Abstract
We recently described a CHO DSBR mutant belonging to the XRCC7 complementation group (XR-C2) that has the interesting phenotype of being radiosensitive, but having only a modest defect in VDJ recombination. This cell line expresses only slightly reduced levels of DNA-PKcs but has undetectable DNA-PK activity. Limited sequence analyses of DNA-PKcs transcripts from XR-C2 revealed a point mutation that results in an amino acid substitution of glutamic acid for glycine six residues from the C-terminus. To determine whether this single substitution was responsible for the phenotype in XR-C2 cells, we introduced the mutation into a DNA-PKcs expression vector. Whereas transfection of this expression vector significantly restores the VDJ recombination deficits in DNA-PKcs-deficient cells, radioresistance is not restored. Thus, expression of this mutant form of DNA-PKcs in DNA-PKcs- deficient cells substantially recapitulates the phenotype observed in XR-C2, and we conclude that this single amino acid substitution is responsible for the non-homologous end joining deficits observed in XR-C2.
INTRODUCTION
Double-strand breaks (DSBs) in DNA are induced by a number of DNA-damaging agents including ionizing radiation and radiomimetic drugs which, if left unrepaired, lead to chromosome loss and cell death. The predominant pathway for the repair of ionizing radiation induced DSBs in mammalian cells is non-homologous end joining (NHEJ) (1–3). The NHEJ machinery also resolves the DNA breaks that are generated during the site-specific recombination process (VDJ recombination) that assembles immune receptor genes from their disparate component gene segments during lymphocyte development (4–7).
Six different molecules have been shown to be required for NHEJ: Ku70, Ku86, DNA-PKcs, XRCC4, DNA ligase IV and Artemis. Animals, humans and cell lines deficient in any of these factors display radiosensitivity as well as deficits in VDJ recombination that result in the disease, severe combined immunodeficiency (SCID) (8–12).
Three of the six known NHEJ factors comprise the DNA-PK complex, a serine/threonine protein kinase that must be physically associated with DNA to be active (reviewed in 13). The complex contains two subunits. The ∼465 kDa catalytic subunit (DNA-PKcs) possesses weak DNA binding activity as well as protein kinase activity (14,15). The regulatory subunit, the Ku heterodimer, directs DNA-PKcs to DNA ends and stabilizes its DNA binding so that it is efficiently activated (16,17). Recent data suggest that Artemis possesses both exonucleolytic and endonucleolytic activities that may be important for DNA end processing prior to resolution of DSBs (18). XRCC4 is a 37 kDa protein that interacts with, and catalytically stimulates, the activity of DNA ligase IV (19–21). The XRCC4–DNA ligase IV complex carries out the final step of joining DNA ends in NHEJ (22).
DNA-PK is central in the pathway because it initially recognizes and binds to damaged DNA and then targets other repair activities to the site of damage. Recent data demonstrate unequivocally that DNA-PK’s kinase activity is essential during NHEJ (23–25) though definition of DNA-PK’s physiologic target(s) has been elusive. Five of the six known NHEJ factors are known in vitro or in vivo targets of DNA-PK (only DNA ligase IV is not) (18,26–28). Recent data suggest that DNA-PK’s phosphorylation of Artemis alters its nucleolytic activity suggesting that this phosphorylation event may be critical during NHEJ. Still, the physiologic relevance of DNA-PK’s phosphorylation of Artemis or any other factor has yet to be proven, and the precise role of DNA-PK in NHEJ is uncertain.
In the numerous cell lines and animal models previously characterized as being defective in NHEJ, the vast majority show profound deficits in both VDJ recombination and radioresistance (29). Three exceptions to the concordance of VDJ recombination deficiency and radioresistance have been reported. First, Riballo et al. (30) described a mutation in the DNA ligase IV gene of a leukemia patient who dramatically over-responded to radiotherapy. The mutant DNA ligase IV from this patient was specifically defective in the adenylation step of ligation, but the enzyme retained residual DNA ligase activity. Though cell lines derived from this patient are radiosensitive, VDJ recombination (as tested in transient assays) is relatively normal. Recently, several additional human patients have been described who have germline DNA ligase IV mutations and who have similar phenotypes (31). Secondly, we have demonstrated that murine SCID fibroblasts that express minimal levels of wild-type DNA-PKcs from a transfected construct support VDJ recombination, but maintain the radiosensitive phenotype of the parental SCID fibroblast cells (25). These data are consistent with a model whereby minimal NHEJ function can suffice to repair the few breaks (probably only two per cell) associated with VDJ recombination, whereas wild-type NHEJ activity is required to sustain normal radioresistance. The third example of discordant deficits in VDJ recombination and radioresistance is our previous description of the CHO mutant cell line, XR-C2 (32). This cell line belongs to the XRCC7 complementation group, implicating DNA-PKcs as the defective factor. As expected, XR-C2 is markedly sensitive to ionizing radiation; however, VDJ recombination is only modestly reduced. In transient recombination assays with this cell line, coding joint resolution is diminished by ∼2-fold and single joint resolution by ∼3-fold. Furthermore, western analyses revealed slightly reduced levels of DNA-PKcs in XR-C2; however, kinase activity was completely undetectable suggesting a possible mutation within the kinase domain (32).
We have defined a mutation that results in a single amino acid substitution six residues from the C-terminus, introducing a glycine to glutamic acid interchange. To document that this mutation is responsible for the XR-C2 phenotype, a mutational approach was undertaken. We find that DNA-PKcs-deficient rodent cells expressing human DNA-PKcs with the G→E interchange essentially recapitulate the novel phenotype observed in the XR-C2 DSBR mutant cell line.
MATERIALS AND METHODS
cDNA synthesis and sequencing
Total RNA was isolated by the guanidium isothiocyanate method from the XR-C2 cell line as well as the parental line, CHO9. First-strand DNA-PKcs cDNA synthesis was performed as follows: total RNA (1 µg) was added to a reverse transcription solution consisting of 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 5 mM MgCl2, 1 mM dNTPs, 2.5 µM oligo(dT)15 primer, 20 U RNasin and 50 U MuLV (Murine Leukaemia Virus) reverse transcriptase. The reaction mixture (final volume, 20 µl) was incubated for 30 min at 42°C, heated to 95°C for 5 min, and then chilled on ice. The newly generated RNA:cDNA hybrids were amplified by the polymerase chain reaction (PCR) with specific DNA-PKcs primers, 5a and HCs6, resulting in a 1.3 kb product. PCR was performed with 30 cycles consisting of denaturation for 30 s at 94°C, primer annealing at 55°C for 30 s and extension at 72°C for 2 min (DNA thermal cycles; Perkin-Elmer Cetus, Norwalk, CT). Following PCR the amplified 1.3 kb cDNA was gel-purified (Qiaex; Qiagen, Chatsworth, CA), and cloned into a TA cloning vector (Promega, Madison, WI). DNA sequence was obtained with a T7 sequence kit (Pharmacia Biotech, Uppsala, Sweden), using [γ-32P]dATP. The samples were resolved in a 6% polyacrylamide gel at 40 W. Gels were exposed to X-ray film.
Oligonucleotides
The following oligonucleotides were used for amplification and sequencing of hamster DNA-PKcs from both the XR-C2 cell line and the parental line, CHO9: 5a, 5′-GACTCAAAGCCACCTGGGAACCTG; HSC6, 5′-CTGCTGTCAGTGAAGGTCTAGGAG; SCID3, 5′-GATTGAAGGAGAATCGTAT CGC; SCID4, 5′-ACATTTATTTCTTGAATCCA. The following oligonucleotides were used for constructing the mutant DNA-PKcs expression construct: 3′mut, 5′-TTCCCGGGTCACATCCAGGGCTCCCACTCCTCCCAGGTTCTGCC; 5′Eco721, 5′-TATGACGGTAGGGGAAAGCCATTG.
Cell lines and culture
The XR-C2 cell line has been described previously (32). V3, a Chinese hamster ovary DSBR mutant cell line known to be defective in DNA-PKcs was the generous gift of Drs Joanna Hesse and Martin Gellert (National Institutes of Health, Bethesda, MD). Sf19, a SCID mouse fibroblast cell line was generously provided by Dr Mel Bosma (Fox Chase Cancer Center, Philadelphia, PA). Clonal transfectants of the Sf19 cell line expressing wild-type or mutant human DNA-PKcs, or vector alone were derived by co-transfecting 25 µg of PvuI linearized DNA-PKcs expression constructs and 1 µg of NotI linearized pcDNAI/Neo plasmid (Invitrogen, Carlsbad, CA) with FuGENE 6 Transfection Reagent (Roche Molecular Biochemicals, Indianapolis, IN). Forty-eight hours after each transfection, cells were placed under selection conditions (400 µg/ml G418). Individual colonies were isolated and were screened for DNA-PKcs expression by immunoblot analysis. The following Sf19 clonal transfectants were utilized. Clone 2-2 and clone 2-7 expresses wild-type DNA-PKcs and were described previously (25). Clones C3-8 and C-3-18 express the mutant DNA-PKcs. Clone pc-3 was transfected with vector only (25).
Plasmids
The VDJ recombination substrates pJH201 and pJH290 (33,34), and the RAG-1 and RAG-2 expression plasmids pJH548 and pJH549, respectively (35), were a gift from Dr M. Gellert (National Institutes of Health, Bethesda, MD). The wild-type DNA-PKcs expression vector was described earlier (36). The mutant DNA-PKcs construct was generated by PCR mutagenesis. Briefly, an oligonucleotide (3′mut) that encodes a glycine to glutamic acid substitution at amino acid position 4122 of human DNA-PKcs and includes an XmaI restriction site, was used in conjunction with oligonucleotide 5′Eco721 to amplify 1.18 kb of the DNA-PKcs coding region. This fragment was subcloned (TA cloning kit; Invitrogen) and sequenced. Subsequently, an Eco721 to XmaI restriction fragment was subcloned into the wild-type DNA-PKcs expression construct from which the analogous fragment had been previously excised.
Assessment of radiosensitivity
3000 cells were resuspended in serum-free media and then exposed to 0, 1, 2 or 4 G ionizing radiation, using a 32Co source and immediately plated in 100 mm2 tissue culture dishes in complete media. After 7–10 days, cell colonies were fixed and stained with crystal violet. Within each experiment, irradiation sensitivities were determined in triplicate, and all experiments were repeated at least three times.
VDJ recombination assay
To assess VDJ recombination in V3 cells, RAG1 and RAG2 expression constructs (3 µg each), wild-type or mutant DNA-PKcs expression plasmid or the pCMV6 vector control (6 µg), and recombination substrates (1 µg of pJH201 or pJH290) were transiently introduced into cells using FuGENE 6 (Roche Molecular Biochemicals). Forty-eight hours later, plasmid substrates were rescued from the cells by alkaline lysis, treated with DpnI, and a portion transformed into chemically competent Escherichia coli (Max efficiencyDH5α™; Life Technologies, Gaithersburg, MD). Transformed bacteria were spread onto two LB agar plates containing either 100 µg/ml ampicillin or 100 µg/ml ampicillin and 22 µg/ml chloramphenicol.
To examine the fine structure of coding joints, plasmid DNA from a fraction of the pJH290 recombinants was prepared and sequenced. To examine the fidelity of signal joining, plasmid DNA was prepared from a fraction of the pJH201 recombinants and restricted with HindIII plus ApaLI. A novel ApaLI site is created by a perfectly fused pJH201 signal joint.
Protein extract preparation
Whole cell extracts were prepared by a modification of the method of Finnie et al. (37). Briefly, 20 × 106 cells were harvested and washed. Cell pellets were resuspended in 20 µl of extraction buffer [20 mM HEPES, pH 7.8; 50 mM NaF; 450 mM NaCl; 25% (v/v) glycerol; 0.2 mM EDTA; 0.5 mM DTT; and protease inhibitors (Complete, EDTA free; Roche Molecular Biochemicals)]. The resuspended cell pellets were subjected to three freeze/thaw cycles (liquid nitrogen/37°C), and centrifuged at 4°C. Supernatants were stored at –80°C prior to use and concentrations were determined by Bradford analysis using BSA as a standard.
Immunoblotting
Indicated amounts of extracts were electrophoresed in 5% SDS–PAGE and transferred to poly(vinylidene difluoride) membranes. A mixture of the mouse monoclonal anti-DNA-PKcs antibodies 42-27 and 18.2 (gift of Dr T. Carter, St John’s University, Jamaica, NY) (38) was used as the primary antibody (1:300), and a goat anti-mouse IgG conjugated to horseradish peroxidase as the secondary antibody. Membranes were then incubated with a chemiluminescent substrate (ECL; Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer’s recommendations.
DNA-PK microfractionation and measurement of kinase activity
The SignaTECT DNA-PK assay system (Promega) was used to assay DNA-PK activity with the following modifications. Cell extracts (250 or 50 µg as indicated) were absorbed onto 20 µl of pre-swollen double-stranded DNA-cellulose beads (Amersham Pharmacia Biotech) for 30 min at 4°C. Subsequently, the DNA cellulose was washed three times with 1 ml of buffer A [25 mM HEPES, pH 7.9; 50 mM KCl; 10 mM MgCl2; 10% (v/v) glycerol; 1 mM EDTA; 1 mM EGTA; and 1 mM DTT] and then resuspended in 20 µl of DNA-PK reaction buffer containing 100 µg/ml BSA. Kinase reactions were conducted with 10 µl aliquots of the resuspended DNA-PK absorbed cellulose beads and were performed in both the presence and absence of a biotinylated DNA-PK p53-derived substrate peptide. Terminated reactions were analyzed by spotting onto SAM2™ membrane, washing, and counting in a scintillation counter as per the manufacturer’s instructions. At least three different extract preparations were tested for each cell line, and each assay was performed in duplicate.
To assess autophosphorylation of DNA-PKcs in V3 cells, 250 µg of whole cell extracts were absorbed onto 20 µl of DNA cellulose for 30 min on ice and then washed three times in buffer A. DNA cellulose fractions were then incubated in buffer A with 32P-labeled ATP and incubated for 15 min and then analyzed by SDS–PAGE and autoradiography or western blotting.
RESULTS
Sequence analysis of the 3′-terminal region of the DNA-PKcs cDNA from XR-C2 cells reveals a single amino acid substitution
Previously, several groups have described mutations in the extreme 3′ region of the DNA-PKcs coding region. The murine C.B-17 SCID mutation in DNA-PKcs is a T→A transition that introduces an ochre stop codon at amino acid 4045 resulting in an 83 amino acid C-terminal truncation (39). Similarly, the V3 mutant CHO cell line has been shown to have a mutation in one allele that introduces a premature termination codon that would result in a 104 amino acid C-terminal truncation (40). In the irs-20 mutant cell line, a mutation in the fourth codon from the termination codon (analogous to position 4124 in human DNA-PKcs) results in a substitution of lysine for glutamic acid (40). To determine whether the XR-C2 cell line also harbors a mutation(s) in this region, we performed RT–PCR using primers 5a and HCs6. The resulting 1.3 kb PCR product was cloned from both XR-C2 and the parental cell line, CHO9. Sequence analysis revealed a G→A transition in XR-C2 cells in the sixth codon prior to the termination codon (analogous to 4122 in human DNA-PKcs). As can be seen in Figure 1, this mutation results in the substitution of glutamic acid for glycine in the sixth residue from the C-terminus.
Figure 1.
A single nucleotide substitution in DNA-PKcs in XR-C2 introduces a G→E interchange six amino acids from the C-terminus. A 1.3 kb RT–PCR product from XR-C2 was cloned and sequenced. A single substitution was defined and compared to DNA-PKcs sequenced from the parental cell line CHO9. The substitution results in a glycine to glutamic acid substitution at six amino acids from the C-terminus as indicated.
Transiently expressed mutant DNA-PKcs partially restores VDJ recombination in DNA-PKcs-deficient cells
Because of the difficulties in sequencing the entire coding region from both DNA-PKcs alleles in XR-C2, we decided to employ a mutational approach to determine whether the glycine to glutamic acid substitution six amino acids from the C-terminus could explain the NHEJ deficiency in XR-C2. To that end, we generated an expression construct that includes the glycine to glutamic acid substitution in human DNA-PKcs (see Materials and Methods). We then assessed the ability of both wild-type and the G4122E mutant DNA-PKcs to support VDJ recombination in transient assays. As can be seen in Table 1, the wild-type DNA-PKcs construct substantially complements both the signal joint deficit (2.2% recombination versus 0.09% in transfections without DNA-PKcs expression vector) and the coding joint deficit (0.56% recombination versus 0% in transfections without DNA-PKcs expression vector). These data are in good agreement with previous complementation studies with the V3 cell line showing significant restoration of both signal and coding end joining by introducing wild-type human DNA-PKcs (24,41). In contrast, the G4122E DNA-PKcs mutant only partially restores VDJ recombination in V3 cells. Coding end joining is restored 6-fold less well than in transfections using wild-type DNA-PKcs (0.56 versus 0.09%). Still, coding joint formation is substantially more frequent in transfections with the mutant DNA-PKcs construct than with no DNA-PKcs expression vector (0.09% versus none). Similarly, signal end joining is restored ∼4-fold less well than in transfections using wild-type DNA-PKcs, but still 5-fold better than in transfections using RAG expression vectors but no DNA-PKcs expression vector. Thus, in the V3 mutant cell line, the G4122E mutant DNA-PKcs can only facilitate reduced levels of VDJ recombination. This is similar to the XR-C2 cell line in which coding and signal joints are diminished by ∼2- and ∼3-fold, respectively, as compared to the parental cell line (32).
Table 1. G4122E mutant DNA-PKcs supports reduced levels of VDJ recombination.
| Expression vectors | Experiment | Signal (pJH201) AmpR + CamR / AmpR | Ratea (%) | Fidelityb (%) | Coding (pJH290) AmpR + CamR / AmpR | Ratea (%) |
|---|---|---|---|---|---|---|
| No RAGS | 1 | 0/5560 | 0.00 | 0/22 380 | 0.00 | |
| 2 | 0/660 | 0.00 | 0/0 | 0.00 | ||
| 3 | 0/91 080 | 0.00 | 1/114 960 | 0.00 | ||
| 4 | 0/85 840 | 0.00 | 1/40 368 | 0.00 | ||
| RAGS | 1 | 11/6380 | 0.17 | 0/44 000 | 0.00 | |
| 2 | 0/840 | 0.00 | 0/0 | 0.00 | ||
| 3 | 31/63 120 | 0.05 | 0/53 760 | 0.00 | ||
| 4 | 40/22 736 | 0.18 | 0/79 576 | 0.00 | ||
| 0.09 ± 0.04c | ||||||
| Wild-type DNA-PKcs | 1 | 127/3300 | 3.85 | 70 (7/10) | 114/21 000 | 0.54 |
| 2 | 7/720 | 0.90 | ND | 10/3120 | 0.32 | |
| 3 | 496/32 520 | 1.52 | 80 (8/10) | 402/60 360 | 0.67 | |
| 4 | 230/9976 | 2.31 | 100 (7/7) | 345/49 300 | 0.70 | |
| 2.2 ± 0.64c | 86.7 ± 7.6d | 0.56 ± 0.09c | ||||
| G4122E DNA-PKcs | 1 | 34/3760 | 0.90 | 90 (9/10) | 8/16 080 | 0.05 |
| 2 | 2/1560 | 0.13 | ND | 4/2640 | 0.16 | |
| 3 | 140/31 080 | 0.45 | 89 (8/9) | 50/100 800 | 0.05 | |
| 4 | 188/35 380 | 0.53 | 90 (9/10) | 54/57 072 | 0.09 | |
| 0.50 ± 0.16c | 89.0 ± 0.3d | 0.09 ± 0.03c |
aRate is the percentage of replicated substrates that underwent V(D)J recombination and is derived from the formula [AmpR + CamR / AmpR] × 100.
bFidelity is expressed as the percentage of recovered signal joints that were susceptible to ApaLI digestion.
cAverage recombination rate ± SEM.
dAverage fidelity ± SEM.
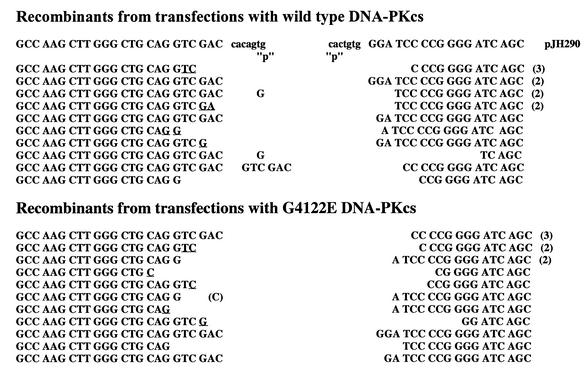
We next examined the fidelity of both coding and signal end joining in V3 cells complemented with wild-type or mutant DNA-PKcs expression vectors. To examine signal joining, plasmid DNA prepared from individual chloramphenicol-resistant colonies was restricted with HindIII and ApaLI. (A novel ApaLI site is created by head-to-head fusion of the two heptamers in the recombination signal sequence of pJH201.) As can be seen in Table 1, the fidelity of signal joints is indistinguishable between joints facilitated by wild-type or G4122E mutant DNA-PKcs. To examine the fidelity of coding joints, plasmid DNA was prepared and sequenced from individual chloramphenicol-resistant colonies. As can be seen in Figure 2, coding joints isolated from transfections using the G4122 mutant DNA-PKcs construct are structurally normal. The degree of nucleotide deletion and P segment retention is not different from recombinants isolated from transfectants using wild-type versus G4122E mutant DNA-PKcs constructs. In sum, whereas the frequency of recombination events is 4–6-fold lower in transfections using the mutant construct as compared to the wild-type DNA-PKcs construct, both coding and signal joints facilitated by the G4122E mutant are structurally normal. Thus, we conclude that the G4122E mutant DNA-PKcs expression vector can only partially complement the VDJ recombination deficits in V3 cells.
Figure 2.
Coding joints facilitated by the G4122E mutant DNA-PKcs are structurally normal. The sequence of coding ends and heptamers, as they are in the pJH290 substrate, are shown above the sequences of the recombinant junctions. Numbers to the right of sequences indicate the number of observations of that sequence. ‘P’ denotes palindromic nucleotides added to each junction. Nucleotides that cannot unequivocally be assigned to a particular coding end are underlined and listed in the more 5′ location.
Sf19 cells that stably express G4122E mutant DNA-PKcs are radiosensitive and lack DNA-PK activity
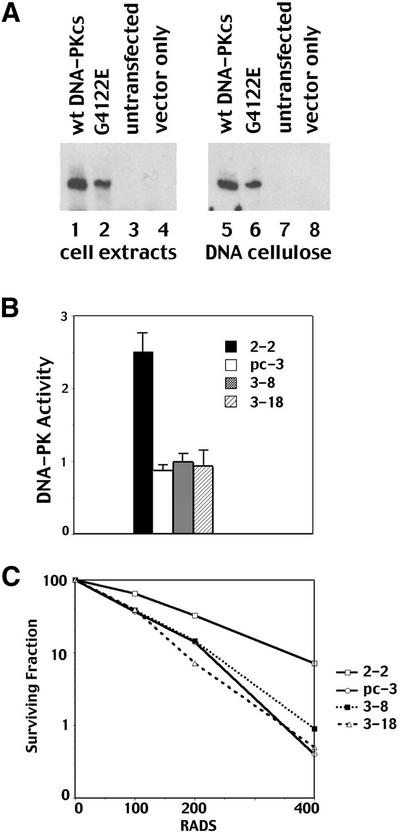
To determine whether the G4122E mutant DNA-PKcs could reverse radiosensitivity, stable transfectants were derived using a murine SCID fibroblast cell line, Sf19. To that end, the mutant DNA-PKcs construct was co-transfected into the SF19 cell line with a plasmid encoding neomycin resistance. After selection, individual colonies were selected and screened for DNA-PKcs expression by immunoblotting. Two individual clones expressing G4122E were expanded for further study (clones 3-8 and 3-18) and compared to a previously established SF19 clonal transfectant expressing wild-type DNA-PKcs (clone 2-2) (25) or vector only control transfectant (clone pc-3). The G4122E mutant protein is stably expressed in clone 3-18 murine (Fig. 3A, lane 2). (Clone 3-8 expresses slightly lower levels of the mutant protein; data not shown.) Furthermore, DNA cellulose fractionation of whole cell extracts demonstrates that the mutant protein co-localizes with Ku (not shown) onto DNA cellulose as efficiently as wild-type DNA-PKcs (Fig. 3A, lane 6). Thus, the glycine to glutamic acid interchange does not interfere with the interaction of DNA-PKcs with Ku. Expression of the G4122E mutant protein in murine SCID fibroblasts does not restore kinase activity as ascertained by a standard pulldown assay (Fig. 3B). Whereas DNA-PK activity is readily detectable from the SF19 transfectant expressing wild-type DNA-PKcs, DNA-PK activity in the two clones expressing the G4122E mutant protein is indistinguishable from the vector only control. Consistent with previous reports from our laboratory and others (24,25), complementation of the radiosensitivity of murine SCID cells is only observed in transfectants expressing the kinase competent form of DNA-PKcs (Fig. 3C). Though expression of the mutant protein in clones 3-8 and 3-18 is slightly less than wild-type DNA-PKcs in clone 2-2, it is substantially higher than in another wild-type transfectant (2-7; data not shown). This transfectant has easily detectable kinase activity and is fully complemented for both radioresistance and VDJ recombination (25). Thus, we conclude that the G4122E mutant DNA-PKcs cannot substantially restore DNA-PK activity or radioresistance, but it can significantly facilitate VDJ recombination.
Figure 3.
G4122E mutant DNA-PKcs does not restore kinase activity or radioresistance to murine SCID fibroblasts. (A) Immunoblot analyses of whole cell extracts (lanes 1–4) or DNA cellulose fractions (lanes 5–8) from SF19 cells stably transfected with the following: wild-type human DNA-PKcs expression vector (clone 2-2, lanes 1 and 5); G4122E mutant DNA-PKcs expression vector (clone 3-18, lanes 2 and 6); not transfected (lanes 3 and 7); empty expression vector (clone pc-3, lanes 4 and 8). (B) Whole cell extracts (250 µg) prepared from SF19 transfectants 2-2 (wild-type DNA-PKcs, black bar), pc-3 (vector only, white bar), 3-8 (G4122E, stippled bar) and 3-18 (G4122E, striped bar) were assayed for DNA-PK activity as described in Materials and Methods. Each cell extract was tested in duplicate, and at least four independent extracts were tested for each cell line. Bars indicate the SD. (C) Radiation resistance of SF19 transfectants 2-2 (wild-type DNA-PKcs), pc-3 (vector only), 3-8 (G4122E) and 3-18 (G4122E) was assessed as described in Materials and Methods. Data is presented as percent survival of unirradiated controls and is a representative experiment from four different assays.
V3 cells that substantially overexpress the G4122E mutant DNA-PKcs have minimal DNA-PK activity but are still substantially radiosensitive
As discussed above, the phenotypes of both the XR-C2 cell line and the Sf19 transfectants expressing the G4122E mutant protein are very similar to murine SCID cells expressing minimal levels of kinase competent DNA-PKcs. Thus, a reasonable hypothesis is that the G4122E mutant protein possesses minimal kinase activity—undetectable in either the XR-C2 cell line (32) or in the Sf19 transfectants described above. In ongoing work, we have succeeded in expressing considerably higher levels of human DNA-PKcs in the DNA-PKcs-deficient, CHO DSBR mutant cell line V3 than in murine SCID fibroblasts. We considered that weak DNA-PK activity might be more readily detectable in cells overexpressing the G4122E mutant protein. Thus, stable V3 transfectants expressing either wild-type human DNA-PKcs or the G4122E mutant protein were derived. Cell clones expressing high levels of human DNA-PKcs were selected for further study. As can be seen in Figure 4A, the wild-type and G4122 expressing clones have similar levels of DNA-PKcs. The amount is substantially higher than the amount detectable in the XR-C2 cell line although it is possible that the antibody used for DNA-PKcs detection recognizes the human protein better than hamster DNA-PKcs. We next assessed DNA-PK activity in the V3 transfectants. In a standard assay (using 250 µg of extract and incubating the reaction for 60 min), no significant activity could be detected in the transfectants expressing the G4122E mutant protein (data not shown). We considered that the mutant protein might be activated more slowly than wild-type DNA-PKcs. Thus, we next performed kinase assays with different incubation periods (Fig. 4B). As can be seen after 10 or 30 min, DNA-PK activity in the G4122E transfectants is not significantly different from vector only transfectants. However, a small amount of DNA-PK activity is consistently detected in extracts from transfectants expressing G4122E after 120 min. Thus, the G4122E mutant protein possesses weak DNA-PK activity as tested in the DNA-PK pulldown assay.
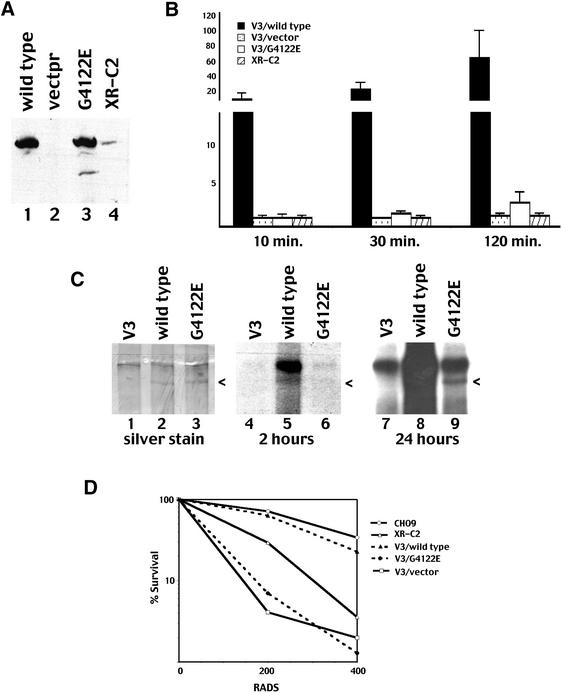
Figure 4.
G4122E mutant DNA-PKcs possesses weak DNA-PK activity. (A) Immunoblot analyses of whole cell extracts from V3 cells stably transfected with the following: wild-type human DNA-PKcs expression vector (lane 1); vector alone (lane 2); G4122E mutant DNA-PKcs expression vector (lanes 3); XR-C2 (lane 4). (B) Whole cell extracts (250 µg) prepared from V3 transfectants wild-type DNA-PKcs (black bar), pc-3 vector only (stippled bar), G4122E mutant protein (white bar) or XR-C2 (striped bar) were assayed for DNA-PK activity. Reactions were allowed to proceed for 10, 30 or 120 min as indicated. Each cell extract was tested in duplicate, and three independent extracts were tested for each cell line. Bars indicate the SD. (C) DNA cellulose fractions of 250 µg whole cell extracts from the following V3 transfectants: empty vector (lane 1); wild-type DNA-PKcs (lane 2); and G4122E mutant (lane 3) were incubated in kinase buffer with 32P-labeled ATP. Subsequently, bound proteins were analyzed by silver staining of a 6% SDS–PAGE gel (left) and autoradiography (middle and right). (D) Radiation resistance of V3 transfectants expressing wild-type DNA-PKcs; vector only, G4122E mutant protein was compared to that of the CHOK9 and XR-C2 cell lines. Data is presented as percent survival of unirradiated controls and is a representative experiment from three different assays.
It has been shown previously that DNA-PK autophosphorylates on all three component polypeptides. We have recently described the autophosphorylation sites within DNA-PKcs (42); an emerging consensus is that autophosphorylation is important for DNA-PK’s function (42). Autophosphorylation of both wild-type and G4122E mutant DNA-PKcs was next assessed by autoradiography of silver stained SDS–PAGE of DNA cellulose fraction of kinase reactions. As can be seen (Fig. 4C), transfected DNA-PKcs can be visualized in silver stained SDS–PAGE gels. (The identity of the indicated bands as DNA-PKcs was confirmed by immunoblotting; data not shown.) Phosphorylated wild-type DNA-PKcs is apparent after 2 h of autoradiography. After longer exposure, weak phosphorylation of the G4122E mutant protein is also apparent. Thus, the G4122E mutant protein is capable of weak phosphorylation of both the standard p53 peptide substrate and itself, and we conclude that the G4122E mutant protein possesses weak protein kinase activity.
The V3 transfectants also afforded us the opportunity to functionally compare the human DNA-PKcs mutant protein expressed in hamster cells with the original hamster cell lines. As can be seen (Fig. 4D), though wild-type human DNA-PKcs substantially complements the V3 cell line, the wild-type transfectant is still slightly more radiosensitive than CHO9 (the parental line of XR-C2). V3 cells expressing the G4122E mutant protein display similar radiosensitivity as empty vector control transfectants. The XR-C2 cell line is slightly more radioresistant than the V3 transfectants expressing the G4122E mutant protein. This might represent cell line variation between V3 and CHO9, potential species incompatibility between human DNA-PKcs and hamster Ku, or may be an artifact of the high DNA-PKcs expression levels in these transfectants. In summary, stable expression of the G4122E mutant protein in V3 cells essentially reiterates the phenotype of the XR-C2 cell line.
DISCUSSION
The original description of the XR-C2 cell line suggested that DNA-PK’s kinase activity is not essential for VDJ recombination. However, mutational strategies published from two laboratories (24,25) have clearly documented that kinase inactive forms of DNA-PKcs are incapable of reversing either the DNA repair or VDJ recombination defects in DNA-PKcs-deficient cells. We have shown previously that the phenotype of murine SCID cells complemented with the G4122E mutant protein is indistinguishable from cells expressing barely detectable levels of wild-type DNA-PKcs (at least 10-fold lower than the level required to fully complement SCID mouse fibroblasts) (25). In these cells, DNA-PK activity is undetectable, the cells maintain the radiosensitive phenotype of the parental DNA-PKcs-deficient cells, but VDJ recombination is partially restored. Thus, an emerging consensus is that wild-type levels of NHEJ function are required to facilitate repair of numerous DNA breaks (as is the case in experimental conditions commonly utilized to assess radioresistance), but more minimal NHEJ can function in VDJ recombination when fewer DNA breaks need to be resolved. Here we have documented that the G4122E mutant protein possesses weak DNA-dependent protein kinase activity. Thus, these data are completely consistent with the conclusion that minimal DNA-PK kinase activity can sustain VDJ recombination but not radioresistance. Furthermore, the finding of weak kinase activity in the mutant protein explains the novel phenotype of XR-C2.
To date, spontaneous germline mutations within the DNA-PKcs gene have been identified in three species (mice, horses and dogs) (39,43,44), and five spontaneous DNA-PKcs mutations have been defined in cell lines (four rodent cell lines and one human cell line) (40,45,46). With the exception of the recently documented mutation in MO59J (DNA-PKcs-deficient human glioma cell line), all of the DNA-PKcs mutations are in the C-terminal 1000 amino acids (46). The murine SCID mutation, and the mutations in irs20, V3 and XR-C2 are all located in the extreme C-terminus (last 104 of 4127 amino acids) of DNA-PKcs.
Two of these four extremely C-terminal substitutions are in the last six amino acids suggesting that the extreme C-terminus of DNA-PKcs is functionally critical. Sequence analyses of DNA-PKcs from divergent vertebrates has identified four highly conserved regions of DNA-PKcs (47). The PI3 kinase region is one of the highly conserved regions, and this domain includes approximately 300 amino acids at the C-terminus. The extreme C-terminus (including the last 35 amino acids) does not share homology with the lipid kinases within this family; however, this region is well conserved when comparing the protein kinases of the PI3 kinase family. Thus, it is reasonable to hypothesize that this conserved C-terminal region is required for protein kinase activity, potentially explaining why the murine SCID, irs20, V3 and XR-C2 mutations are so deleterious.
The last 35 amino acids of DNA-PKcs are extremely homologous between the seven available DNA-PKcs sequences. An alignment of the C-terminal 35 amino acids from available vertebrate AT-like PI3 kinase family members is presented in Figure 5. Of note, the glycine, six amino acids from the C-terminus, is present in all seven DNA-PKcs sequences, and is also present in both ATM and ATR from all species sequenced to date (human, mouse, Xenopus and Arabidopsis) (Fig. 5). Though homology within this region is not as dramatic in other AT-related kinases (Tor, esr1, Frap, arabidopsis ATM), the glycine six amino acids from the C-terminus is completely conserved even in these less related proteins. Glycine residues often induce bends in both α helices and β sheets. Thus, it seems possible that the glycine at position 4122 is particularly critical to the structural conformation of the extreme C-terminus.
Figure 5.
Glycine 4122 is uniformly conserved in the AT-like PI3 kinase family members. An alignment of the C-terminal 35 amino acids of available vertebrate DNA-PKcs, AT and ATR sequences is presented. Sequences were retrieved from the NCBI database.
Since the G4122E mutant protein can still colocalize with Ku onto DNA-cellulose, it seems unlikely that this region of DNA-PKcs is critical for the interaction between Ku and DNA-PKcs. Similarly, Priestley et al. (40) and Beamish et al. (48) demonstrated that mutant irs-20 (E4124K mutant) and the murine SCID mutant both assemble with Ku onto DNA ends. Furthermore, we have recently studied two splice variants of human DNA-PKcs. One of the splice variants encodes a protein with a 244 C-terminal truncation; this truncated protein also assembles with Ku onto DNA ends (K.Meek, unpublished data). Thus, it is very unlikely that the C-terminus of DNA-PKcs is important for its interaction with Ku. Since in murine SCID cells, in XR-C2, and in irs-20, the protein kinase motifs of DNA-PKcs are intact, it is intriguing to speculate that the C-terminus of DNA-PKcs is important in kinase activation, perhaps by interacting with DNA.
Recent studies from DeFazio et al. (49) demonstrate that DNA-PK’s kinase activity is not activated without synapses of two DNA-PK complexes. Another possibility that must be considered is that the G4122E mutation impairs the ability of DNA-PK to synapse with a second complex, thus explaining the severely decreased kinase activity of the mutant protein. Studies are currently underway to address these two potential explanations for the defective kinase activity of the G4122E mutant protein.
It is well established that (with few exceptions) deficiencies in NHEJ result in both radiosensitivity and impaired VDJ recombination. One such exception is our previous description of the XR-C2 mutant cell line that is radiosensitive (because of a deficiency in DNA-PK activity) but only modestly impaired in the ability to support VDJ recombination. Here, we have defined a single nucleotide substitution in the expressed DNA-PKcs allele of XR-C2. This mutation results in a single amino acid substitution six residues from the C-terminus, introducing a glycine to glutamic acid interchange. To examine whether the G4122E mutation is indeed responsible for the novel phenotype of the XR-C2 mutant cell line, a mutational approach was undertaken. In sum, we find that the mutant G4122E protein possesses weak DNA-PK activity and that expression of DNA-PKcs with the G→E interchange in DNA-PKcs-deficient rodent cells reiterates the novel phenotype observed in the XR-C2 cell line. These data suggest a critical role for the extreme C-terminus of DNA-PKcs in kinase activation. Furthermore, these data substantiate an emerging consensus: that minimal DNA-PK activity can suffice for VDJ recombination, but normal levels are required to repair IR-induced DSBs.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by Public Health Service grants AI32600 and AI42938 from the National Institutes of Health (K.M.). This work was supported by the European Union grant FIGH-CT1999-00010 and the Fund 2000 of the Leiden University Medical Center (M.Z.Z.).
REFERENCES
- 1.Critchlow S.E. and Jackson,S.P. (1998) DNA end-joining: from yeast to man. Trends Biochem. Sci., 23, 394–398. [DOI] [PubMed] [Google Scholar]
- 2.Pastink A., Eeken,J.C. and Lohman,P.H. (2001) Genomic integrity and the repair of double-strand DNA breaks. Mutat. Res., 480–481, 37–50. [DOI] [PubMed] [Google Scholar]
- 3.Lieber M.R. (1999) The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes Cells, 4, 77–85. [DOI] [PubMed] [Google Scholar]
- 4.Schlissel M.S. (2002) Does artemis end the hunt for the hairpin-opening activity in V(D)J recombination? Cell, 109, 1–4. [DOI] [PubMed] [Google Scholar]
- 5.Grawunder U., West,R.B. and Lieber,M.R. (1998) Antigen receptor gene rearrangement. Curr. Opin. Immunol., 10, 172–180. [DOI] [PubMed] [Google Scholar]
- 6.Lewis S.M. (1994) The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv. Immunol., 56, 27–150. [DOI] [PubMed] [Google Scholar]
- 7.Gellert M. (1997) Recent advances in understanding V(D)J recombination. Adv. Immunol., 64, 39–64. [DOI] [PubMed] [Google Scholar]
- 8.Meek K., Kienker,L., Dallas,C., Wang,W., Dark,M.J., Venta,P.J., Huie,M.L., Hirschhorn,R. and Bell,T. (2001) SCID in Jack Russell terriers: a new animal model of DNA-PKcs deficiency. J. Immunol., 167, 2142–2150. [DOI] [PubMed] [Google Scholar]
- 9.Wiler R., Leber,R., Moore,B.B., VanDyk,L.F., Perryman,L.E. and Meek,K. (1995) Equine severe combined immunodeficiency: a defect in V(D)J recombination and DNA-dependent protein kinase activity. Proc. Natl Acad. Sci. USA, 92, 11485–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieber M.R., Hesse,J.E., Lewis,S., Bosma,G.C., Rosenberg,N., Mizuuchi,K., Bosma,M.J. and Gellert,M. (1988) The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell, 55, 7–16. [DOI] [PubMed] [Google Scholar]
- 11.Moshous D., Callebaut,I., de Chasseval,R., Corneo,B., Cavazzana-Calvo,M., Le Deist,F., Tezcan,I., Sanal,O., Bertrand,Y., Philippe,N. et al. (2001) Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell, 105, 177–186. [DOI] [PubMed] [Google Scholar]
- 12.Taccioli G.E., Gottlieb,T.M., Blunt,T., Priestley,A., Demengeot,J., Mizuta,R., Lehmann,A.R., Alt,F.W., Jackson,S.P. and Jeggo,P.A. (1994) Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science, 265, 1442–1445. [DOI] [PubMed] [Google Scholar]
- 13.Lees-Miller S.P. (1996) The DNA-dependent protein kinase, DNA-PK: 10 years and no ends in sight. Biochem. Cell Biol., 74, 503–512. [DOI] [PubMed] [Google Scholar]
- 14.Hartley K.O., Gell,D., Smith,G.C., Zhang,H., Divecha,N., Connelly,M.A., Admon,A., Lees-Miller,S.P., Anderson,C.W. and Jackson,S.P. (1995) DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell, 82, 849–856. [DOI] [PubMed] [Google Scholar]
- 15.Yaneva M., Kowalewski,T. and Lieber,M.R. (1997) Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J., 16, 5098–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottlieb T.M. and Jackson,S.P. (1993) The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell, 72, 131–142. [DOI] [PubMed] [Google Scholar]
- 17.Suwa A., Hirakata,M., Takeda,Y., Jesch,S.A., Mimori,T. and Hardin,J.A. (1994) DNA-dependent protein kinase (Ku protein–p350 complex) assembles on double-stranded DNA. Proc. Natl Acad. Sci. USA, 91, 6904–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Y., Pannicke,U., Schwarz,K. and Lieber,M.R. (2002) Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell, 108, 781–794. [DOI] [PubMed] [Google Scholar]
- 19.Li Z., Otevrel,T., Gao,Y., Cheng,H.L., Seed,B., Stamato,T.D., Taccioli,G.E. and Alt,F.W. (1995) The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell, 83, 1079–1089. [DOI] [PubMed] [Google Scholar]
- 20.Modesti M., Hesse,J.E. and Gellert,M. (1999) DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. EMBO J., 18, 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grawunder U., Wilm,M., Wu,X., Kulesza,P., Wilson,T.E., Mann,M. and Lieber,M.R. (1997) Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature, 388, 492–495. [DOI] [PubMed] [Google Scholar]
- 22.Grawunder U., Zimmer,D., Kulesza,P. and Lieber,M.R. (1998) Requirement for an interaction of XRCC4 with DNA ligase IV for wild-type V(D)J recombination and DNA double-strand break repair in vivo. J. Biol. Chem., 273, 24708–24714. [DOI] [PubMed] [Google Scholar]
- 23.Baumann P. and West,S.C. (1998) DNA end-joining catalyzed by human cell-free extracts. Proc. Natl Acad. Sci. USA, 95, 14066–14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurimasa A., Kumano,S., Boubnov,N.V., Story,M.D., Tung,C.S., Peterson,S.R. and Chen,D.J. (1999) Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol., 19, 3877–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kienker L.J., Shin,E.K. and Meek,K. (2000) Both V(D)J recombination and radioresistance require DNA-PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res., 28, 2752–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leber R., Wise,T.W., Mizuta,R. and Meek,K. (1998) The XRCC4 gene product is a target for and interacts with the DNA dependent protein kinase. J. Biol. Chem., 273, 1794–1801. [DOI] [PubMed] [Google Scholar]
- 27.Chan D.W. and Lees-Miller,S.P. (1996) The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem., 271, 8936–8941. [DOI] [PubMed] [Google Scholar]
- 28.Douglas P., Moorhead,G.B., Ye,R. and Lees-Miller,S.P. (2001) Protein phosphatases regulate DNA-dependent protein kinase activity. J. Biol. Chem., 276, 18992–18998. [DOI] [PubMed] [Google Scholar]
- 29.Zdzienicka M. (1999) Mammalian X-ray-sensitive mutants which are defective in non-homologous (illegitimate) DNA double-strand break repair. Biochimie, 81, 107–116. [DOI] [PubMed] [Google Scholar]
- 30.Riballo E., Critchlow,S.E., Teo,S.H., Doherty,A.J., Priestley,A., BroughtonB., Kysela,B., Beamish,H., Plowman,N., Arlett,C.F. et al. (1999) Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol., 9, 699–702. [DOI] [PubMed] [Google Scholar]
- 31.O’Driscoll M., Cerosaletti,K.M., Girard,P.M., Dai,Y., Stumm,M., Kysela,B., Hirsch,B., Gennery,A., Palmer,S.E., Seidel,J. et al. (2001) DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Cell, 8, 1175–1185. [DOI] [PubMed] [Google Scholar]
- 32.Errami A., Overkamp,W.J., He,D.M., Friedl,A.A., Gell,D.A., Eckardt-Schupp,F., Jackson,S.P., Hendrickson,E.A., Lohman,P.H. and Zdzienicka,M.Z. (2000) A new X-ray sensitive CHO cell mutant of ionizing radiation group 7,XR-C2, that is defective in DSB repair but has only a mild defect in V(D)J recombination. Mutat. Res., 461, 59–69. [DOI] [PubMed] [Google Scholar]
- 33.Hesse J.E., Lieber,M.R., Gellert,M. and Mizuuchi,K. (1987) Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell, 49, 775–783. [DOI] [PubMed] [Google Scholar]
- 34.Lewis S.M., Hesse,J.E., Mizuuchi,K. and Gellert,M. (1988) Novel strand exchanges in V(D)J recombination. Cell, 55, 1099–1107. [DOI] [PubMed] [Google Scholar]
- 35.Sadofsky M.J., Hesse,J.E. and Gellert,M. (1994) Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res., 22, 1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin E.K., Rijkers,T., Pastink,A. and Meek,K. (2000) Analyses of TCRB rearrangements substantiate a profound deficit in recombination signal sequence joining in SCID foals: implications for the role of DNA-dependent protein kinase in V(D)J recombination. J. Immunol., 164, 1416–1424. [DOI] [PubMed] [Google Scholar]
- 37.Finnie N.J., Gottlieb,T.M., Blunt,T., Jeggo,P.A. and Jackson,S.P. (1995) DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proc. Natl Acad. Sci. USA, 92, 320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirchgessner C.U., Patil,C.K., Evans,J.W., Cuomo,C.A., Fried,L.M., Carter,T., Oettinger,M.A. and Brown,J.M. (1995) DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science, 267, 1178–1183. [DOI] [PubMed] [Google Scholar]
- 39.Blunt T., Gell,D., Fox,M., Taccioli,G.E., Lehmann,A.R., Jackson,S.P. and Jeggo,P.A. (1996) Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc. Natl Acad. Sci. USA, 93, 10285–10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Priestley A., Beamish,H.J., Gell,D., Amatucci,A.G., Muhlmann-Diaz,M.C., Singleton,B.K., Smith,G.C., Blunt,T., Schalkwyk,L.C., Bedford,J.S. et al. (1998) Molecular and biochemical characterisation of DNA-dependent protein kinase-defective rodent mutant irs-20. Nucleic Acids Res., 26, 1965–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin J.Y., Muhlmann-Diaz,M.C., Stackhouse,M.A., Robinson,J.F., Taccioli,G.E., Chen,D.J. and Bedford,J.S. (1997) An ionizing radiation-sensitive CHO mutant cell line: irs-20. IV. Genetic complementation, V(D)J recombination and the scid phenotype. Radiat. Res., 147, 166–171. [PubMed] [Google Scholar]
- 42.Douglas P., Sapkota,G.P., Morrice,N., Yu,Y., Goodarzi,A.A., Merkle,D., Meek,K., Alessi,D.R. and Lees-Miller,S.P. (2002) Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem. J., 368, 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin E.K., Perryman,L.E. and Meek,K. (1997) A kinase-negative mutation of DNA-PK(CS) in equine SCID results in defective coding and signal joint formation. J. Immunol., 158, 3565–3569. [PubMed] [Google Scholar]
- 44.Ding Q., Bramble,L., Yuzbasiyan-Gurkan,V., Bell,T. and Meek,K. (2002) DNA-PKcs mutations in dogs and horses: Allele frequency and association with neoplasia. Gene, 283, 263–269. [DOI] [PubMed] [Google Scholar]
- 45.Fukumura R., Araki,R., Fujimori,A., Mori,M., Saito,T., Watanabe,F., Sarashi,M., Itsukaichi,H., Eguchi-Kasai,K., Sato,K. et al. (1998) Murine cell line SX9 bearing a mutation in the dna-pkcs gene exhibits aberrant V(D)J recombination not only in the coding joint but also in the signal joint. J. Biol. Chem., 273, 13058–13064. [DOI] [PubMed] [Google Scholar]
- 46.Anderson C.W., Dunn,J.J., Freimuth,P.I., Galloway,A.M. and Allalunis-Turner,M.J. (2001) Frameshift mutation in PRKDC, the gene for DNA-PKcs, in the DNA repair-defective, human, glioma-derived cell line M059J. Radiat. Res., 156, 2–9. [DOI] [PubMed] [Google Scholar]
- 47.Fujimori A., Araki,R., Fukumura,R., Ohhata,T., Takahashi,H., Kawahara,A., Tatsumi,K. and Abe,M. (2000) Identification of four highly conserved regions in DNA-PKcs. Immunogenetics, 51, 965–973. [DOI] [PubMed] [Google Scholar]
- 48.Beamish H.J., Jessberger,R., Riballo,E., Priestley,A., Blunt,T., Kysela,B. and Jeggo,P.A. (2000) The C-terminal conserved domain of DNA-PKcs, missing in the SCID mouse, is required for kinase activity. Nucleic Acids Res., 28, 1506–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeFazio L.G., Stansel,R.M., Griffith,J.D. and Chu,G. (2002) Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J., 21, 3192–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]