Abstract
In view of their perceived chemopreventive activities against hormone-dependent cancers, cardiovascular disease, and postmenopausal ailments, there is considerable interest in engineering plants to contain isoflavone phytoestrogens. However, attempts to date have only resulted in low levels of isoflavone accumulation in non-legumes. Introducing soybean isoflavone synthase (IFS) into Arabidopsis thaliana leads to accumulation of low levels of genistein glycosides. Leaves of wild-type A. thaliana contain high levels of similar conjugates of the flavonols quercetin and kaempferol, which could be increased by threefold on introduction of an alfalfa chalcone isomerase transgene. Levels of genistein were not increased by expressing both IFS and alfalfa chalcone isomerase, but levels of flavonol conjugates were reduced to a greater extent than could be accounted for by flux into isoflavone. Introduction of IFS into the tt6/tt3 double mutant blocked in flavonol, and anthocyanin synthesis resulted in high levels of genistein. The bottleneck for constitutive isoflavone production in Arabidopsis is, therefore, competition for flavanone between IFS and endogenous flavonol synthesis, and the flavonol pathway is reciprocally but disproportionately affected by IFS.
Keywords: flavonol, phytoestrogen, antioxidant, metabolic engineering, transgenic plants
Isoflavonoids are limited primarily to leguminous plants, where they function as preformed or inducible antimicrobial or anti-insect compounds, as inducers of the nodulation genes of symbiotic Rhizobium bacteria, or as allelopathic agents (1). Many reports link the dietary consumption of the simple isoflavones daidzein and genistein (Fig. 1) to a range of potential health benefits (2–4). Major dietary sources of isoflavones for humans are soybean and chickpea seed products.
Fig 1.
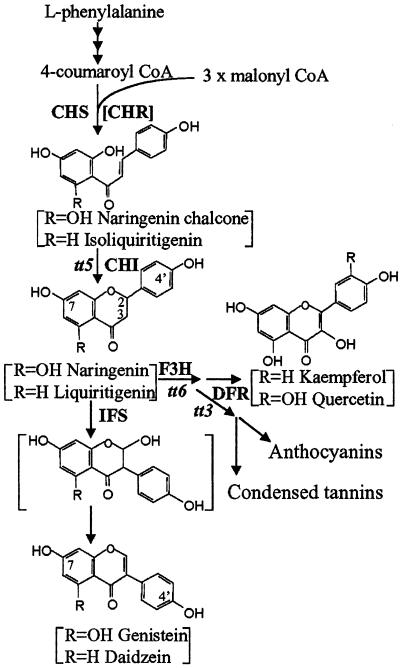
Biosynthesis of flavonoids and isoflavonoids, indicating competing pathways for naringenin in transgenic Arabidopsis expressing IFS. Formation of liquiritigenin requires the activity of a chalcone reductase (CHR) that co-acts with chalcone synthase (CHS) to form isoliquritigenin. CHR is present in legumes, but not in Arabidopsis. Daidzein cannot be made in Arabidopsis without introduction of CHR and a legume CHI. Steps blocked in the tt3, tt5, and tt6 mutations are shown. F3H, flavonoid 3-hydroxylase; DFR, dihydroflavonol reductase.
Epidemiological studies have demonstrated a link between consumption of soy isoflavones and reduced risks of breast and prostate cancers in humans (3–5); genistein and 4′-O-methylgenistein prevent development of chemically induced cancers in mammalian cell systems (6–9). Isoflavones may possess other health-promoting activities, including chemoprevention of osteoporosis and cardiovascular disease (10–12). There is, therefore, considerable interest in the possibility of introducing them into vegetables, grains, and fruits for dietary disease prevention.
Isoflavonoids originate from a flavanone intermediate that is ubiquitously present in plants. For entry into the isoflavonoid pathway, flavanone undergoes migration of the B-ring to the 3-position, followed by hydroxylation at the 2-position, which is catalyzed by the cytochrome P450 enzyme CYP93C1 [2-hydroxyisoflavanone synthase, commonly termed isoflavone synthase (IFS)]. The resulting 2-hydroxyisoflavanone is unstable and dehydrates to yield the corresponding isoflavone (ref. 13; Fig. 1).
cDNAs encoding IFS have been cloned from soybean and other species (14–16), and IFS has been introduced into Arabidopsis thaliana, corn, and tobacco. However, in all cases, only small amounts of genistein were formed, and in most cases, the nature of potential glycoconjugates was not determined (15, 17). Many flavonoids and isoflavonoids are glycosylated, and the sugar residues may be further modified by acylation (18, 19). Conjugation may decrease the bioavailability of isoflavone phytoestrogens (20).
Here, we identify the isoflavone glycoconjugates that accumulate after expression of IFS in A. thaliana, and use transgenic approaches and Arabidopsis mutants to define the bottleneck(s) for efficient isoflavone production in a plant that does not naturally possess this pathway.
Materials and Methods
Plant Materials and Transformation.
Seeds of Arabidopsis tt mutants in the Landsberg Erecta (Ler) background were obtained from the Ohio State University Arabidopsis Biological Resource Center. Plants were grown in Mix 200 soil mixture (Scotts, Marysville, OH) under a 12-h light/12-h dark regime (≈170 μE fluorescent light) at 22°C and 50% relative humidity. Plants for transformation were maintained under 16-h light/8-h dark. Plants of the same or different generations used for comparison of flavonoid/isoflavonoid profiles were always grown together under identical conditions.
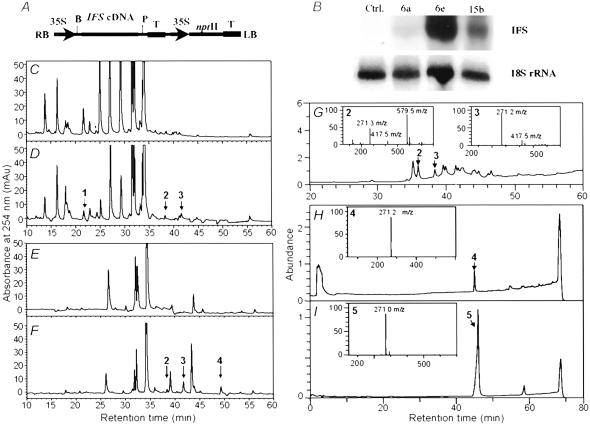
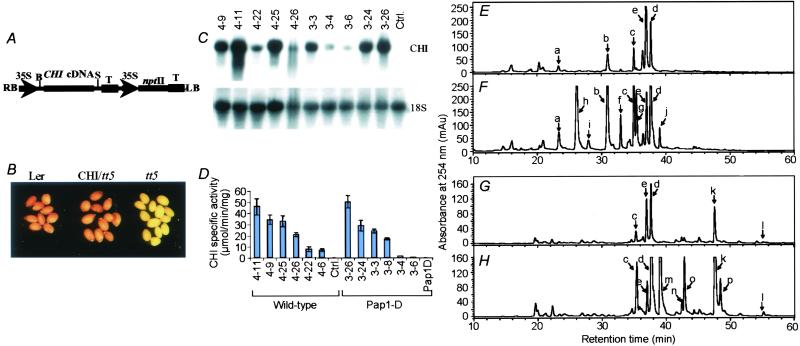
The soybean CYP93C1v2 cDNA (14) was cloned into the BamHI and PstI sites of binary vector pCHF3 to create pCHFIFS (Fig. 2A). The alfalfa chalcone isomerase (CHI) cDNA was excised from pBSK with BamHI and XhoI, pCHF3 was digested with BamHI and SalI, and the CHI insert was ligated into pCHF3 to give pCHFCHI (for example, see Fig. 4A).
Fig 2.
Expression of IFS and accumulation of genistein conjugates in transgenic Arabidopsis. (A) Binary soybean IFS construct. 35S, cauliflower mosaic virus 35S promoter; B = BamHI; P = PstI; T, pea rubisco 3′ terminator. (B) RNA gel blot analysis of soybean IFS transcripts in Arabidopsis transformants and empty vector controls. (C and D) HPLC profiles of nonhydrolyzed extracts from leaves of an empty vector control plant (C) and line 6E expressing soybean IFS (D). (E and F) HPLC profiles of β-glucosidase-treated extracts from leaves of an empty vector plant (E) and line 6E (F). Most peaks correspond to flavonol glycosides. Genistein conjugates are labeled 1, 2, and 3; free genistein is labeled 4. (G–I) Mass spectrometry total ion chromatographs (TIC) and corresponding mass spectra (Insets) of partially purified genistein conjugate peaks 2 and 3 (G), partially purified genistein conjugate peak 4 (H), and authentic genistein (I).
Fig 4.
Expression of alfalfa CHI in Arabidopsis. (A) Binary alfalfa CHI construct. B, BamHI; S, SalI; T, rubisco 3′ terminator. (B) Complementation of the tt5 mutation by transformation with alfalfa CHI. (C) RNA gel blot analysis of alfalfa CHI transcripts in independent transformants in WT (Col-0) and pap1-D backgrounds. (D) Transgene CHI activities in independent transformants in Col-0 or Col-0/pap1-D backgrounds. (E and F) HPLC profiles of nonhydrolyzed extracts from leaves of an empty vector plant (E) and from lines 4–11 expressing alfalfa CHI (F). (G and H) HPLC profiles of β-glucosidase-treated extracts from leaves of an empty vector plant (G) and from lines 4–11 (H). Compounds are: a, Rha-Gluc-Rha-quercetin; b, Rha-Gluc-Rha-kaempferol; c, Rha-Rha-Q; d, Rha-Rha-K; e, Sinapic acid; f, Gluc-Rha-Q; g, rutin; l, kaempferol, p, quercetin; k and n, unidentified kaempferol conjugates; o, unidentified quercetin conjugate; h, i, j, and m, other unknown flavonol conjugates.
Arabidopsis was transformed with Agrobacterium tumefaciens strain C58C1 harboring pCHFIFS or pCHFCHI by the floral dip method (21). Seeds were selected on plates containing either kanamycin (for selection of transgenes in the Col-0 and tt mutant background) or kanamycin and Basta (for selection of transgenes in the pap1-D background).
Genetic Crosses.
T3 homozygous lines containing CHI or IFS transgenes were used for crosses. Hand emasculated CHI transgenic lines were used as females. Seeds from cross-pollinated flowers were germinated on solid medium under suitable antibiotic selection, and leaves from F1 or F2 progenies were collected for PCR screening for the presence of the transgene.
DNA Extraction and PCR Screening.
Genomic DNA was isolated from T1 antibiotic-resistant plants or individual progeny of crosses by a standard procedure (22). One μl of DNA solution was used for PCR analysis with 35 cycles of 94°C for 30 s, 54°C (for CHI and IFS) or 50°C (for pap1-D) for 30 s, and 72°C for 45 s. Primers were for alfalfa CHI (CHI forward: 5′-CTTCATCAAGTTCACTGCCA-3′ and CHI reverse: 5′-AAATGCAGTGATATAGGAGTA-3′), soybean CYP93c1v2 (IFS forward: 5′-CTTCATAGGACACCTTCATC-3′ and IFS reverse: 5′-GTCAACTTCGTCCACAAGTC-3′) or PAP1 (PAP1 reverse: 5′-ATCAAGAGAGGAAGA-3′) coupled with a T7 universal primer for the activation tag.
RNA Extraction and Gel Blot Analysis.
Total RNA was isolated from 100 mg of leaves with the Tri reagent extraction kit (Molecular Research Center, Cincinnati). Twenty micrograms of RNA was used for Northern hybridization with 32P-labeled CHI or IFS cDNA fragments in Church buffer (23) at 65°C. The hybridized membrane was washed in phosphate buffer I [40 mM NaHPO4, pH 7.2/5% (wt/vol) SDS/1% (wt/vol) BSA] and buffer II [40 mM NaHPO4, pH 7.2/1% (wt/vol) SDS] twice at 65°C and was then used to expose x-ray film for 16 h.
Enzymatic Assays.
For assay of alfalfa CHI, 2′,4,4′-trihydroxychalcone was used as substrate and the reaction was monitored by the decrease in absorbance at 400 nm, as described (24). For measurement of IFS activity, microsomes were prepared as described (25), and assays measuring NADPH-dependent conversion of [3H]liquiritigenin to daidzein were performed as described (24). Reaction mixtures were spotted with daidzein standard on an Si250(F) silica gel TLC plate (J.T. Baker) which was developed with chloroform:methanol (10:1). Spots were observed under UV light and scraped from the plates for determination of radioactivity by scintillation counting.
Metabolite Extraction and Analysis.
For analysis of flavonoid and isoflavonoid glycoconjugates, leaves were extracted sequentially with iced acetone and acetone:methanol (1:1). A portion of the extract was digested with almond β-glucosidase, as described (25). Samples were applied to an ODS2 reverse-phase column (5 μm particle size, 4.6 × 250 mm) and eluted in 1% (vol/vol) phosphoric acid with an increasing concentration gradient of acetonitrile [0–5 min, 5% (vol/vol); 5–10 min, 5–10%; 10–15 min, 10–15%; 15–20 min, 15%; 20–25 min, 15–17%; 25–30 min, 17–23%; 30–65 min, 23–50% at a flow rate of 1 ml/min. UV absorption was monitored with a photodiode array detector. Levels of flavonols were quantified from standard curves by using authentic kaempferol, quercetin, and genistein standards.
Identification of genistein conjugates was by liquid chromatography/mass spectrometry using a Hewlett Packard 1100 liquid chromatograph (C18 reverse-phase column, 5 μm, 4.6 mm × 250 mm, J.T. Baker) coupled to a Bruker Esquire Ion-Trap MS equipped with an electrospray source. Samples were eluted as above but with 0.1% aqueous acetic acid replacing the phosphoric acid. Positive mass spectra were acquired by using an ion source voltage of 4.0 kV and a capillary offset of 70.7 V. Nebulization was achieved by using a nitrogen sheath gas at 70 psi, and desolvation was achieved by using nitrogen heated to 360°C at 10.5 psi. Ion charge control was set at 30,000 with a max acquire time of 100 ms.
For analysis of genistein, flavonol, and anthocyanidin aglycones, leaves were extracted with 80% methanol (containing 0.5% HCl if anthocyanins were being analyzed) and hydrolyzed in 1 N HCl at 90°C for 2 h. Hydrolysates were extracted twice with ethyl acetate, and the organic phase was removed, concentrated, and dissolved in methanol for HPLC analysis by using a linear gradient of acetonitrile (0–5 min, 27%; 5–45 min, 30%; 45–46 min, 30–100%; at a flow rate of 1 ml/min). The gradient used for analysis of flavonoids from the tt6/tt3 lines was 0–5 min, 11%; 2–10 min, 23%; 10–69 min, 55%. The aqueous phase was lyophilized and dissolved in methanol for HPLC analysis for anthocyanidins as above. Absorbance values at 510 nm were used for the quantification of anthocyanidins.
Statistical Analysis.
Statistical analysis was performed by one-way ANOVA using SAS programs (SAS Institute, Cary, NC). A Student's t test also was included to compare each set of data.
Results
Production of Isoflavone Conjugates in Arabidopsis.
Soybean IFS in the constitutive expression vector pCHF3 (Fig. 2A) was transformed into A. thaliana ecotype Columbia (Col-0). Eighteen independent transgenic lines were recovered. Expression of IFS transcripts in T2 progeny plants that were PCR positive for the IFS construct was confirmed by RNA gel blot analysis (Fig. 2B); plants were further analyzed for levels of genistein and genistein conjugates.
The HPLC profile of leaf extracts from empty vector-transformed plants shows a series of glycosides of the flavonols kaempferol and quercetin (ref. 26; Fig. 2C). Their identity was confirmed by UV-absorption characteristics and shifts in peak positions after pretreatment of extracts with β-glucosidase (Fig. 2E; see below). In contrast, extracts from leaves expressing IFS contained three extra peaks (compounds 1–3) with UV-absorption spectra similar to that of authentic genistein (Fig. 2D). After treatment with β-glucosidase, compound 1 disappeared and a new peak (compound 4) appeared (Fig. 2F). Compound 1, therefore, is a β-glucoside of genistein, and compounds 2 and 3 are conjugates in which a sugar other than glucose is directly linked to the isoflavone. This result was confirmed by liquid chromatography/mass spectrometry. The fragmentation patterns were consistent with the loss of one or more sugars from a genistein aglycone (Figs. 2 G–I); they identify compound 1 as a glucoside of genistein, compound 2 as glucose-rhamnose-genistein, and compound 3 as rhamnose-genistein. Compound 4 is free genistein. No bathochromic shift in absorption maximum diagnostic of an unsubstituted 7-hydroxyl group (27) was observed for compounds 1, 2, or 3 after treatment of ethanolic solutions with sodium acetate (data not shown), confirming the expected linkage of the glycans to the 7-position.
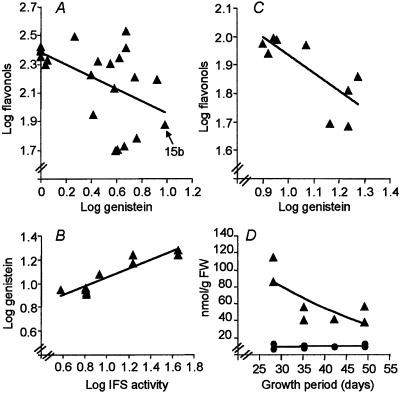
Of the IFS transformants, line 15b produced the highest level of genistein, ≈7.4 nmol/g fresh weight after acid hydrolysis of conjugates, a level very much lower than the levels of endogenous flavonols (Fig. 3A). Isoflavones fed to plant cell cultures can be incorporated into the insoluble cell-wall fraction (28). However, analysis of alkaline hydrolysates revealed no evidence for the presence of genistein in the cell-wall fraction.
Fig 3.
Relations between IFS activity and genistein and flavonol levels in transgenic Arabidopsis. (A) Genistein and total flavonols (kaempferol plus quercetin) in T2 plants from 18 independent IFS primary transformants. Regression analysis, R2 = 0.264. (B) IFS activity and genistein levels in T3 progeny of line 15b. R2 = 0.843. (C) Genistein and total flavonols in the same lines as analyzed in B, R2 = 0.584. (D) Time course of genistein (•) and kaempferol (▴) levels in a homozygous 15b line.
To determine whether IFS activity might be limiting for isoflavone formation, microsomes were isolated from independent T3 homozygous 15b plants, and in vitro IFS activity was determined. Leaf extracts from the same plants also were analyzed for genistein levels after acid hydrolysis. There was a log-linear relationship between genistein levels and IFS activity in the various plants (Fig. 3B). However, this was not a 1:1 relationship (compare PAL activity and chlorogenic acid production in tobacco; ref. 29), which indicates there are additional factors controlling genistein accumulation. Interestingly, there was a negative relationship between levels of genistein and flavonol conjugates in the T2 lines (Fig. 3A) that was confirmed for a series of homozygous T3 plants from line 15b (Fig. 3C).
Detached leaves of untransformed Arabidopsis were fed genistein through their cut petioles. Approximately 122 nmol/g of fresh weight of free genistein and about 2.45 nmol/g of fresh weight rhamnose-genistein were detected after 4 h. The rate of conjugation (about 0.6 nmol/g/h) suggests that glycosylation is not a rate-limiting factor for accumulation of genistein in Arabidopsis.
To determine whether accumulation of genistein was stable, leaves of a T3 homozygous 15b transformant were analyzed at different growth stages from appearance of the rosette leaves to the beginning of seed set. Levels of genistein conjugates remained constant for up to 49 days after germination, whereas kaempferol levels declined to less than 60% of their maximum levels by 49 days (Fig. 3D)
Expression of Alfalfa CHI Increases Flavonol Accumulation in Arabidopsis.
To increase genistein production, we attempted to increase flux into the provision of substrate (naringenin) for IFS. CHI precedes IFS in isoflavonoid biosynthesis and is rate-limiting for flavonoid biosynthesis in tomato fruit peel (30). CHI from legumes is active toward both naringenin chalcone and isoliquiritigenin (2′,4,4′-trihydroxychalcone), whereas non-legume CHIs generally do not use isoliquiritigenin (31). Maize CHI can complement the tt5 CHI-null mutation in Arabidopsis (32), but it is not known whether legume CHIs, which might couple better with legume IFS than the endogenous Arabidopsis CHI, are functional in Arabidopsis. Therefore, we transformed the Arabidopsis tt5 mutant (in the Ler ecotype) with alfalfa CHI by using the construct shown in Fig. 4A. This transformation resulted in restoration of the seed-coat coloration (Fig. 4B), indicating functionality of alfalfa CHI in the biosynthesis of the seed-coat condensed tannins. Alfalfa CHI also was introduced into WT Col-0, and into an activation-tagged Col-0 line overexpressing the PAP1 MYB gene that exhibits an over 80-fold increase in anthocyanin levels resulting from a strong, coordinated induction of the enzymes of the general phenylpropanoid and anthocyanin pathways (33). There was a strong correlation between CHI transgene transcript levels (Fig. 4C) and CHI activity toward isoliquiritigenin (Fig. 4D), with lines 4–11 and 3–26 having the highest expression among the WT and pap1-D background plants, respectively.
Leaves from untransformed and CHI transgenic plants were extracted, and unhydrolyzed and β-glucosidase-treated extracts were analyzed for flavonoid composition by HPLC (Fig. 4 E–H). Expression of alfalfa CHI increased the levels of rhamnose-glucose-rhamnose-quercetin, rhamnose-glucose-rhamnose-kaempferol, and quercetin and kaempferol di-rhamnosides (Fig. 4 F and H); glucose-rhamnose-quercetin, rutin (quercetin 3-β-d-rutinoside) and three yet-to-be-identified flavonol conjugates also were detected. No naringenin or naringenin conjugates were detected in WT plants, or in transgenic plants expressing alfalfa CHI.
Expression of IFS Reduces Levels of Flavonoid Conjugates in Arabidopsis.
To test whether expression of alfalfa CHI would result in an increase in genistein production, IFS transgenic line 15b was crossed with line 4–11 harboring the alfalfa CHI transgene, and F2 progeny plants were screened for retention of IFS and CHI transgenes by PCR. Positive progeny then were analyzed for genistein (after hydrolysis of leaf extracts) and flavonols. Homozygous 15b and 4–11 plants (T4 generation) were included for comparison. The apparently higher levels of genistein in F2 progeny lines expressing both transgenes compared with 15b T4 plants were not statistically significant (Table 1; for details of individual lines, see Table 3, which is published as supporting information on the PNAS web site, www.pnas.org). However, as predicted from the data in Fig. 3, the lines expressing both transgenes contained lower flavonol levels than did line 4–11.
Table 1.
Genistein and flavonol levels in transgenic Arabidopsis expressing soybean IFS and/or alfalfa CHI
| Line | Generation | Alfalfa CHI activity, nmol/min/mg | Quercetin, nmol/g FW | Kaempferol, nmol/g FW | Total flavonols, nmol/g FW | Genistein, nmol/g FW |
|---|---|---|---|---|---|---|
| Wild-type control | T4 | 0 | 32.4 ± 5.9 | 285.8 ± 35.8 | 318.2 ± 36.7† | 0† |
| CHI 4-11 | T4 | 49.6 ± 3.0 | 79.6 ± 36.8 | 475.3 ± 77.8 | 555.0 ± 84.5‡ | 0† |
| IFS 15b | T4 | 0 | 29.4 ± 7.3 | 162.7 ± 37.4 | 192.1 ± 43.9§∥ | 11.9 ± 1.9‡¶ |
| IFS/CHI | F2 | ND | 60 ± 12.1 | 300.1 ± 52.5 | 360 ± 61.5†§¶ | 22.3 ± 6.4‡§ |
| IFS/CHI | F3 | 46.2 ± 2.3 | 43.5 ± 4.7 | 287.4 ± 35.5 | 330.9 ± 39.6†§¶ | 10.6 ± 1.0¶ |
| IFS/CHI | F3 | 0 | 28.7 ± 4.1 | 78.2 ± 7.0 | 106.8 ± 8.8∥ | 18.5 ± 1.2§ |
Plants were grown under 12 hr light/12 hr dark, ∼150 μE light intensity, for 35 days. Data for T4 plants are means ± SE from at least five replicate plants with 2 or 3 independent analyses for each plant. Individual progeny plants were analyzed three times (see supporting information).
†‡§¶∥ Symbols represent pairwise statistical analyses. Different symbols represent significantly different values at P ≤ 0.05; numbers with the same symbols are not significantly different at P > 0.05.
Not determined.
F3 progeny that had retained both IFS and CHI transgene (7 plants).
F3 progeny that had lost the CHI transgene through segregation (2 plants).
This reduction in flavonol levels was confirmed in a larger number of F3 progeny plants (Tables 1 and 3). For most F3 lines, levels of flavonol conjugates were between 13 and 70% of the average value for the 4–11 line. However, there was no relation between genistein levels and CHI activity. The two lines with highest genistein levels (3–3–1 and 3–4–1) had lost the CHI transgene through segregation, as determined by both PCR (data not shown) and the lack of enzyme activity with isoliquiritigenin as substrate (Tables 1 and 3). Furthermore, in the F3 progeny that had retained both transgenes, genistein levels were, overall, no higher than in line 15b.
pap1-D plants expressing alfalfa CHI had high levels of both flavonols and anthocyanin (Table 2 and Table 4, which is published as supporting information on the PNAS web site). Progeny of a cross between IFS line 6E and CHI:pap1-D line 3–26 were selected for double resistance to kanamycin and bialaphos, and the genotype was confirmed by PCR. F2 plants had similar genistein levels to the 6E parent, but anthocyanin levels were reduced by as much as 80% in confirmed CHI:IFS:PAP1 lines. Flavonol levels were not reduced, however, and two lines, 1–1 and 1–2, had strongly increased levels of kaempferol conjugates (Tables 2 and 4).
Table 2.
Genistein, flavonol, and anthocyanin levels in transgenic Arabidopsis expressing soybean IFS, alfalfa CHI, and over-expressing PAP1
| Line | Generation | Quercetin, nmol/g FW | Kaempferol, nmol/g FW | Total flavonols, nmol/g FW | Anthocyanins, Au/g FW | Genistein, nmol/g FW |
|---|---|---|---|---|---|---|
| Wild-type | >T3 | 21.0 ± 5.7 | 418.9 ± 33.3 | 439.9 ± 30.9‡ | 3.1 ± 0.4‡ | 0‡ |
| Pap1-D | >T3 | 200 ± 8.3 | 345.5 ± 48.3 | 545.7 ± 40.3‡§ | 192.0 ± 7.1§ | 0‡ |
| CHI/Pap1-D (3–26) | >T3 | 89.3 ± 6.1 | 666.6 ± 103.0 | 755.9 ± 103.7§∥ | 131.1 ± 10.6¶ | 0‡ |
| IFS 6E | >T3 | 20.8 ± 6.6 | 323.5 ± 4.3 | 344.3 ± 9.5¶ | 1.1 ± 0.2∥ | 3.5 ± 0.2§ |
| 3–26/6E | F2 | 72.8 ± 12.1 | 1029.8 ± 84.6 | 1102.6 ± 91.1∥ | 30.7 ± 5.7 | 4.8 ± 2.4§ |
Plants were grown under 12 h light/12 h dark, ∼200 μE light intensity. Data for >T3 plants are means ± SE from three replicate plants with three independent analyses for each plant. Data for F2 progeny represent triplicate analyses of five plants (see supporting information).
UV absorbance at 510 nm.
‡§¶∥
Symbols represent pairwise statistical analyses. Different symbols represent significantly different values at P ≤ 0.05; numbers with the same symbols are not significantly different at P > 0.05.
Production of Genistein in the tt6/tt3 Mutant.
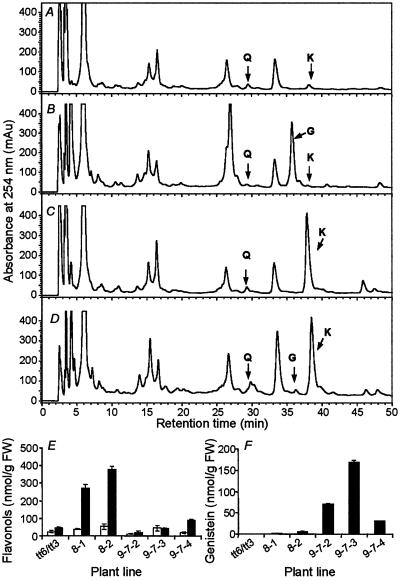
The tt6/tt3 double mutant of Arabidopsis in ecotype Ler is impaired in expression of both flavanone 3-β-hydroxylase (F3H), the entry point enzyme for flavonol/anthocyanin synthesis and dihydroflavonol reductase (DFR; ref. 34). Soybean IFS was transformed into both WT Ler and tt6/tt3 to determine the effects of the blockage in flavonol synthesis on genistein production, and T3 plants were produced. Surprisingly, both kaempferol and quercetin were detected in all tt6/tt3 lines, although kaempferol levels were at least one order of magnitude lower than in WT Ler (Fig. 5 A, B, and E). Expression of IFS in WT Ler resulted in low levels of genistein (up to 5.4 nmol/g fresh wt), similar to those in the Col-0 background (Fig. 5 D and F). However, expression of IFS in tt6/tt3 resulted in much greater amounts of genistein, with levels from 31–169 nmol/g fresh wt in three T3 progeny plants (Fig. 5 B and F).
Fig 5.
Isoflavone levels in Arabidopsis impaired in flavonol production. (A–D) HPLC profiles of hydrolyzed extracts from leaves of the tt6/tt3 mutant and WT Ler with and without expression of soybean IFS (T3 generation). (A) tt6/tt3. (B) tt6/tt3 expressing IFS. (C) Ler WT. (D) Ler expressing IFS. Compounds are: G, genistein; K, kaempferol; Q, quercetin. (E and F) Levels of flavonols (E) and genistein (F) in individual T3 plants of Ler (eight designations) and tt6/tt3 (nine designations) expressing IFS. White bars, quercetin; black bars, kaempferol.
Discussion
Factors Affecting Flux into Isoflavones in Arabidopsis.
There have been few reports on the introduction of new natural product pathways into plants through genetic manipulation. A notable example is the introduction of the provitamin A pathway into rice, a process that involved the introduction of three genes (35). Introduction of genistein into plants can theoretically be achieved by transformation with a single enzyme, IFS. However, this new pathway could interfere with endogenous flavonoid biosynthesis and/or fail to “plug in” to the endogenous flavonoid precursor pathway.
Isoflavones usually exist as glucosides and malonyl glucosides in legumes (36, 37). Arabidopsis conjugates genistein to the same sugars that are attached to kaempferol and quercetin, although with different position specificity. However, levels of genistein conjugates in IFS-only transgenics were similar to the levels reported from previous attempts to engineer genistein in Arabidopsis and other species (15, 17) and were approximately two orders of magnitude less than levels in soybean seeds. Genistein production could be limited by low in vivo IFS activity or by preferential channeling of naringenin into flavonol biosynthesis. Genistein may be a poor substrate for endogenous glycosyltransferases, and the aglycone may be turned over. Our results strongly support the second hypothesis. In particular, the observation that expression of IFS in the tt6/tt3 line with reduced flavonol biosynthesis resulted in the highest levels of genistein accumulation confirms that IFS activity was not itself limiting for genistein production, and that the major problem for isoflavone engineering is the partition of flux between flavonol and isoflavone biosynthesis at the level of naringenin, the substrate for both IFS and F3H. A physical complex may exist involving CHS, CHI, and F3H (38), and lack of access of IFS to such a complex could explain the present results.
Alfalfa CHI functionally complements the Arabidopsis tt5 CHI-null mutation for condensed tannin production in the seed coat. The elevated CHI activity in leaves expressing alfalfa CHI resulted in an approximately threefold increase in the levels of flavonol glycosides. Increased CHI expression also leads to increased flavonol content in tomato fruit peel (30) but not, however, in the flesh of the fruit or in the leaves. Unusually, the peel of WT tomato fruit contains high levels of naringenin chalcone but very low levels of flavonols, suggesting limitation of CHI activity (30). In Arabidopsis, there is no significant pool of naringenin chalcone, and this intermediate is effectively processed into naringenin and downstream flavonols via endogenous CHI.
Legume CHIs are equally active with naringenin chalcone and isoliquiritigenin, the precursor of the 5-deoxy class of isoflavonoids commonly found in legumes (31). According to recent proposals for the involvement of multienzyme complexes in flavonoid biosynthesis (38), soybean IFS might be able to “couple” better to a legume CHI than to the endogenous Arabidopsis CHI. However, increasing flux into naringenin production by up-regulation of CHI expression increased flavonol biosynthesis but not genistein production. Inducing flavonoid biosynthesis in Arabidopsis by ultraviolet-B illumination led to a 3-fold increase in genistein accumulation (17). It will be interesting to determine whether further improvements in genistein production can be made by exposing IFS/tt3/tt6 transgenics to UV irradiation or by introducing alfalfa CHI into this background.
IFS Expression Affects Flavonol and Anthocyanin Biosynthesis.
Genistein production in transgenic Arabidopsis expressing IFS is associated with disproportionate reductions in flavonols in the CHI over-expressing lines and in anthocyanins in the pap1-D background. This result cannot be solely explained by channeling of flux from endogenous pathways into the newly introduced isoflavonoid pathway. Alternative explanations include partial disruption of metabolic complexes involving F3H or flavonoid 3′-hydroxylase (F3′H; ref. 38) by the membrane-bound IFS, inhibition of flavonol glycosylation and sequestration by genistein (possibly by competition for glycosyltransferases) or other biological activities of genistein such as tyrosine kinase inhibition (39).
Biotechnological Implications.
All of the genistein produced in Arabidopsis is glycosylated. Studies on uptake of dietary genistein have suggested that the free aglycone is highly bioavailable (40), but uptake in both the stomach and intestinal tract is lower for glycosides (41, 42). Nevertheless, isoflavone glycosides are hydrolyzed to their aglycones by lactobacilli, Bacteroides and Bifidobacteria in the intestinal flora, and by lactase phlorizin hydrolase in the small intestinal brush border cells (43). Glycosylation may be a favorable trait for engineered isoflavonoid nutraceuticals, as it results in their storage in the vacuole (44), away from further potential metabolism.
There are currently no values available for the levels of dietary genistein that should be achieved for health benefits. However, it seems likely, based on in vitro studies, that the levels obtained in the present model system in the absence of down-regulation of the flavonol pathway would be too low. Flavonols such as quercetin themselves have health-promoting activities, including the ability to reduce plasma LDL levels (45, 46), and reduced flavonol levels could easily outweigh benefits from expression of low levels of isoflavones. The unexpected impact of genistein production on flavonol levels highlights the need for a better understanding of how metabolic flux is channeled into the various branch pathways of flavonoid biosynthesis. Future biotechnological applications of isoflavone engineering in vegetative tissues will require a solution to the problem of flux partitioning between IFS and F3H.
Supplementary Material
Acknowledgments
We thank David Huhman and Dr. Lloyd Sumner for assistance with MS analysis, Dr. Bryan Unruh for help with statistics, Darla Boydston for artwork, and Drs. Gregory D. May and Deyu Xie for critical reading of the manuscript. This work was supported by the Samuel Roberts Noble Foundation and Oklahoma Center for the Advancement of Science and Technology Health Research Program Grant HR02-040R (to R.A.D.).
Abbreviations
IFS, isoflavone synthase
CHI, chalcone isomerase
F3H, flavanone 3-β-hydroxylase
References
- 1.Dixon R. A. (1999) in Comprehensive Natural Products Chemistry, ed. Sankawa, U. (Elsevier, New York), Vol. 1, pp. 773–823. [Google Scholar]
- 2.Palevitz B. A. (2000) The Scientist 14, 8-9. [Google Scholar]
- 3.Setchell K. D. R. & Cassidy, A. (1999) J. Nutr. 129, 758S-767S. [DOI] [PubMed] [Google Scholar]
- 4.Lamartiniere C. A. (2000) Am. J. Clin. Nutr. 71, 1705S-1707S. [DOI] [PubMed] [Google Scholar]
- 5.Adlercreutz M. (1998) Baillieres Clin. Endocrinol. Metab. 12, 605-623. [DOI] [PubMed] [Google Scholar]
- 6.Rauth S., Kichina, J. & Green, A. (1997) Br. J. Cancer 75, 1559-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fritz W. A., Coward, L., Wang, J. & Lamartiniere, C. A. (1998) Carcinogenesis 19, 2151-2158. [DOI] [PubMed] [Google Scholar]
- 8.Hsu J. T., Hung, H. C., Chen, C. J., Hsu, W. L. & Ying, C. W. (1999) J. Nutr. Biochem. 10, 510-517. [DOI] [PubMed] [Google Scholar]
- 9.Pollard M. & Wolter, W. (2000) Prostate 45, 101-105. [DOI] [PubMed] [Google Scholar]
- 10.Alekel D. L., StGermain, A., Pererson, C. T., Hanson, K. B., Stewart, J. W. & Toda, T. (2000) Am. J. Clin. Nutr. 72, 844-852. [DOI] [PubMed] [Google Scholar]
- 11.MerzDemlow B. E., Duncan, A. M., Wangen, K. E., Xu, X., Carr, T. P., Phipps, W. R. & Kurzer, M. S. (2000) Am. J. Clin. Nutr. 71, 1462-1469. [DOI] [PubMed] [Google Scholar]
- 12.Uesugi T., Toda, T., Tsuji, K. & Ishida, H. (2001) Biol. Pharm. Bull. 24, 368-372. [DOI] [PubMed] [Google Scholar]
- 13.Kochs G. & Grisebach, H. (1986) Eur. J. Biochem. 155, 311-318. [DOI] [PubMed] [Google Scholar]
- 14.Steele C. L., Gijzen, M., Qutob, D. & Dixon, R. A. (1999) Arch. Biochem. Biophys. 367, 147-150. [DOI] [PubMed] [Google Scholar]
- 15.Jung W., Yu, O., Lau, S.-M. C., O'Keefe, D. P., Odell, J., Fader, G. & McGonigle, B. (2000) Nat. Biotechnol. 18, 208-212. [DOI] [PubMed] [Google Scholar]
- 16.Akashi T., Aoki, T. & Ayabe, S. (1999) Plant Physiol. 121, 821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu O., Jung, W., Shi, J., Croes, R. A., Fader, G. M., McGonigle, B. & Odell, J. T. (2000) Plant Physiol. 124, 781-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara H., Tanaka, Y., Yonekura-Sakakibara, K., Fukuchi-Mizutani, M., Nakao, M., Fukui, Y., Yamaguchi, M., Ashikari, T. & Kusumi, T. (1998) Plant J. 16, 421-431. [DOI] [PubMed] [Google Scholar]
- 19.Coward L., Smith, M., Kirk, M. & Barnes, S. (1998) Am. J. Clin. Nutr. 68, 1486S-1491S. [DOI] [PubMed] [Google Scholar]
- 20.Izumi T., Piskula, M. K., Osawa, S., Obata, A., Tobe, K., Saito, M., Kataoka, S., Kubota, Y. & Kikuchi, M. (2000) J. Nutr. 130, 1695-1699. [DOI] [PubMed] [Google Scholar]
- 21.Clough S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 22.Dellaporta S. L., Wood, J. & Hicks, J. B. (1983) Plant Mol. Biol. Rep. 1, 19-21. [Google Scholar]
- 23.Church G. H. & Gilbert, W. (1984) Proc. Natl. Acad. Sci. USA 81, 65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards R. & Kessmann, H. (1992) in Molecular Plant Pathology–A Practical Approach, eds. Gurr, S. J., McPherson, M. J. & Bowles, D. J. (IRL, Oxford), Vol. 2, pp. 45–62. [Google Scholar]
- 25.Liu C.-J. & Dixon, R. A. (2001) Plant Cell 13, 2643-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham T. L. (1998) Plant Physiol. Biochem. (Paris) 36, 135-144. [Google Scholar]
- 27.Mabry T. J., Markham, K. R. & Thomas, M. B., (1970) The Systematic Identification of the Flavonoids (Springer, New York).
- 28.Park H. H., Hakamatsuka, T., Sankawa, U. & Ebizuka, Y. (1995) Phytochemistry 38, 373-380. [Google Scholar]
- 29.Bate N. J., Orr, J., Ni, W., Meroni, A., Nadler-Hassar, T., Doerner, P. W., Dixon, R. A., Lamb, C. J. & Elkind, Y. (1994) Proc. Natl. Acad. Sci. USA 91, 7608-7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muir S. R., Collins, G. J., Robinson, S., Hughes, S., Bovy, A., De Vos, C. H. R., van Tunen, A. J. & Verhoeyen, M. E. (2001) Nat. Biotechnol. 19, 470-474. [DOI] [PubMed] [Google Scholar]
- 31.Dixon R. A., Blyden, E. R., Robbins, M. P., van Tunen, A. J. & Mol, J. N. M. (1988) Phytochemistry 27, 2801-2808. [Google Scholar]
- 32.Dong X., Braun, E. L. & Grotewald, E. (2001) Plant Physiol. 127, 46-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borevitz J., Xia, Y., Blount, J. W., Dixon, R. A. & Lamb, C. (2001) Plant Cell 12, 2383-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkel-Shirley B. (2001) Plant Physiol. 126, 485-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye X., Al-Babili, S., Kloti, A., Zhang, J., Lucca, P., Beyer, P. & Potrykus, I. (2000) Science 287, 303-305. [DOI] [PubMed] [Google Scholar]
- 36.Graham T. L., Kim, J. E. & Graham, M. Y. (1990) Mol. Plant–Microbe Interact. 3, 157-166. [Google Scholar]
- 37.Park H.-H., Hakamatsuka, T., Noguchi, H., Sankawa, U. & Ebizuka, Y. (1992) Chem. Pharm. Bull. 40, 1978-1980. [DOI] [PubMed] [Google Scholar]
- 38.Winkel-Shirley B. (1999) Physiol. Plant. 107, 142-149. [Google Scholar]
- 39.Akiyama T., Ishida, J., Nakagawa, S., Ogawara, H., Watanabe, S., Itoh, N., Shibuya, M. & Fukami, Y. (1987) J. Biol. Chem. 262, 5592-5595. [PubMed] [Google Scholar]
- 40.Sfakianos J., Coward, L., Kirk, M. & Barnes, S. (1997) J. Nutr. 127, 1260-1268. [DOI] [PubMed] [Google Scholar]
- 41.Piskula M. K., Yamakoshi, J. & Iwai, Y. (1999) FEBS Lett. 447, 287-291. [DOI] [PubMed] [Google Scholar]
- 42.Steensma A., Noteborn, H. P. J. M., vanderJagt, R. C. M., Polman, T. H. G., Mengelers, M. J. B. & Kuiper, H. A. (1999) Environ. Toxicol. Pharmacol. 7, 209-211. [DOI] [PubMed] [Google Scholar]
- 43.Day A. J., Canada, F. J., Diaz, J. C., Kroon, P. A., Mclauchlan, R., Faulds, C. B., Plumb, G. W., Morgan, M. R. A. & Williamson, G. (2000) FEBS Lett. 468, 166-170. [DOI] [PubMed] [Google Scholar]
- 44.Mackenbrock U., Vogelsang, R. & Barz, W. (1992) Z. Naturforsch., C 47, 815-822. [Google Scholar]
- 45.Hertog M. G. L., Feskens, E. J. M., Hollman, P. C. H., Katan, M. B. & Kromhout, D. (1993) Lancet 342, 1007-1011. [DOI] [PubMed] [Google Scholar]
- 46.Arai Y., Watanabe, S., Kimira, M., Shimoi, K., Mochizuki, R. & Kinae, N. (2000) J. Nutr. 130, 2243-2250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.