the present study is that SeMet differentially affected cells wild-type or null for p53 genes (Fig. 5A). SeMet protected p53 wild-type cells from DNA damage, whereas p53-null cells were unprotected. We used UV radiation as an exogenous source of DNA damage, because endogenous levels of DNA damage were too low to detect. The implication is that SeMet may, in a chemopreventive context, promote the repair of damage arising from endogenous sources, although the level of induced DNA damage in response to UV radiation is higher than would be encountered under normal conditions.
Among the DNA repair genes that are p53-regulated are Gadd45a and p48XPE. Both gene products participate in nucleotide excision repair (NER), the pathway for repair of UV damage, as well as important carcinogens, including benzo(a)pyrene (30). It is mainly the global genomic repair (GGR) subpathway of NER that is compromised in cells lacking p53 or Gadd45a genes (16, 31). Notably, GGR defects are often associated with carcinogenesis. Human patients defective for the xeroderma pigmentosum XPC gene exhibit faulty GGR but normal transcription-coupled DNA repair (TCR) and are cancer prone (discussed in ref. 32). Mice lacking XPC or Gadd45a genes were prone to chemical carcinogenesis (33, 34) and showed increased mutagenesis of a germline LacZ reporter gene (34). Hence, modulating p53 for NER would be predicted to enhance lesion removal relevant to carcinogenesis initiation protocols such as using dimethylbenzathracine or carcinogens associated with human carcinogenesis including benzo(a)pyrene, aflatoxins, or 2-acetylaminofluorene (33). On the other hand, endogenous mutagens associated with oxygen-free radicals may play a more prominent role in human cancers (35). A subset of lesions induced by reactive oxygen species are repaired by NER (36), and a GGR defect in repair of this type of lesion would likely contribute to a cancer-prone condition. Promoting the GGR branch of NER would be predicted to counter carcinogenesis.
It is noteworthy that we observed no apoptosis nor cell-cycle arrest in this study and a previous one (ref. 18 and results not shown). One possible explanation for this finding is that additional modifications such as phosphorylation may be required to activate p53 for apoptosis, inasmuch as the redox state is only one level of p53 regulation (24). We did not observe any detectable p53 phosphorylation (results not shown), although other possible modifications cannot be excluded. Hence, promoting the reduced state in the absence of other signals might selectively activate the DNA-repair branch of the p53 pathway. Although cell type differences also contribute to cellular endpoints, as lymphoid cells have a lower threshold for apoptosis and are more prone to apoptosis than the epithelial and fibroblast cells used in this study (discussed in ref. 37). We do not exclude the possibility of other p53-dependent endpoints in other cell types.
It is perhaps surprising that SeMet affected cell survival in a p53-dependent manner (Fig. 5A), inasmuch as GGR contributes little to cell survival after DNA damage (28). Of course, cell survival is a complex endpoint. Although SeMet alone did not cause cell-cycle arrest (18), UV irradiation would elicit a cell-cycle checkpoint response in p53 wild-type cells, lacking in p53-null cells, that may contribute to protection from DNA damage (16). To address the role of NER and GGR in protection from UV radiation, we used human fibroblasts carrying defective NER genes. SeMet treatment did not protect xeroderma pigmentosum type A fibroblast cells from UV radiation, evidence that NER was the key pathway affected (results not shown). We also used fibroblasts from Cockayne's syndrome (CSB) patients, which are defective for TCR but functional for GGR. SeMet treatment did not protect CSB cells from UV radiation (Fig. 7, which is published as supporting information on the PNAS web site), evidence that TCR is required for protection from DNA damage. Hence, the connection of the cell survival results to GGR is indirect (16). However, in the context of chemoprevention, these findings demonstrate the differential effects of selenium on p53 wild-type and p53-null cells.
In conclusion, we show that SeMet can activate p53 by a redox mechanism independent of DNA damage. At the selenium concentrations used, within the physiological range of clinical studies (38), p53-dependent DNA repair was activated. The DNA-repair branch of the p53 pathway is well documented (15, 16, 39, 40), yet less is known of the mechanism. Deletion of either Gadd45a or p48XPE genes recapitulates the NER defect observed in p53-null cells (16, 40), implicating these gene products in the process. Hence, the mechanisms that govern the expression of this subset of p53-dependent genes will be important to ascertain and may differ from other p53-regulated genes. Indeed, the Gadd45a gene is regulated not only by p53 but also by the BRCA1 tumor suppressor (41). Augmentation of DNA repair is emerging as an important paradigm in tumor suppression and cancer chemoprevention.
Supplementary Material
Acknowledgments
This work was supported by American Cancer Society Grant RSG-02-028-01 CNE (to M.L.S.).
Abbreviations
SeMet, selenomethionine
Ref1, bifunctional protein encoding redox factor-1
MEF, mouse embryo fibroblasts
PDTC, pyrrolidine dithiocarbamate
Ref-DN, dominant–negative mutant redox-factor-1
CMV, cytomegalovirus
NEM, N-ethylmaleimide
MPB, 3-(maleimido-propioryl)-biocytin
CAT, chloramphenicol acetyltransferase
HCR, host-cell reactivation
PDTC, pyrrolidine dithiocarbamate
NER, nucleotide excision repair
GGR, global genomic repair
AP, alkaline phosphatase
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 13969.
References
- 1.Ip C. (1981) Cancer Res. 41, 4386-4390. [PubMed] [Google Scholar]
- 2.Reddy B. S., Sugie, S., Maryyama, H., el-Bayoumy, K. & Marra, P. (1987) Cancer Res. 47, 5901-5904. [PubMed] [Google Scholar]
- 3.Nelson M. A., Porterfield, B. W., Jacobs, E. T. & Clark, L. C. (1999) Semin. Urol. Oncol. 17, 91-96. [PubMed] [Google Scholar]
- 4.Lu J., Jiang, C., Kaeck, M., Ganther, H., Vadhanavikit, S., Ip, C. & Thompson, H. J. (1995) Biochem. Pharmacol. 50, 213-219. [DOI] [PubMed] [Google Scholar]
- 5.Sinha R., Said, T. K. & Medina, D. (1996) Cancer Lett. (Shannon, Irel.) 107, 277-284. [DOI] [PubMed] [Google Scholar]
- 6.Stewart M. S., Spallholtz, J. E., Neldner, K. H. & Pence, B. C. (1999) Free Radical Biol. Med. 26, 42-48. [DOI] [PubMed] [Google Scholar]
- 7.Fiala E. S., Staretz, M. E., Pandya, G. A., El-Bayoumy, K. & Hamilton, S. R. (1998) Carcinogenesis 19, 597-604. [DOI] [PubMed] [Google Scholar]
- 8.Jiang C., Jiang, W., Ip, C., Ganther, H. & Lu, J. (1999) Mol. Carcinog. 26, 213-225. [DOI] [PubMed] [Google Scholar]
- 9.Spolar M. R., Schaffer, E. M. N., Beelman, R. B. & Milner, J. A. (1999) Cancer Lett. (Shannon, Irel.) 138, 145-150. [DOI] [PubMed] [Google Scholar]
- 10.Allan C. B., Lacourciere, G. M. & Stadtman, T. C. (1999) Annu. Rev. Nutr. 19, 1-16. [DOI] [PubMed] [Google Scholar]
- 11.Gaiddon C., Moorthy, N. C. & Prives, C. (1999) EMBO J. 16, 5609-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueno M., Masutani, H., Arai, R. J., Yamauchi, A., Hirota, K., Sakai, T., Inamoto, T., Yamaoka, Y., Yodoi, J. & Nikaido, T. (1999) J. Biol. Chem. 274, 35809-35815. [DOI] [PubMed] [Google Scholar]
- 13.Evans A. R., Limp-Foster, M. & Kelley, M. R. (2000) Mutat. Res. 461, 83-108. [DOI] [PubMed] [Google Scholar]
- 14.Kastan M. B., Zhan, Q., el-Deiry, W. S., Carrier, F., Jacks, T., Walsh, W. V., Plunkett, B. S., Vogelstein, B. & Fornace, A. J., Jr. (1992) Cell 71, 587-597. [DOI] [PubMed] [Google Scholar]
- 15.Smith M. L., Chen, I. T., Zhan, Q., O'Connor, P. M. & Fornace, A. J., Jr. (1995) Oncogene 10, 1053-1059. [PubMed] [Google Scholar]
- 16.Smith M. L., Ford, J. M., Hollander, M. C., Bortnick, R. A., Amundson, S. A., Seo, Y. R., Deng, C., Hanawalt, P. C. & Fornace, A. J., Jr. (2000) Mol. Cell. Biol. 20, 3705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan S., Smith, M. L., Rivet, D. J., Duba, D., Zhan, Q., Kohn, K. W., Fornace, A. J., Jr. & O'Connor, P. M. (1995) Cancer Res. 55, 1649-1654. [PubMed] [Google Scholar]
- 18.Seo Y. R., Sweeney, C. & Smith, M. L. (2002) Oncogene 21, 3663-3669. [DOI] [PubMed] [Google Scholar]
- 19.Wu H. H. & Momand, J. (1998) J. Biol. Chem. 273, 18898-18905. [DOI] [PubMed] [Google Scholar]
- 20.Jayaraman L., Murthy, K., Zhu, C., Curran, T., Xanthoudakis, S. & Prives, C. (1997) Genes Dev. 11, 558-570. [DOI] [PubMed] [Google Scholar]
- 21.Makmura L., Hamann, M., Areopagita, A., Furuta, S., Munoz, A. & Momand, J. (2001) Antioxid. Redox Signal. 3, 1105-1118. [DOI] [PubMed] [Google Scholar]
- 22.Pluquet O. & Hainaut, P. (2001) Cancer Lett. (Shannon, Irel.) 174, 1-15. [DOI] [PubMed] [Google Scholar]
- 23.Chao C., Saito, S., Anderson, C. W., Appella, E. & Xu, Y. (2000) Proc. Natl. Acad. Sci. USA 97, 11936-11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rainwater R., Parks, D., Anderson, M. E., Tegtmeyer, P. & Mann, K. (1995) Mol. Cell. Biol. 15, 3892-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmgren A. & Bjornstadt, M. (1995) Methods Enzymol. 252, 199-211. [DOI] [PubMed] [Google Scholar]
- 26.Redman C., Scott, J. A., Baines, A. T., Basye, J. L., Clark, L. C., Calley, C., Payne, C. M. & Nelson, M. A. (1998) Cancer Lett. (Shannon, Irel.) 125, 103-110. [DOI] [PubMed] [Google Scholar]
- 27.Hawkins D. S., Demers, G. W. & Galloway, D. A. (1996) Cancer Res. 56, 892-898. [PubMed] [Google Scholar]
- 28.Ford J. M., Baron, E. L. & Hanawalt, P. C. (1998) Cancer Res. 58, 599-603. [PubMed] [Google Scholar]
- 29.Cistulli C. A. & Kaufman, W. K. (1998) Cancer Res. 58, 1993-2002. [PubMed] [Google Scholar]
- 30.Lloyd D. R. & Hanawalt, P. C. (2000) Cancer Res. 60, 517-521. [PubMed] [Google Scholar]
- 31.Zhu Q., Wani, M. A., El-Mahdy, M. & Wani, A. A. (2000) J. Biol. Chem. 275, 11492-11497. [DOI] [PubMed] [Google Scholar]
- 32.Smith M. L. & Seo, Y. R. (2002) Mutagenesis 17, 149-156. [DOI] [PubMed] [Google Scholar]
- 33.Cheo D. L., Burns, D. K., Meira, L. B., Houle, J. F. & Friedberg, E. C. (1999) Cancer Res. 59, 771-775. [PubMed] [Google Scholar]
- 34.Hollander M. C., Kovalsky, O., Salvador, J. M., Kim, K. E., Patterson, A. D., Haines, D. C. & Fornace, A. J., Jr. (2001) Cancer Res. 61, 2487-2491. [PubMed] [Google Scholar]
- 35.Ames B. & Gold, L. S. (1997) Environ. Health Perspect. 105, Suppl. 14, 865-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuraoka I., Bender, C., Romieu, A., Cadet, J., Wood, R. D. & Lindahl, T. (2000) Proc. Natl. Acad. Sci. USA 97, 3832-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown J. M. & Wouters, B. G. (1999) Cancer Res. 59, 1391-1399. [PubMed] [Google Scholar]
- 38.Clark L. C., Combs, G. F., Jr., Turnbull, B. W., Slate, E. H., Chalker, D. K., Chow, J., Davis, L. S., Glover, R. A., Graham, G. F., Gross, E. G., et al. (1996) J. Am. Med. Assoc. 276, 1957-1963. [PubMed] [Google Scholar]
- 39.Eller M. S., Maeda, T., Magnoni, C., Atwal, D. & Gilchrest, B. (1997) Proc. Natl. Acad. Sci. USA 94, 12627-12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang B. J., Ford, J. M., Hanawalt, P. C. & Chu, G. (1999) Proc. Natl. Acad. Sci. USA 96, 424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan W., Jin, S., Tong, T., Zhao, H., Fan, F., Antinore, M. J., Rajasekaran, B., Wu, M. & Zhan, Q. (2002) J. Biol. Chem. 277, 8061-8067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.