Abstract
A long-term genetic legacy of refugial isolation has been postulated and was demonstrated for maternal refugial lineages for numerous plant and animal species. The lineages were assumed to have remained separated from each other for several glacial periods. The conifer Abies alba Miller, silver fir, is an excellent model to test whether pollen-mediated gene flow may eliminate the genetic imprints of Pleistocene refugial isolation. Two DNA markers with contrasting modes of inheritance were applied to 100 populations covering the entire range of silver fir in Europe. The markers exhibited each two highly conserved alleles based on an insertion/deletion of 80 bp in the fourth intron of the mitochondrial nad5 gene and on a synonymous substitution in the chloroplast psbC gene. The geographical distribution of the maternally inherited mitochondrial variation supported the existence of at least two refugia with two recolonizing maternal lineages remaining largely separated throughout the range. The cline of the nad5 allele frequencies was much steeper than the one of the two psbC alleles. The psbC cline was as wide as the whole range of the species. Our results provide striking evidence that even a species with very long generation times and heavy pollen grains was able to establish a highly efficient pollen-mediated gene flow between refugia. Therefore we postulate that an exchange of genetic information between refugia by range-wide paternal introgression is possible in wind-pollinated plant species.
The general cooling of the earth's climate through the Tertiary was accompanied by frequent oscillations between cold and warm periods. Since the beginning of the Quaternary, the frequencies of these oscillations increased, leading to a series of ice ages (1).
In recent years the study of the genetic and evolutionary consequences of these past events has become a focus of attention. With advances in technology, DNA markers have emerged that enable us to identify genetic lineages and discern their phylogenetic relationships.
A number of European animal species, such as Chorthippus parallelus (2, 3), Ursus arctos (4), and Sorex araneus (5, 6), have been investigated by using mainly maternally inherited DNA markers. Glacial refugia and postglacial recolonization routes were identified. Similar investigations were carried out on European tree species by using maternally inherited DNA markers as well. In both angiosperm and gymnosperm species the postglacial history was reconstructed—e.g., in beech (7), white oaks (8), black alder (9), Scots pine (10), and Norway spruce (11). Comparative phylogeographic studies have recognized that different European plant and animal species shared postglacial common migration routes (12, 13). Three general types of recolonization patterns were defined by different speed and routes of migration from refugia south of the Alps and the Pyrenees, and from the Balkan Peninsula (13). Analyses of sequence divergence between maternal lineages originating from different refugia led to the conclusion that they have been isolated for several glacial periods (1). Therefore, Hewitt (13) postulates that the climatic oscillations of the Quaternary had an impact on speciation because of long-term isolation in refugial areas. Most forest trees of the temperate climate are wind-pollinated with the capacity for long-distance pollen dispersal. The question arises, whether gene flow through pollen confounds long-term genetic effects of refugial isolation.
Wind-pollinated conifers of the family Pinaceae are very suitable to answer that question. According to Wagner (14) their organellar DNA follows contrasting modes of inheritance, chloroplast DNA (cpDNA) being paternally inherited and mitochondrial DNA (mtDNA) being maternally inherited. Recently, studies have emerged that contrasted seed- vs. pollen-mediated gene flow in conifers. While many of them operate on a higher taxonomic level, so far only few shed light on intraspecific organellar gene flow. Within wind-pollinated conifer species an asymmetry was observed, with pollen being the main agent of spatiotemporal gene flow (15–17).
In the present study, the Pinaceae species Abies alba Miller was an excellent model for differential gene flow because two highly conserved alleles were detected both in a cpDNA locus and in an mtDNA locus which could be assigned to either of two far distant glacial refugial areas. Thus, without any assumption on intraspecific phylogeny, the postglacial dispersal of genes through seeds and pollen could be directly contrasted at a range-wide scale. Furthermore, human influence on A. alba is fairly low, the sampled populations are considered to be natural, and the reconstructed processes are thus supposed to be authentic. European Abies species are not sympatric, so that interspecific hybridization events, which could intermingle with the dynamics of intraspecific gene flow, are not likely to occur in the study area.
The two contrasting markers were screened in 100 populations from the entire range of A. alba. On the basis of allele frequencies, centers and widths of the fitted nad5-4 and psbC clines were calculated. The observed patterns led to the hypothesis that wind-pollinated plants are able to establish a highly efficient pollen-mediated gene flow among refugia during interglacial periods. These insights into past gene flow of wind-pollinated plants are discussed for evolutionary consequences.
Materials and Methods
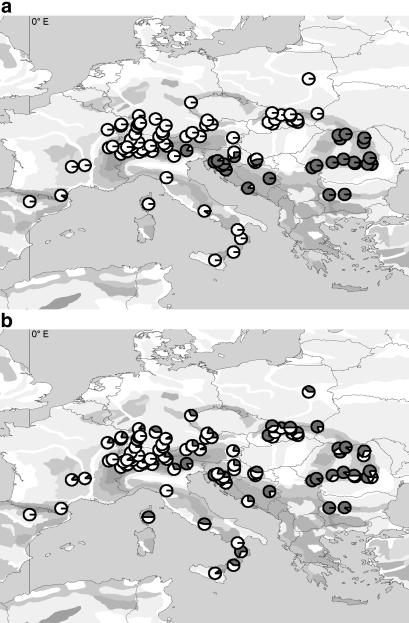
At the present time, silver fir covers a broad mountainous area across central Europe, ranging from ≈0° to 27°E and 38° to 52°N. We obtained samples from 1,062 individuals of 100 silver fir populations covering the entire natural range of the species (Fig. 1). Seventeen populations were included that had been analyzed for cpDNA variation in former studies (18, 19). From each population between 3 and 25 individuals were analyzed. In 86 populations 10 or more individuals were sampled. Given the high number of populations and assuming high fixation values, the optimal sample size would have been 2 or 3 individuals per population to minimize variance (20). Needles from adult trees or from seedlings, and occasionally embryos from seeds, were used. Populations and the distribution of haplotype frequencies were mapped by using the ArcView GIS 3.0 software (ESRI, Redlands, CA).
Fig 1.
Geographic maps of Europe showing all sampled A. alba populations. The circle sectors display the relative frequencies of the observed alleles of the respective marker. (a) Maternally inherited nad5-4 marker. White circle sectors mark allele 1 and dark sectors mark allele 2. (b) Paternally inherited psbC marker. Dark circle sectors mark allele A and white sectors mark allele B.
For inheritance analysis of the newly developed mtDNA marker, we sampled parents and offspring from the following controlled interspecific crosses: A. alba × Abies nordmanniana with 10 offspring, A. nordmanniana × A. alba with 20 offspring, A. alba × Abies pinsapo with 3 offspring and A. alba × Abies cephalonica with 14 offspring.
DNA Extraction.
Genomic DNA was isolated from about 50 mg of fresh needle material or one freshly prepared embryo. DNA was extracted by following a miniaturized CTAB (cetyltrimethylammonium bromide) protocol (21), and DNA concentration was measured in a fluorometer.
mtDNA Marker.
A screening for mtDNA variation with a range of available primers resulted in the detection of one intraspecific polymorphism. This was found in the fourth intron of the mitochondrial NAD dehydrogenase subunit 5 gene (nad5). Using universal primers (22), we obtained sequences from nine A. alba individuals of different geographic origin with an automated sequencer (ABI Prism 310, Applied Biosystems). We used the obtained sequences to design internal diagnostic primers to remove noninformative border regions and to reduce homoplasic effects. The fragment amplified by this new primer pair was named nad5-4. The primer sequences are 5′-GGACAATGACGATCCGAGATA-3′ and 5′-CATCCCTCCCATTGCATTAT-3′.
The PCR mixture (25 μl) contained 1× PCR buffer, 1.6 mM MgCl2, 0.2 μM each primer, 0.1 mM each dNTP, 0.5 unit of Taq DNA polymerase (Eurogentec, Cologne, Germany), and 20 ng of template DNA. The initial denaturation for 3 min at 94°C was followed by 30 cycles of denaturation (1 min at 92°C), annealing (1 min at 52.5°C), and extension (1 min 20 sec at 72°C), and a final extension step of 8 min at 72°C. A 5-μl sample of each PCR product was subjected to gel electrophoresis in a 1.2% agarose gel for 2 h at 6 V/cm.
cpDNA Marker.
We applied a paternally inherited cpDNA marker (18) to all sampled silver fir populations. This marker is a restriction site polymorphism in the photosystem II CP43 protein gene (psbC).
Primers A (5′-GGTCGTGACCAAGAAACCAC-3′) and E (5′-GGACAGGTTCGAAATCACGA-3′) were used to amplify a large portion of the chloroplast psbC gene. The sequence of primer A was published by Demesure et al. (23). Primer E was designed from the sequence of the chloroplast genome of Pinus thunbergii (24). PCR was performed in a volume of 25 μl (see above for ingredients). The initial denaturation (4 min at 94°C) was followed by 35 cycles of denaturation (1 min at 93°C), annealing (1 min at 57°C), and extension (2 min at 72°C), and a final extension step of 10 min at 72°C. A 2.5-μl sample of PCR product was incubated overnight at 37°C with 1 unit of HaeIII restriction endonuclease (MBI Fermentas, St. Leon-Rot, Germany) and 2 μl of buffer Y+-Tango (MBI) in a total volume of 20 μl. Restriction fragments were separated on a 1.5% agarose gel for 3 h at 5.5 V/cm. We sequenced the two psbC variants from each of three individuals by primer walking (see above for sequencing method).
Data Analysis.
To get an impression of the strength of the gradual transition from one refugium to the other, we analyzed allele frequency clines of the corresponding gene markers. Center and width of geographic clines were estimated in the following way. Allele frequencies for each population and locus were calculated. For each locus the allele frequencies were plotted against the geographic longitude of the populations and fitted with the sigmoid function (25)
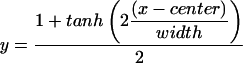 |
by applying the nonlinear least square method of the R-Project (26).
The data were additionally analyzed by geostatistics. Variograms were calculated for the data of each locus with the geo-R package (27) of the R-project. A semivariogram displays the semivariance at different distance classes. Semivariance is defined as the average of the squared differences between observations separated by a specific lag distance. Stationarity is assumed—i.e., the difference between the values at any two points is a function of the distance between the points only (28). The function is valid under the assumption that no selection is acting on the marker and that gene flow is equal for the entire observation area. The variogram is estimated as a discrete function of given distance classes (28):
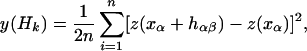 |
where n is the number of pairs in the given distance class Hk. hαβ is a vector belonging to the distance class Hk and k is the number of distance classes.
The shape of a semivariogram may be fitted with a model (such as linear, exponential, spherical, and Gaussian). Typically, the range and sill are two parameters of semivariograms used to describe data. With an increasing distance the difference between the compared entities becomes larger. At some distance, the semivariogram runs into a flat region called the sill. The distance at which the sill is reached is called range. The range generally describes the distance at which two points in space are considered independent. The height of the sill often implies the variability of data.
Results
We applied the maternally inherited mtDNA marker, nad5-4, and the paternally inherited cpDNA marker, psbC, to 1,062 A. alba individuals from 100 populations covering the entire range of the species. We detected two variants in each marker and characterized the nature of the variation by sequence analysis. The distribution of allele frequencies was mapped, centers and widths of the observed clines were calculated, and the data were analyzed with geostatistics.
Geographical Distribution of mtDNA Variation.
We observed an intraspecific length polymorphism with two length variants in nad5-4 of A. alba (data not shown). Alleles 1 and 2 had a size of 230 and 150 bp, respectively, when amplified with the diagnostic primers. Sequence analysis revealed a single insertion/deletion of 80 bp (GenBank accession nos. AY147793 and AY147794). We detected no further mutations. A blast search confirmed that our sequences indeed represented the fourth intron of the mitochondrial nad5 gene. nad5-4 exhibited interspecific length differences that were used to determine the mode of inheritance in controlled interspecific crosses. We were able to confirm uniparentally maternal inheritance of the marker (data not shown).
The distribution of the two alleles throughout Europe showed a very strong subdivision of the natural range (Fig. 1a). Populations from the western part of the range exclusively contained allele 1 and populations from the eastern part contained allele 2. Mixed populations were observed in Croatia, Slovenia, and northeastern Italy (Fig. 1a). East and west of these locations populations were fixed for one allele, except for three populations of the western part, in each of which one individual exhibited allele 2. Those populations were located in the eastern Pyrenees, in the Black Forest, and in central Italy. In total, a greater part of the range was recolonized by the western than by the eastern lineage.
Geographical Distribution of cpDNA Variation.
In the psbC gene two different HaeIII restriction patterns were observed, in accordance with previous findings (18). Allele A exhibited three restriction fragments and allele B exhibited four restriction fragments (data not shown).
The results of the sequence analysis were consistent with the observed restriction fragment patterns. We identified three HaeIII restriction sites for allele A resulting in four restriction fragments. Two of the restriction sites were located so close together that the resulting DNA fragment with a length of 8 bp could not be observed in agarose gels. The sequence of allele B revealed an additional HaeIII restriction site, caused by an A-to-G transition (GenBank accession no. AY147792). In the reading frame this is a silent mutation, causing no change in the amino acid sequence of the translated protein. Both alleles were present throughout most of the analyzed populations of A. alba although they were not evenly distributed. A geographical cline was visible, with allele A being more frequent in the east and allele B being more frequent in the west of the distribution range. Populations that were fixed for one cpDNA variant were found in northwestern Italy (allele B) and in Bulgaria (allele A), marking the putative western and eastern refugial areas.
Comparison of Geographical Clines Between the Two Markers.
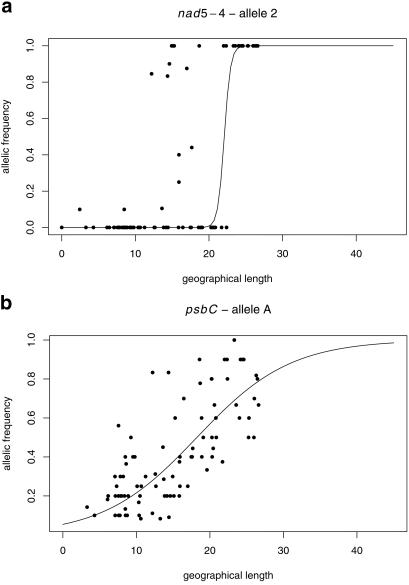
The centers and widths of the east–west clines are displayed in Table 1. The cline of chloroplast psbC alleles is as wide as the natural range of A. alba, whereas the cline of mitochondrial nad5-4 alleles spreads over less than 2° of longitude (Fig. 2). The centers of the two clines are close together. A Kolmogorov–Smirnov test confirmed that the differences in allele frequency distributions of the two markers were highly significant (α < 0.001).
Table 1.
Center and width of east–west clines of nad5-4 and psbC alleles
| Gene
|
Mode of inheritance
|
Longitude, ° | |
|---|---|---|---|
| Center | Width | ||
| nad5-4 | ♀ | 22.0749 | 1.7233 |
| psbC | ♂ | 18.2291 | 25.4734 |
Fig 2.
Fitted clines for mitochondrial nad5-4 (a) and chloroplast psbC (b) variation. Allele frequencies were plotted against geographic longitude.
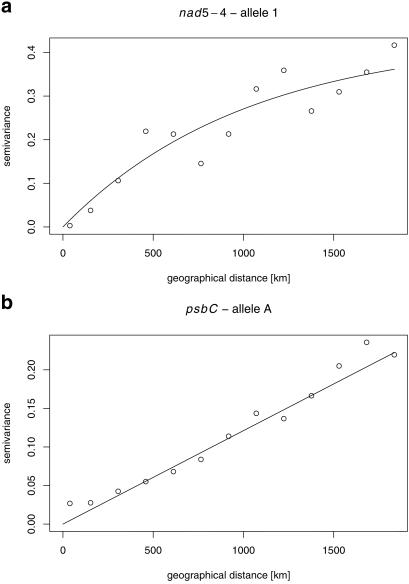
The results of the variogram analysis are displayed in Fig. 3. The shape of the variograms differs between the two markers. The variogram curve of nad5-4 is of an exponential type (Fig. 3a) and reaches a value close to the sill of 0.5. In contrast, the variogram curve of psbC has a linear shape (Fig. 3b), is much shallower, and reaches only ≈0.25. These results indicate a limited range of seed-mediated gene flow (nad5-4) and a continuous pollen-mediated gene flow (psbC) across the whole analyzed region.
Fig 3.
Variograms of mitochondrial nad5-4 (a) and chloroplast psbC (b) allele frequencies.
Discussion
We know of no previous study that has explicitly demonstrated that genetic contact between Pleistocene refugia of A. alba was established through pollen-mediated gene flow. These results will be discussed for general conclusions on gene flow of wind-pollinated species.
Selection or Secondary Contact?
Clines can be interpreted as the result of secondary contact between populations from different refugia or of selection acting along ecological gradients (29–32). To decide this question, a basic knowledge about the location of putative refugia is necessary. The geographical distribution of the two nad5-4 alleles revealed that recent populations of A. alba were divided into two maternal lineages with different geographic origin. It seems likely, that, after the last ice age, the maternal lineage carrying allele 1 originated from a western Mediterranean refugium, whereas the other lineage carrying allele 2 originated from an eastern Mediterranean refugium. This conclusion is in accordance with former allozyme studies indicating putative glacial refugia in the Apennines and the Balkan Peninsula (33). There is supporting evidence for this also from fossil pollen deposits (34). When our maternally inherited marker was used, an introgression zone between the eastern and western lineages was identified in Croatia and Slovenia (Fig. 1a). Pollen maps give evidence for an early contact in this area, ≈7,000–7,500 years ago (34). The fairly low amount of introgression between the maternal lineages points to a small extent of gene flow through seeds after populations became established. This is probably not a matter of selection, but rather one of colonization time and population density. After colonization was completed and the populations reached their optimal density the latter acted as a barrier to gene flow. This was demonstrated by a theoretical study (35). Evidence for another contact zone, in the northern Carpathians, comes also from pollen maps (34). This contact was established between 4,000 B.P. and 1,000 B.P., which may be the reason why no introgression zone was observed in our mtDNA study. Maybe a narrow introgression zone would be visible if the spatial resolution of sampling was increased. With allozyme data (33) two introgression zones were observed, a large one in the Carpathians and a small one in Croatia and Slovenia. Because these markers are biparentally inherited they have also a pollen-mediated component. Thus, using the cpDNA marker, we expected to detect a similar range of gene flow.
From the psbC allele frequencies, there is evidence that the geographical origin of the two maternal refugial lineages corresponds to the origin of the two paternal lineages. Populations are fixed for the respective alleles in the areas where the glacial refugia were postulated. Because the mtDNA marker is located in an intron of the nad5 gene and the mutation underlying the cpDNA marker is a synonymous point mutation, we assume that they are selectively neutral. Of course, a hitch-hiking effect cannot be ruled out with certainty. Altogether, there is stronger evidence for the forming of the psbC cline after secondary contact between two refugial lineages rather than through selection. We may assume that the cline began to form after populations from eastern and western refugia had merged, which was about 7,500 years ago in southern Europe and 1,000–4,000 years ago in the northern Carpathians.
The observed range-wide cline showed that pollen-mediated gene flow has spanned a much greater distance than suggested from allozyme data. This finding is noteworthy because A. alba has very large pollen grains (36), long generation times (≈40 years), and a very long lifespan (>300 years). All these life history traits act against high rates of gene flow.
The Power of the Model.
A. alba may serve as a well-suited model for other wind-pollinated plant species for several reasons. First, the applied markers have ideal properties because they are highly conserved and not prone to homoplasy like, for instance, microsatellite markers (37). There are only two alleles for each marker, and each allele corresponds to a single lineage that can therefore be identified without further phylogenetic analysis. Second, the life history traits of A. alba are bound to encumber pollen-mediated gene flow. Thus, generalizations from our model will rather underestimate than overestimate the effect of pollen-mediated gene flow in other wind-pollinated species.
Therefore we formulate the hypothesis that genetic contact between glacial refugia by means of reproductively effective pollen dispersal is possible during interglacial periods.
Is there evidence from other studies? Markers with opposite modes of inheritance have also been applied to North American conifers such as Pinus flexilis (15), Pinus ponderosa (16), and Pinus albicaulis (17). A higher amount of gene flow through pollen than through seeds was recognized in all three studies. The studies on P. flexilis (15) and P. ponderosa (16) do not allow generalization on range-wide gene flow. The former study is not a range-wide survey, which would be necessary to identify clines. The latter study is on a hybridization zone between two subspecies of P. ponderosa that had a lowered fertility in crossing experiments. This means that there is already a partial reproductive barrier, which may explain the rather narrow cline observed for cpDNA haplotypes. The results from the range-wide study on P. albicaulis (17) generally agree with our observations. However, the applied chloroplast microsatellite markers with their large number of haplotypes combined with relatively small sample sizes make it difficult to discover the explicit range of gene flow. Furthermore, an overestimation of gene flow may not be excluded because of the higher mutation rate and the possible cases of homoplasy in microsatellites.
Evolutionary Consequences.
It has been debated whether gene flow is a restrictive or creative force in evolution (38). In the case of A. alba both forces may act, indeed. On the one hand, high rates of pollen-mediated gene flow may slow down or prevent speciation, although from our data we were not able to judge the amount of introgression at the nuclear level. On the other hand, there could also be a creative aspect (38), because superior alleles or allele combinations can spread throughout the species, enhancing its adaptive potential. This might be true even if nuclear introgression is not significant, because both nucleus and chloroplast contain genes that code for subunits of chloroplast proteins (e.g., ribulose bisphosphate decarboxylase). If these evolved independently for a longer period, new combinations could result.
The application of contrasting uniparentally inherited markers displayed the highly effective dynamics of wind pollination. Some interesting conclusions can be drawn for species where such markers are not available. This is the case in most angiosperm species, where chloroplast and mitochondrial DNA are both maternally inherited. For example, in the white oak species complex many species hybridize (8) and thus form a huge gene pool, where genes can be exchanged widely through paternal gene flow. This exchange may enhance the adaptive potential of populations, although it will not erase all genetic differences that have accumulated between refugia, as can be observed in studies using nuclear markers (e.g., refs. 39 and 40). Thus, from a paternal point of view, ice ages might not leave the deep genetic imprints in wind-pollinated plants that were found so far with maternally inherited markers.
The analysis of differential gene flow will substantially increase knowledge about the impact of different life history traits on evolutionary key processes such as speciation, hybridization, and adaptation.
Acknowledgments
We are very grateful to G. M. Hewitt and R. J. Petit for helpful comments on the manuscript. We thank S. Jelkmann, V. Kuhlenkamp, and I. Schulze for the excellent work in the lab. Further, we thank I. Barbu, H. Braun, W. Eder, B. Fady, A. Franke, L. Llamas Gómez, J. Gracan, E. Hussendörfer, M. Konnert, A. Kormuták, L. Paule, D. Peev, E. Ritter, P. Rotach, I. Strohschneider, W. van der Knaap, G. G. Vendramin, and G. J. Wilhelm for kindly providing us with sample material. We also thank two anonymous reviewers for helpful comments. This work was funded by the European Union Project FOSSILVA (CT-1999-00036).
Abbreviations
cpDNA, chloroplast DNA
mtDNA, mitochondrial DNA
References
- 1.Hewitt G. M. (2000) Nature 405, 907-913. [DOI] [PubMed] [Google Scholar]
- 2.Cooper S. J. B., Ibrahim, K. M. & Hewitt, G. M. (1995) Mol. Ecol. 4, 49-60. [DOI] [PubMed] [Google Scholar]
- 3.Lunt D. H., Ibrahim, K. M. & Hewitt, G. M. (1998) Heredity 80, 633-641. [DOI] [PubMed] [Google Scholar]
- 4.Taberlet P. & Bouvet, J. (1994) Proc. R. Soc. London B 255, 195-200. [DOI] [PubMed] [Google Scholar]
- 5.Taberlet P., Fumagalli, L. & Hausser, J. (1994) Evolution 48, 623-636. [DOI] [PubMed] [Google Scholar]
- 6.Fumagalli L., Hausser, J., Taberlet, P., Gielly, L. & Stewart, D. T. (1996) Hereditas 125, 191-199. [Google Scholar]
- 7.Demesure B., Comps, B. & Petit, R. J. (1996) Evolution 50, 2515-2520. [DOI] [PubMed] [Google Scholar]
- 8.Dumolin-Lapegue S., Demesure, B., Fineschi, S., Le Corre, V. & Petit, R. J. (1997) Genetics 146, 1475-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King R. A. & Ferris, C. (1998) Mol. Ecol. 7, 1151-1161. [Google Scholar]
- 10.Sinclair W. T., Morman, J. D. & Ennos, R. A. (1999) Mol. Ecol. 8, 83-88. [Google Scholar]
- 11.Sperisen C., Büchler, U., Gugerli, F., Mátyás, G. & Geburek, T. (2001) Mol. Ecol. 10, 257-263. [DOI] [PubMed] [Google Scholar]
- 12.Taberlet P., Fumagalli, L., Wust-Saucy, A.-G. & Cosson, J.-F. (1998) Mol. Ecol. 7, 453-464. [DOI] [PubMed] [Google Scholar]
- 13.Hewitt G. M. (1999) Biol. J. Linn. Soc. 68, 87-112. [Google Scholar]
- 14.Wagner D. B. (1992) New Forests 6, 373-390. [Google Scholar]
- 15.Latta R. G. & Mitton, J. B. (1997) Genetics 146, 1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latta R. G. & Mitton, J. B. (1999) Evolution 53, 769-776. [DOI] [PubMed] [Google Scholar]
- 17.Richardson B. A., Brunsfeld, S. J. & Klopfenstein, N. B. (2002) Mol. Ecol. 11, 215-227. [DOI] [PubMed] [Google Scholar]
- 18.Ziegenhagen B., Kormutak, A., Schauerte, M. & Scholz, F. (1995) Forest Genet. 2, 99-107. [Google Scholar]
- 19.Vendramin G. G., Degen, B., Petit, R. J., Anzidei, M., Madaghiele, A. & Ziegenhagen, B. (1999) Mol. Ecol. 8, 1117-1126.10447853 [Google Scholar]
- 20.Pons O. & Petit, R. J. (1995) Theor. Appl. Genet. 90, 462-470. [DOI] [PubMed] [Google Scholar]
- 21.Dumolin S., Demesure, B. & Petit, R. J. (1995) Theor. Appl. Genet. 91, 1253-1256. [DOI] [PubMed] [Google Scholar]
- 22.Wu J., Krutovskii, K. V. & Strauss, S. H. (1998) Genetics 150, 1605-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demesure B., Sodzi, N. & Petit, R. J. (1995) Mol. Ecol. 4, 129-131. [DOI] [PubMed] [Google Scholar]
- 24.Wakasugi T., Tsudzuki, J., Ito, S., Shibata, M. & Sugiura, M. (1994) Plant Mol. Biol. Rep. 12, 227-241. [Google Scholar]
- 25.Buño I., López-Fernández, C., Butlin, R. K., Hewitt, G. M. & Gosálvez, J. (1994) Heredity 73, 625-634. [Google Scholar]
- 26.Ihaka R. & Gentleman, R. (1996) J. Comput. Graphical Stat. 5, 299-314. [Google Scholar]
- 27.Ribeiro P. J. & Diggle, P. J. (2001) R News (ISSN 1609-3631) 1, 15-18.. Available at http://cran.r-project.org/doc/Rnews/. [Google Scholar]
- 28.Wackernagel H., (1995) Multivariate Geostatistics (Springer, Berlin).
- 29.Barton N. H. & Hewitt, G. M. (1985) Annu. Rev. Ecol. Syst. 16, 113-148. [Google Scholar]
- 30.Hewitt G. M. (1989) in Speciation and Its Consequences, eds. Otte, D. & Endler, J. A. (Sinauer, Sunderland, MA), pp. 85–110.
- 31.Hewitt G. M. (1993) in Evolutionary Patterns and Processes, eds. Lees, D. R. & Edwards, D. (Academic, London), pp. 97–123.
- 32.Le Corre V., Roussel, G., Zanetto, A. & Kremer, A. (1998) Heredity 80, 464-473. [Google Scholar]
- 33.Konnert M. & Bergmann, F. (1995) Plant Syst. Evol. 196, 19-30. [Google Scholar]
- 34.Huntley B. & Birks, H. J. B., (1983) An Atlas of Past and Present Pollen Maps for Europe: 0–13,000 Years Ago (Cambridge Univ. Press, Cambridge, U.K.).
- 35.Nichols R. A. & Hewitt, G. M. (1994) Heredity 72, 312-317. [Google Scholar]
- 36.Stanley R. G. & Linskens, H. F., (1974) Pollen (Springer, Heidelberg).
- 37.Liepelt S., Kuhlenkamp, V., Anzidei, M., Vendramin, G. G. & Ziegenhagen, B. (2001) Mol. Ecol. Notes 1, 332-335. [Google Scholar]
- 38.Slatkin M. (1987) Science 236, 787-792. [DOI] [PubMed] [Google Scholar]
- 39.Zanetto A. & Kremer, A. (1995) Heredity 75, 506-517. [Google Scholar]
- 40.Comps B., Gomory, D., Letouzey, J., Thiebaut, B. & Petit, R. J. (2001) Genetics 157, 389-397. [DOI] [PMC free article] [PubMed] [Google Scholar]