Abstract
The Drosophila caudal homeobox gene is required for definition of the anteroposterior axis and for gut development, and CDX1 and CDX2, human homologs, play crucial roles in the regulation of cell proliferation and differentiation in the intestine. Most studies have indicated tumor suppressor functions of Cdx2, with inhibition of proliferation, while the effects of Cdx1 are more controversial. The influence of Drosophila Caudal on cell proliferation is unknown. In this study, we found three potential Caudal binding sequences in the 5′-flanking region of the Drosophila E2F (DE2F) gene and showed by transient transfection assays that they are involved in Caudal transactivation of the dE2F gene promoter. Analyses with transgenic flies carrying an E2F-lacZ fusion gene, with and without mutation in the Caudal binding site, indicated that the Caudal binding sites are required for expression of dE2F in living flies. Caudal-induced E2F expression was also confirmed with a GAL4-UAS system in living flies. In addition, ectopic expression of Caudal with heat-shock promotion induced melanotic tumors in larvae. These results suggest that Caudal is involved in regulation of proliferation through transactivation of the E2F gene in Drosophila.
INTRODUCTION
Homeobox genes generally encode for transcription factors involved in developmental, proliferation and differentiation processes (1,2). The Drosophila caudal example is required for definition of the anteroposterior axis during early embryogenesis (3), and for gut development (4). When caudal activity is completely lacking, the hindgut and anal pads are almost absent.
In vertebrates, Drosophila caudal-related homeobox genes have also been characterized: Xcad-1, Xcad-2 and Xcad-3 in Xenopus laevis; CdxA, CdxB and CdxC in chicken; Cdx-1, Cdx-2 and Cdx-4 in mice; and CDX1 and CDX2 in the human (5). Cdx1 and Cdx2 are known to play crucial roles in the regulation of cell proliferation and differentiation in the intestine (6). Cdx1 is mainly expressed in undifferentiated crypt cells, while Cdx2 is expressed at a higher level in the differentiated villus tip (7). Most studies have indicated a tumor suppressor function of Cdx2, which inhibits proliferation (8,9). However, the effects of Cdx1 on cell proliferation are controversial and it has been reported to exert oncogenic potential by upregulating ras (10). Such ras activation is considered to be an early event in genesis of colorectal cancers and results in altered cell growth and differentiation (11,12). Recently, however, Cdx1 was also reported to inhibit proliferation of intestinal epithelial cells, by down-regulation of D-type cyclins (13).
Whether Drosophila caudal plays a role in the regulation of proliferation is unknown. Interestingly, we found three putative Caudal binding site-related sequences, A/CTTTATA/G (14), in the 5′-flanking region of the Drosophila E2F (dE2F) gene, encoding a transcription factor playing important roles in cell proliferation (15). Well known targets of E2F are critical for cell-cycle progression and DNA synthesis. These include cell-cycle regulators, such as cyclin E, cyclin A, cdc2, cdc25 and Myc, and DNA replication-related genes, such as DNA polymerase α, thymidine kinase, PCNA and ribonucleotide reductase (16). The basic unit of E2F is a heterodimer of E2F and DP. In mammals, at least, six E2F family members (E2F-1 to -6) and two dimer partners (DP1 and DP2) have been characterized (17–21). Drosophila contains two E2Fs, dE2F and dE2F2, and one dimer partner, dDP (22–24). It has been reported that promoters of Drosophila genes encoding DNA polymerase α, PCNA and Raf contain the E2F recognition sequence (5′-TTTCGCGC), which is essential for promoter activity (25–28).
In the present study, since putative Caudal binding sites are located in the 5′-flanking region of the dE2F gene, we investigated involvement of the protein in transcriptional regulation. The results indicate that it can directly transactivate the dE2F gene promoter in vitro and in vivo.
MATERIALS AND METHODS
Oligonucleotides
The following double-stranded oligonucleotides containing a 6 bp linker sequence, recognized by BglII and BamHI, were chemically synthesized. Sequences containing three Caudal sites or base substitutional mutants in the E2F promoter region were as follows: E2F-Cad1 wild-type (wt), 5′-gatccTTTGGGATATTTATGAGTACAAACa-3′ and 3′-gAAA CCCTATAAATACTCATGTTTGtctag-5′; E2F-Cad1 mutant (mut), 5′-gatccTTTGGGATATGGCTGAGTACAAACa-3′ and 3′-gAAACCCTATACCGACTCATGTTTGtctag-5′; E2F- Cad2 wt, 5′-gatccACTTTTCTCTATAAAGTTGCAAATa-3′ and 3′-gTGAAAAGAGATATTTCAACGTTTAtctag-5′; E2F-Cad2 mut, 5′-gatccACTTTTCTCTAGCCAGTTGCAAATa- 3′ and 3′-gTGAAAAGAGATCGGTCAACGTTTAtctag-5′; E2F-Cad3 wt, 5′-gatccGGCATAATTTTTATGATACGTAGAa-3′ and 3′-gCCGTATTAAAAATACTATGCATCTtctag-5′; E2F-Cad3 mut, 5′-gatccGGCATAATTTGCCTGATACGTAGAa-3′ and 3′-gCCGTATTAAACGGACTATGCATCTtctag-5′.
Double-stranded oligonucleotides for site-directed mutagenesis were as follows: E2F-Cad1-SM, 5′-GCGATTTT GGGATATGGCTGAGTACAAACT-3′ and 3′-CACTTA AACCCTATACCGACTCATGTTTGA-5′; E2F-Cad2-SM, 5′-CTAACTTTTCTCTAGCCAGTTGCAAATCTT-3′ and 3′-GATTGAAAAGAGATCGGTCAACGTTTAGAA-5′; E2F- Cad3-SM, 5′-TCAGGCATAATTTGGCTGATACGTAGA AAT-3′ and 3′-AGTCCGTATTAAACCGACTATGCATCT TTA-5′. Mutated bases are underlined and lower case letters indicate the linker sequences recognizable by BglII and BamHI.
Plasmid construction
The promoter region of the dE2F gene was cloned by polymerase chain reaction (PCR) using Drosophila genomic DNA. Sequence analysis was performed to confirm the nucleotide sequence. The primers used, containing the linker sequences 5′-KpnI and 3′-XhoI were as follows: 5′-GCGG GTACCATGTGTTGGGCAAGGGAT-3′ and 3′-GCGGAG CTCGCACTGCAGTCGTCAAGC-5′. An amplified 2401 bp fragment containing the dE2F promoter region (–2184 to +217) was inserted into the KpnI and XhoI sites of pGL2-Basic (Promega) and the resulting reporter construct was designated as pE2F-Luc. To construct the plasmids pE2F-lacZ and pE2Fcadmut-lacZ for transgenic flies, the dE2F promoter region (–2184 to +217) with and without base-substituted mutations in the Caudal binding site were inserted into the KpnI site of the plasmid pCaSpeR-AUG-βgal (29).
The expression plasmid pAc-cad containing the Drosophila caudal coding region (1419 bp) placed under control of the Drosophila acin 5C gene promoter was constructed by inserting the coding region fragment into the XhoI site of pAc5.1/V5-HisA (Invitrogen). In the case of the GST-cad fusion plasmid, a caudal DNA fragment containing a DNA binding region of 729–976 bp downstream of the transcription start site (30) was inserted into the pGEX 4T-2 vector with XhoI. For construction of the plasmid pUAS-cad, the caudal coding region (1419 bp) was inserted into the XbaI and EcoRI sites of the pUAST vector (31).
Expression of GST fusion proteins and gel-mobility shift assays
Expression of GST-Cad fusion proteins in Escherichia coli was carried out as described previously (32). The E.coli were collected and suspended in a solution containing 25 mM HEPES pH 7.9, 1 mM EDTA, 0.02% 2-mercaptoethanol, 10% glycerol, 0.1% Tween-80 and 0.2 M KCl, sonicated and then centrifuged. The supernatant was loaded onto a glutathione–Sepharose column equilibrated with buffer A (20 mM Tris–HCl pH 7.8, 6% glycerol, 0.1 M KCl, 0.1 mM EDTA, 0.2% Tween-80 and 5 mM DTT) and washed with 20 ml of buffer A and 20 mM WE buffer (20 mM Tris–HCl pH 7.8, 2 mM MgCl2 and 1 mM DTT). Then the proteins were eluted with 20 ml of buffer G (50 mM Tris–HCl pH 9.6 and 5 mM glutathione), concentrated using Centricon 20 (Amicon) and dialyzed against PC buffer (50 mM Tris–HCl pH 7.8, 1 mM DTT and 0.1 mM EDTA) containing 0.15 M KCl and 50% glycerol. The purified proteins were stored at –20°C. Gel-mobility shift assays were performed as described previously (33). 32P-labeled probes were incubated for 15 min on ice in a reaction mixture containing 20 mM HEPES pH 7.5, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 0.5 mM DTT, 0.1 M KCl, 500 ng of sonicated herring sperm DNA and 500 ng of poly(dI–dC). For this step, unlabeled oligonucleotides were added as competitors. Then, 20 µg of purified GST fusion proteins were introduced followed by incubation for 10 min on ice. DNA–protein complexes were electrophoretically resolved on 6% polyacrylamide gels in 50 mM Tris–borate pH 8.3, 1 mM EDTA and 2.5% glycerol at 25°C. Gels were dried and then autoradiographed or analyzed with a BAS2000 (Fuji Film) imaging analyzer.
Cell culture, DNA transfection and luciferase assay
Drosophila S2 cells were grown at 25°C in M3 (BF) medium (Sigma) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Gibco BRL). Transfection of DNA mixtures into S2 cells was performed using dimethyldioctadecyl ammonium bromide (DDAB) (34) and the cells were harvested at 48 h thereafter. The luciferase assay was carried out by means of a Luciferase Assay System (Promega), as described previously (35). Normalized luciferase activities were calculated by determining the luciferase/β-galactosidase activity ratios.
Site-directed mutagenesis
To obtain mutant reporter plasmids carrying base-substitution mutations in the Caudal binding sites in the 5′-flanking region of the Drosophila E2F gene, mutagenesis reactions were carried out on double-stranded DNA of pE2F-Luc using a QuickChange™ Site-Directed Mutagenesis Kit (Stratagene). The reactions were set up essentially as recommended by the manufacturer. The mutations, as well as the fidelity of the rest of the DNA were confirmed by sequencing or enzymatic manipulation.
Establishment of transgenic flies and fly stocks
For ectopic expression experiments, the GAL4-UAS system was used with Hsp70-GAL4 flies (hs-GAL4) provided by the Bloomington Stock Center. Seven independent transgenic lines carrying UAS-cad were generated with the plasmid pUAS-cad by P-element-mediated transformation (36). Although only data with the line carrying UAS-cad in the second chromosome are shown in this report, essentially the same results were obtained with the other lines. To examine the role of the Caudal binding sites for dE2F gene expression in vivo, three independent transgenic lines carrying E2F-lacZ and two independent lines carrying the E2Fcadmut-lacZ fusion gene were generated with pE2F-lacZ and pE2Fcadmut-lacZ constructs, respectively. The lines carrying the same fusion genes showed the same lacZ expression patterns. An E2Frm729 line in which P-element-lacZ was inserted upstream of the initiation site of the dE2F gene in the same orientation as dE2F gene was also used (27).
Ectopic expression of Caudal by heat-shock induction
Females carrying homozygous hs-GAL4 in the third chromosome were crossed with males carrying homozygous UAS-cad in the second chromosome. The progeny third larvae were heat shocked at 37°C for 30 min and then returned to 25°C.
RT–PCR
Total RNA from larvae was isolated with Trizol Reagent (Molecular Research Center, Inc.) according to the protocol furnished by the manufacturer and cDNAs were synthesized with M-MLV-RT (reverse transcriptase) (Promega). The RT–PCR products were analyzed on agarose gels stained with ethidium bromide.
X-gal staining
The tissues were dissected and fixed for 15 min in phosphate-buffered saline (PBS: 130 mM NaCl, 7 mM Na2HPO4·2H2O, 3 mM NaH2PO4·2H2O, pH 7.5) containing 5% glutaraldehyde, washed in PBS, and immersed in 0.2% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranocyde (X-gal) in staining buffer containing 6.1 mM K4Fe(CN)6, 6.1 mM K3Fe(CN)6, 1 mM MgCl2, 150 mM NaCl, 10 mM Na2HPO4 and 10 mM NaH2PO4. Incubation was in the dark at 37°C.
RESULTS
Potential Caudal recognition sequences in the 5′-flanking region of the Drosophila E2F gene
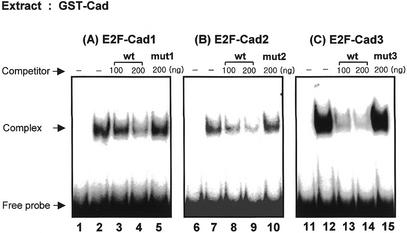
We found three putative Caudal binding sites similar to the Caudal binding consensus sequences, A/CTTTATA/G in the 5′-flanking region of the dE2F gene (Fig. 1). They were named E2F-Cad1, E2F-Cad2 and E2F-Cad3. The E2F-Luc reporter plasmid containing the dE2F gene (–2184 to +217) was constructed as described in the Materials and Methods. To examine whether the Caudal protein can recognize the putative binding sites found in the dE2F promoter region, a gel-mobility shift assay was performed using recombinant GST-Cad fusion proteins. Oligonucleotides E2F-Cad1 wt, E2F-Cad2 wt and E2F-Cad3 wt, were used as probes. Binding of GST-Cad to the oligonucleotide E2F-Cad1 wt was detected and the DNA–protein complexes were diminished effectively by adding unlabeled oligonucleotides E2F-Cad 1 wt, but not oligonucleotide E2F-Cad1 mut carrying base substitutions in the Caudal core binding sequence (Fig. 2A). A complex between GST-Caudal and the oligonucleotide E2F-Cad2 wt was detected (Fig. 2B) and diminished by adding excess amounts of unlabeled E2F-Cad2 wt. Such competition was not evident with E2F-Cad2 mut. The complex was also detected when the assay was performed with the radiolabeled E2F-Cad3 wt oligonucleotide as probe (Fig. 2C). Unlabeled oligonucleotide E2F-Cad3 wt competed with the formation of complex, whereas unlabeled E2F-Cad3 mut did not. The results indicate that GST-Caudal binds to E2F-Cad1, E2F-Cad2 and E2F-Cad3 binding sites with sequence specificity.
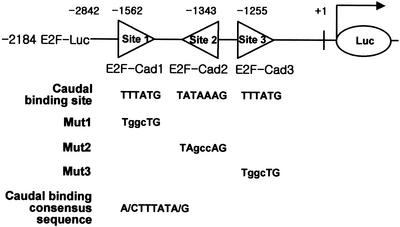
Figure 1.
Potential Caudal homeodomain binding sites in the Drosophila E2F promoter. The three potential Caudal homeodomain binding sites in Drosophila E2F gene promoter, named E2F-Cad1, E2F-Cad2 and E2F-Cad3, are shown. The reporter construct used in the transfection assay contains a more upstream region, up to –2184, and a downstream region, up to +217. Lower case letters indicate mutated bases in the Caudal sites with numbering relative to the transcription initiation site.
Figure 2.
Complex formation between putative Caudal binding sites in the dE2F promoter and GST-Cad. Radiolabeled double-stranded E2F-Cad oligonucleotides were incubated with 20 µg of GST-Cad in the presence or absence of the indicated competitor oligonucleotides. Lanes 1, 6 and 11, GST only. Lanes 2, 7 and 12, binding without competitor. Lanes 3, 4, 8, 9, 13 and 14, unlabeled competitor DNA with the wild-type E2F-Cad sequences (wt). Lanes 5, 10 and 15, unlabeled competitor DNA with mutants having three base changes in E2F-Cad sequence (mut). E2F-Cad1, E2F-Cad2 and E2F-Cad3, oligonucleotides containing the Caudal sites 1–3 of the dE2F gene promoter.
Activation of the dE2F gene promoter by Caudal
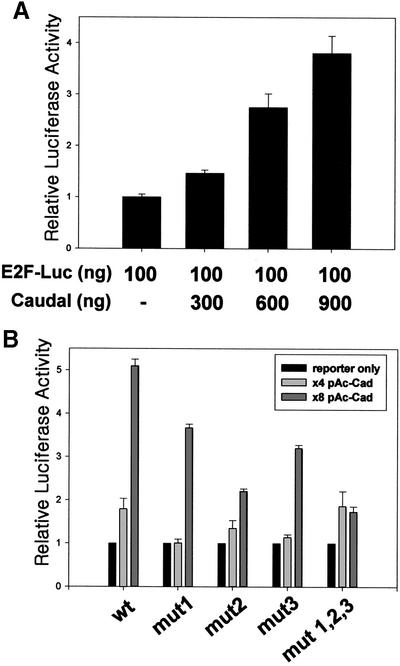
Since GST-Cad binds to the three potential Caudal binding sites in the 5′-flanking region of the dE2F gene, we investigated whether the Caudal homeodomain protein can regulate activity of the dE2F promoter, using a pE2F-Luc reporter plasmid containing the promoter region (–2184 to +217) of the dE2F gene fused with the luciferase reporter gene. Effects of Caudal on dE2F gene promoter activity were examined in transient transfection experiments using pE2F-Luc reporter plasmids and a pAc-cad expression plasmid in the Drosophila S2 cell line. The wild-type dE2F promoter was transactivated depending on the amount, up to nearly 4- or 5-fold, of pAc-cad (Fig. 3A). For further confirmation of Caudal binding sites as targets of Caudal, the mutant reporter plasmids, pE2F-Cad1mut-Luc, pE2F-Cad2mut-Luc and pE2F-Cad3mut-Luc having the same mutation as those used for the gel-mobility shift assay, and the E2F-Cad1,2,3mut-Luc having base-substitution mutations in all three Caudal binding sites were constructed and used for reporter assays. As shown in Figure 3B, mutant reporter plasmids showed little activation with Caudal expression plasmids. A comparison between promoter activities of dE2F mutants and the wild type revealed no alteration in promoter activity (data not shown). These observations can be explained by the results of RT–PCR analysis in Drosophila S2 cells, by which Drosophila caudal mRNA was barely detectable (data not shown). Thus, Caudal apparently activates the dE2F promoter and this is mediated by the Caudal binding sites in the dE2F promoter.
Figure 3.
Effects of Caudal on luciferase expression driven by the dE2F promoter. Transfections were performed with Drosophila S2 cells and promoter activities measured as luciferase activity normalized to β-galactosidase activity at 48 h thereafter. (A) Each transfection mixture contained the E2F-Luc reporter plasmid, the pAc-lacZ internal control plasmid and the indicated amount of Caudal expressing plasmid, pAc-cad. (B) Constructs carrying a point mutation in Caudal binding sites in the dE2F promoter were cotransfected with the indicated amount of pAc-cad. The mean activities ±SE from three or four independent transfections are shown.
Role of the Caudal binding sites in expression of dE2F in living flies
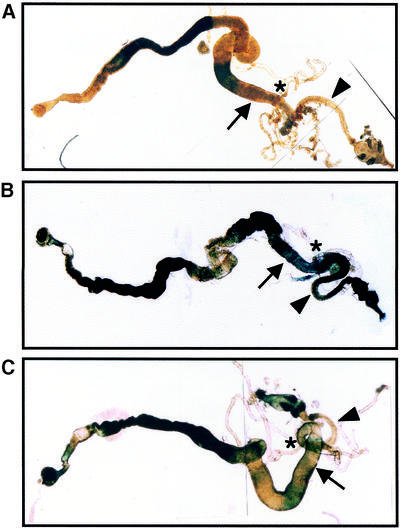
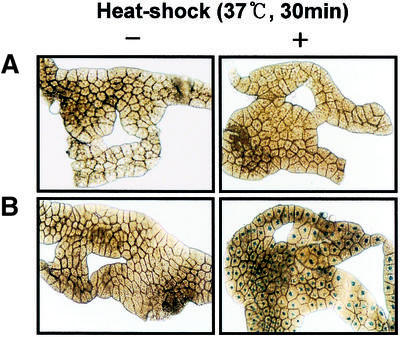
To investigate the role of the Caudal sites for expression of dE2F in living flies, we established transgenic flies carrying the dE2F-lacZ fusion gene, containing the dE2F promoter region (–2184 to +217) fused to lacZ, or E2Fcadmut-lacZ with base-substituted mutations in the Caudal binding sites of the dE2F promoter. Drosophila caudal is known to be expressed in the posterior midgut, Malpighian tubules and hindgut of the embryo (4) and in the anal plates and hindgut of adult flies (37). Therefore, we compared expression patterns of lacZ in the gut of adult flies carrying E2F-lacZ and E2Fcadmut-lacZ by X-gal staining. The lacZ staining signals of adult flies carrying the E2Fcadmut-lacZ were markedly decreased in the posterior midgut and the hindgut (Fig. 4B and C). Endogenous β-galactosidase expression was detected in the anterior part and the central region of the midgut (Fig. 4A). This result indicates that the Caudal binding sites are required for dE2F gene expression in the posterior midgut and the hindgut.
Figure 4.
Effects of Caudal binding sites on dE2F expression in vivo. β-Galactosidase expression in the wild-type (Oregon R) (A) and transgenic flies carrying E2F-lacZ (B) and E2Fcadmut-lacZ (C) was detected by X-gal staining. The adult guts were dissected and stained with 0.2% X-gal solution in the dark. Reduced expression of the E2Fcadmut-lacZ is apparent in the posterior midgut and hindgut, compared with E2F-lacZ. The arrow indicates the posterior midgut and the arrowhead the hindgut. The asterisk indicates the Malpighian tubules.
Effects of ectopic Caudal expression on the expression of dE2F
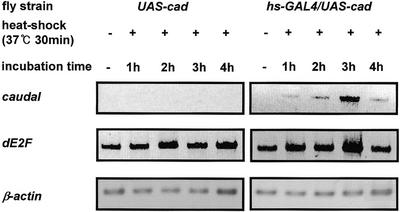
Ectopic expression of Caudal in living flies was performed with the GAL4-UAS system (31,38). The coding region of caudal was subcloned into the pUAST vector, and the resultant plasmid was designated pUAS-cad. We established transgenic flies for ectopic expression of Caudal with the pUAS-cad construct by P-element-mediated transformation (36). Transgenic flies carrying UAS-cad were crossed with transgenic flies carrying GAL4 cDNA placed under control of the hsp70 gene promoter (hs-GAL4). Ectopic expression of Caudal in the transgenic larvae after heat shock was confirmed by RT–PCR (Fig. 5). The caudal mRNA in larvae carrying single copies of hs-GAL4 and UAS-cad was detected at 1 h after heat shock and its level increased with time to reach a maximum at 3 h (Fig. 5). However, caudal mRNA in larvae carrying only UAS-cad was not detected at any time (Fig. 5). As mentioned above, we examined the dE2F expression in larvae carrying hs-GAL4 and UAS-cad by RT–PCR. The expression level of dE2F was increased at 3 h after heat shock (Fig. 5).
Figure 5.
Expression of caudal and dE2F in hs-GAL4/UAS-cad third instar larvae. Total RNA was prepared from the third instar larvae carrying hs-GAL4 and UAS-cad after heat shock at 37°C for 30 min and incubated at 25°C for various time periods. Then, RT–PCR was performed to measure caudal and dE2F mRNA levels. The line carrying UAS-cad was used as a control and processed in the same way as that carrying hs-GAL4/UAS-cad.
To investigate the effects of ectopic Caudal expression on the dE2F gene expression in vivo, we made a UAS-cad/+;E2Frm729/hs-GAL4 line by crossing E2Frm729 flies, in which lacZ is inserted upstream of the initiation site of the dE2F gene, with transgenic flies carrying UAS-cad with the hs-GAL4 line. The third instar larvae of the UAS-cad/+;E2Frm729/hs-GAL4 line were heat shocked at 37°C for 30 min and incubated at 25°C for 8 h. Then, they were dissected and X-gal staining was carried out. Expression of the lacZ gene in the heat-shocked UAS-cad/+;E2Frm729/hs-GAL4 larvae was increased dramatically in fat bodies by overexpressed Caudal (Fig. 6B).
Figure 6.
Effects of ectopic Caudal expression on dE2F gene expression in vivo. The third instar larvae of UAS-cad/+;E2Frm729/hs-GAL4 lines (B) were dissected after heat shock (37°C, 30 min) and incubated at 25°C for 8 h. They were then fixed in 5% glutaraldehyde and stained with 0.2% X-gal solution in the dark at 37°C. Tissues from E2Frm729 larvae were processed in the same way (A).
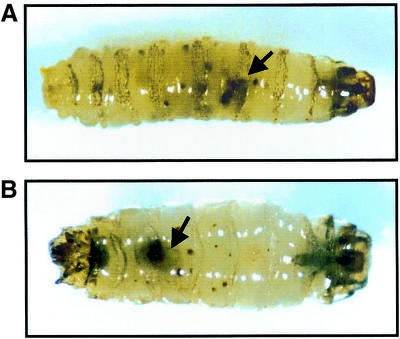
Effects of the ectopic Caudal expression during development were also analyzed with transgenic flies carrying hs-GAL4 and UAS-cad. Heat-shock induction of Caudal caused generation of melanotic tumors (Fig. 7), indicating that overexpression can result in abnormal proliferation and differentiation.
Figure 7.
Formation of melanotic tumors with ectopic Caudal expression under the GAL4-UAS system. Melanotic tumors (arrow) are apparent in third instar larvae at 36 h (A) and 48 h (B) after heat shock. Most of the larvae had died by that time.
DISCUSSION
In this study, we found three Caudal binding sites in the 5′-flanking region of the dE2F gene (Fig. 1) and demonstrated their requirement for in vivo expression of the dE2F gene. Thus, the homeodomain protein Caudal positively regulates the cell proliferation-related gene dE2F at the transcriptional level.
Caudal-related homeodomain proteins bind to an AT-rich sequence of A/CTTTATA/G (14). Drosophila caudal is expressed in the posterior region of the embryo during early embryogenesis and activates the segmentation gene, fushi tarazu, involved in terminal specification (39). Maternal and zygotic caudal products are known to exert their function through activation of folded gastrulation, fork head and wingless (4). Caudal can induce Distal-less gene expression and internal analia development by activating the brachyenteron and even-skipped genes (37). Cdx1 and Cdx2, mammalian homologs of the Drosophila caudal gene, are mainly expressed in the intestine and colon where they appear to be involved in the regulation of cell proliferation and differentiation (40). Cdx1 expression is restricted to the proliferative crypt compartment (41), while Cdx2 is found at high levels in the differentiated cells of villus tip (42). The several targets of Cdx2 include the Hox-C8 homeobox gene (43), the small intestinal genes encoding sucrase-isomaltase (44), lactase-phlorizin hydrolase (45), phospholipase-A (46), calbindin-D9K (47), the colonic gene for carbonic anhydrase I (48), and the genes encoding insulin and glucagon in the pancreas (49). These are all differentiation-related, supporting a tumor suppressor function of Cdx2 inhibiting proliferation (8,9). In the case of Cdx1, only Hox-A7 has been shown as a direct target (50). Overexpression of Cdx1 in non-intestinal cells increases cell growth in vitro and promotes cell tumorigenicity in xenografts (2), which suggests oncogenic potential. Intestinal metaplasia in gastric and esophagal tumors is associated with ectopic expression of Cdx1 (51) and recently we reported CDX1 to have a pro-proliferative effect through transactivation of the PCNA promoter activity (52). Here, we demonstrated that Caudal directly transactivates the dE2F gene promoter activity. E2F is a key regulator of cell proliferation. The well known targets of E2F are critical for cell-cycle progression and DNA synthesis, such as cyclin E, cyclin A, cdc2, cdc25, Myc, DNA polymerase α, thymidine kinase, PCNA and ribonucleotide reductase (16). Therefore, our results indicate that Drosophila Caudal may be involved in cell proliferation by regulating dE2F gene expression so that its function is similar to that of Cdx1 rather than Cdx2.
Recently, it was reported that both E2F and DP mutant larvae exhibit development of melanotic tumors (53). Interestingly, we also observed that ectopic Caudal expression under the control of the heat-shock promoter leads to death in the developmental stage and generation of melanotic tumor in third instar larvae (Fig. 7). It has been proposed that two distinct classes of melanotic tumor mutations exist (54). Class 1 affect autoimmune responses or the normal immune system reaction to abnormal target tissue. Class 2, in contrast, cause obvious defects in the hematopoietic organs or hemocytes, resulting in aberrant immunity, contributing to melanotic tumor formation. In the present case, upregulation of E2F expression by ectopic Caudal induced under hs-GAL4/UAS-cad was detected especially in the fat body (Fig. 6), which is responsible for immune response in Drosophila (55). We have also found that caudal is expressed in the fat body and is involved in the immune response of Drosophila (M.-A. Yoo and W.-J. Lee, unpublished results). Further studies to investigate the roles of Caudal and E2F in the development of melanotic tumors are clearly warranted.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr N. Dyson and Bloomington Stock Center for E2Frm729 and hs-GAL4 and thank Dr M. Moore for valuable comments on the manuscript. This work was supported by a grant (F-4-09) from the 21C Frontier Functional Human Genome Project of the Ministry of Science and Technology of Korea.
REFERENCES
- 1.Kessel M. and Gruss,P. (1990) Murine developmental control genes. Science, 249, 374–379. [DOI] [PubMed] [Google Scholar]
- 2.Maulbecker C.C. and Gruss,P. (1993) The oncogenic potential of deregulated homeobox genes. Cell Growth Differ., 4, 431–441. [PubMed] [Google Scholar]
- 3.McGinnis W. and Krumlauf,R. (1992) Homeobox genes and axial patterning. Cell, 68, 283–302. [DOI] [PubMed] [Google Scholar]
- 4.Wu L.H. and Lengyel,J.A. (1998) Role of caudal in hindgut specification and gastrulation suggests homology between Drosophila amnioproctodeal invagination and vertebrate blastopore. Development, 125, 2433–2442. [DOI] [PubMed] [Google Scholar]
- 5.Freund J.N., Domon-Dell,C., Kedinger,M. and Duluc,I. (1998) The Cdx-1 and Cdx-2 homeobox genes in the intestine. Biochem. Cell Biol., 76, 957–969. [DOI] [PubMed] [Google Scholar]
- 6.Suh E. and Traber,P.G. (1996) An intestine-specific homeobox gene regulates proliferation and differentiation. Mol. Cell. Biol., 16, 619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silberg D.G., Swain,G.P., Suh,E.R. and Traber,P.G. (2000) Cdx1 and Cdx2 expression during intestinal development. Gastroenterology, 119, 961–971. [DOI] [PubMed] [Google Scholar]
- 8.Mallo G.V., Rechreche,H., Frigerio,J.M., Rocha,D., Zweibaum,A., Lacasa,M., Jordan,B.R., Dusetti,N.J., Dagorn,J.C. and Iovanna,J.L. (1997) Molecular cloning, sequencing and expression of the mRNA encoding human Cdx1 and Cdx2 homeobox. Down-regulation of Cdx1 and Cdx2 mRNA expression during colorectal carcinogenesis. Int. J. Cancer, 74, 35–44. [DOI] [PubMed] [Google Scholar]
- 9.Wicking C., Simms,L.A., Evans,T., Walsh,M., Chawengsaksophak,K., Beck,F., Chenevix-Trench,G., Young,J., Jass,J., Leggett,B. and Wainwright,B. (1998) CDX2, a human homologue of Drosophila caudal, is mutated in both alleles in a replication error positive colorectal cancer. Oncogene, 17, 657–659. [DOI] [PubMed] [Google Scholar]
- 10.Lorentz O., Cadoret,A., Duluc,I., Capeau,J., Gespach,C., Cherqui,G. and Freund,J.N. (1999) Downregulation of the colon tumour-suppressor homeobox gene Cdx-2 by oncogenic ras. Oncogene, 18, 87–92. [DOI] [PubMed] [Google Scholar]
- 11.Fearon E.R. and Vogelstein,B. (1990) A genetic model for colorectal tumorigenesis. Cell, 61, 759–767. [DOI] [PubMed] [Google Scholar]
- 12.Baron-Delage S., Mahraoui,L., Cadoret,A., Veissiere,D., Taillemite,J.L., Chastre,E., Gespach,C., Zweibaum,A., Capeau,J., Brot-Laroche,E. and Cherqui,G. (1996) Deregulation of hexose transporter expression in Caco-2 cells by ras and polyoma middle T oncogenes. Am. J. Physiol., 270, G314–G323. [DOI] [PubMed] [Google Scholar]
- 13.Lynch J., Suh,E.R., Silberg,D.G., Rulyak,S., Blanchard,N. and Traber,P.G. (2000) The caudal-related homeodomain protein Cdx1 inhibits proliferation of intestinal epithelial cells by downregulation of D-type cyclins. J. Biol. Chem., 275, 4499–4506. [DOI] [PubMed] [Google Scholar]
- 14.Margalit Y., Yarus,S., Shapira,E., Gruenbaum,Y. and Fainsod,A. (1993) Isolation and characterization of target sequences of the chicken CdxA homeobox gene. Nucleic Acids Res., 21, 4915–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinberg R.A. (1996) E2F and cell proliferation: a world turned upside down. Cell, 85, 457–459. [DOI] [PubMed] [Google Scholar]
- 16.DeGregori J., Leone,G., Miron,A., Jakoi,L. and Nevins,J.R. (1997) Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl Acad. Sci. USA, 94, 7245–7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bandara L.R., Buck,V.M., Zamanian,M., Jhonston,L.H. and La Thangue,N.B. (1993) Functional synergy between DP-1 and E2F-1 in the cell cycle-regulatory transcription factor DRTF1/E2F. EMBO J., 12, 4317–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helin K., Wu,C.L., Fattaey,A.R., Lees,J.A., Dynlacht,B.D., Ngwu,C. and Harlow,E. (1993) Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev., 7, 1850–1861. [DOI] [PubMed] [Google Scholar]
- 19.Hijimans E.M., Voorhoeve,P.M., Beijersbergen,R.L., van’t Veer,L.J. and Bernards,R. (1995) E2F-5, a new family member that interacts with p130 in vivo. Mol. Cell. Biol., 15, 3082–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trimarchi J.M., Fairchild,B., Verona,R., Moberg,K., Andon,N. and Lees,J.A. (1998) E2F-6, a new E2F family that can behave as a transcriptional repressor. Proc. Natl Acad. Sci. USA, 95, 2850–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vairo G., Livingston,D.M. and Ginsberg,D. (1995) Functional interaction between E2F-4 and p130: evidence for distinct mechanisms underlying growth suppression by different retinoblastoma protein family members. Genes Dev., 9, 869–881. [DOI] [PubMed] [Google Scholar]
- 22.Dynlacht B.P., Brook,A., Dembski,M., Yenush,L. and Dyson,N. (1994) DNA-binding and trans-activation properties of Drosophila E2F and DP proteins. Proc. Natl Acad. Sci. USA, 91, 6359–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao X.F., Alphey,L., Bandara,L.R., Lam,E.W., Glover,D. and La Thangue,N.B. (1995) Functional conservation of the cell cycle-regulating transcription factor DRTF1/E2F and its pathway of control in Drosophila melanogaster. J. Cell Sci., 108, 2945–2954. [DOI] [PubMed] [Google Scholar]
- 24.Sawado T., Yamaguchi,M., Nishimoto,Y., Ohno,K., Sakaguchi,K. and Matsukage,A. (1998) dE2F2, a novel E2F-family transcriptional factor in Drosophila melanogaster. Biochem. Biophys. Res. Commun., 251, 409–415. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi M., Hayashi,Y. and Matsukage,A. (1995) Essential role of E2F recognition sites in regulation of the proliferating cell nuclear antigen gene promoter during Drosophila development. J. Biol. Chem., 270, 25159–25165. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi M., Hayashi,Y., Hirose,F., Nishimoto,Y. and Matsukage,A. (1997) Distinct roles of E2F recognition sites as positive or negative elements in regulation of the DNA polymerase α 180 kDa catalytic subunit gene promoter during Drosophila development. Nucleic Acids Res., 25, 3847–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duronio R.J., O’Farrell,P.H., Xie,J.E., Brook,A. and Dyson,N. (1995) The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev., 9, 1445–1455. [DOI] [PubMed] [Google Scholar]
- 28.Kwon E.J., Oh,E.J., Kim,Y.S., Hirose,F., Ohno,K., Nishida,Y., Matsukage,A., Yamaguchi,M. and Yoo,M.A. (2001) E2F-dependent transcription of the raf proto-oncogene during Drosophila development. Nucleic Acids Res., 29, 1808–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thummel C.S., Boulet,A.M. and Lipshitz,H.D. (1988) Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene, 74, 445–456. [DOI] [PubMed] [Google Scholar]
- 30.Mlodzik M. and Gehring,W.J. (1987) Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell, 48, 465–478. [DOI] [PubMed] [Google Scholar]
- 31.Brand A.H. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotype. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 32.Kwon E.J., Park,H.S., Kim,Y.S., Oh,E.J., Nishida,Y., Matsukage,A., Yoo,M.A. and Yamaguchi,M. (2000) Transcriptional regulation of the Drosophila raf proto-oncogene by Drosophila STAT during development and in immune responses. J. Biol. Chem., 275, 19824–19830. [DOI] [PubMed] [Google Scholar]
- 33.Hirose F., Yamaguchi,M., Handa,H., Inomata,Y. and Matsukage,A. (1993) Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase alpha and proliferating cell nuclear antigen. J. Biol. Chem., 268, 2092–2099. [PubMed] [Google Scholar]
- 34.Han K. (1996) An efficient DDAB-mediated transfection of Drosophila S2 cells. Nucleic Acids Res., 24, 4362–4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawado T., Hirose,F., Takahashi,Y., Sasaki,T., Shinomiya,T., Sakaguchi,K., Matsukage,A. and Yamaguchi,M. (1998) The DNA replication-related element (DRE)/DRE-binding factor system is a transcriptional regulator of the Drosophila E2F gene. J. Biol. Chem., 273, 26042–26051. [DOI] [PubMed] [Google Scholar]
- 36.Spradling A.C. (1986) Drosophila: A Practical Approach. IRL Press, Oxford, UK.
- 37.Moreno E. and Moreta,G. (1999) Caudal is the Hox gene that specifies the most posterior Drosophila segment. Nature, 400, 873–877. [DOI] [PubMed] [Google Scholar]
- 38.Fisher J.A., Giniger,E., Maniatis,T. and Ptashne,M. (1988) GAL4 activates transcription in Drosophila. Nature, 332, 853–856. [DOI] [PubMed] [Google Scholar]
- 39.Dearolf C.R., Topol,J. and Parker,C.S. (1989) The caudal gene product is a direct activator of fushi tarazu transcription during Drosophila embryogenesis. Nature, 341, 340–343. [DOI] [PubMed] [Google Scholar]
- 40.James R. and Kazenwadel,J. (1991) Homeobox gene expression in the intestinal epithelium of adult mice. J. Biol. Chem., 266, 3246–3251. [PubMed] [Google Scholar]
- 41.Subramanian V., Meyer,B. and Evans,G.S. (1998) The murine Cdx1 gene product localises to the proliferative compartment in the developing and regenerating intestinal epithelium. Differentiation, 64, 11–18. [DOI] [PubMed] [Google Scholar]
- 42.James R., Erler,T. and Kazenwadel,J. (1994) Structure of the murine homeobox gene cdx-2. Expression in embryonic and adult intestinal epithelium. J. Biol. Chem., 269, 15229–15237. [PubMed] [Google Scholar]
- 43.Shashikant C.S. and Ruddle,F.H. (1996) Combinations of closely situated cis-acting elements determine tissue-specific patterns and anterior extent of early Hoxc8 expression. Proc. Natl Acad. Sci. USA, 93, 12364–12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suh E., Chen,L., Taylor,J. and Traber,P.G. (1994) A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol. Cell. Biol., 14, 7340–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Troelsen J.T., Mitchelmore,C., Spodsberg,N., Jensen,A.M., Noren,O. and Sjostrom,H. (1997) Regulation of lactase-phlorizin hydrolase gene expression by the caudal-related homeodomain protein Cdx-2. Biochem. J., 322, 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor J.K., Boll,W., Levy,T., Suh,E.R., Siang,S., Mantei,N. and Traber,P.G. (1997) Comparison of intestinal phospholipase A/lysophospholipase and sucrase-isomerase genes suggests a common structure for enterocyte-specific promoters. DNA Cell Biol., 16, 1419–1428. [DOI] [PubMed] [Google Scholar]
- 47.Colnot S., Romagnolo,B., Lambert,M., Cluzeaud,F., Porteu,A., Vandewalle,A., Thomasset,M., Kahn,A. and Perret,C. (1998) Intestinal expression of the calbindin-D9K gene in transgenic mice. Requirement for a Cdx2-binding site in a distal activator region. J. Biol. Chem., 273, 31939–31946. [DOI] [PubMed] [Google Scholar]
- 48.Drummond F., Sowden,J., Morrison,K. and Edwards,Y.H. (1996) The caudal-type homeobox protein Cdx-2 binds to the colon promoter of the carbonic anhydrase 1 gene. Eur. J. Biochem., 236, 670–681. [DOI] [PubMed] [Google Scholar]
- 49.German M.S., Wang,J., Chadwick,R.B. and Rutter,W.J. (1992) Synergistic activation of the insulin gene by a LIM-homeodomain protein and a basic helix–loop–helix protein: building a functional insulin mini enhancer complex. Genes Dev., 6, 2165–2176. [DOI] [PubMed] [Google Scholar]
- 50.Subramanian V., Meyer,B.I. and Gruss,P. (1995) Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell, 83, 641–653. [DOI] [PubMed] [Google Scholar]
- 51.Silberg D.G., Furth,E.E., Taylor,J.K., Schuck,T., Chiou,T. and Traber,P.G. (1997) CDX1 protein expression in normal, metaplastic, and neoplatic human alimentary tract epithelium. Gastroenterology, 113, 478–486. [DOI] [PubMed] [Google Scholar]
- 52.Oh E.J., Park,J.H., Cho,M., Lee,W.J., Choi,Y.H. and Yoo,M.A. (2002) The caudal-related homeodomain protein CDX1 activates proliferating cell nuclear antigen expression in hepatocellular and colorectal carcinoma cells. Int. J. Oncol., 20, 23–29. [PubMed] [Google Scholar]
- 53.Royzman I., Whittaker,A.J. and Orr-Weaver,T.L. (1997) Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes Dev., 11, 1999–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watson K.L., Johonson,T.K. and Denell,R.E. (1991) Lethal (l) aberrant immune response mutations leading to melanotic tumor formation in Drosophila melanogaster. Dev. Genet., 12, 173–187. [DOI] [PubMed] [Google Scholar]
- 55.Georgel P., Kappler,C., Langley,E., Gross,I., Nicolas,E., Reichhart,J.M. and Hoffmann,J.A. (1995) Drosophila immunity. A sequence homologous to mammalian interferon consensus response element enhances the activity of the diptericin promoter. Nucleic Acids Res., 23, 1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]