Abstract
Synthesis of the Gag-Pol protein of the human immunodeficiency virus type 1 (HIV-1) requires a programmed –1 ribosomal frameshifting when ribosomes translate the unspliced viral messenger RNA. This frameshift occurs at a slippery sequence followed by an RNA structure motif that stimulates frameshifting. This motif is commonly assumed to be a simple stem–loop for HIV-1. In this study, we show that the frameshift stimulatory signal is more complex than believed and consists of a two-stem helix. The upper stem–loop corresponds to the classic stem–loop, and the lower stem is formed by pairing the spacer region following the slippery sequence and preceding this classic stem–loop with a segment downstream of this stem–loop. A three-purine bulge interrupts the two stems. This structure was suggested by enzymatic probing with nuclease V1 of an RNA fragment corresponding to the gag/pol frameshift region of HIV-1. The involvement of the novel lower stem in frameshifting was supported by site-directed mutagenesis. A fragment encompassing the gag/pol frameshift region of HIV-1 was inserted in the beginning of the coding sequence of a reporter gene coding for the firefly luciferase, such that expression of luciferase requires a –1 frameshift. When the reporter was expressed in COS cells, mutations that disrupt the capacity to form the lower stem reduced frameshifting, whereas compensatory changes that allow re-formation of this stem restored the frameshift efficiency near wild-type level. The two-stem structure that we propose for the frameshift stimulatory signal of HIV-1 differs from the RNA triple helix structure recently proposed.
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) uses a programmed –1 ribosomal frameshift to produce the Gag-Pol polyprotein, the precursor of its enzymes, when ribosomes translate the full-length viral messenger RNA. Conventional translation of this RNA by the majority of ribosomes produces the Gag polyprotein, the precursor of the structural proteins of the virus, while the frameshift allows a small number of ribosomes to change the reading frame at a specific sequence and extend their reading over the stop codon of the 0 frame gag gene, until the stop codon of the pol gene in the –1 frame is encountered (1). The ratio of Gag-Pol to Gag is critical for viral assembly and replication, and increasing or decreasing the frameshifting efficiency interferes with the formation of infectious viral particles (2–4). Programmed –1 ribosomal frameshift has also been reported in several other retroviruses, coronaviruses, plant viruses (5,6), in a yeast virus (7,8), in bacteria (9–12) and, recently, in humans (13).
A programmed –1 ribosomal frameshift requires two elements in the mRNA: (i) a heptanucleotide where the frameshift occurs, called the slippery sequence, with an X XXY YYZ consensus sequence (where the 0 frame is indicated by spaces), and (ii) a secondary structure located downstream of the slippery sequence that stimulates the frameshift. These two elements are separated in most cases by a short spacer sequence (5). When ribosomes bearing two tRNAs, the peptidyl-tRNA and the aminoacyl-tRNA, in the P and A site, respectively, encounter the slippery sequence, a minority of them shift the reading frame. This recoding event can be described as follows: the two tRNAs whose anticodons interact with the codons of the mRNA in the 0 frame (X XXY YYZ) unpair from this mRNA, the ribosome shifts backward by 1 nt and the tRNAs re-pair in the –1 frame (XXX YYY) (14). Peptide bond formation then occurs and translation resumes in the new reading frame (reviewed in 15). The stimulatory secondary structure downstream of the slippery sequence makes the ribosome pause at the slippery sequence (16–18). However, this pausing is necessary but not sufficient for promoting the frameshift and little correlation was found between the extent of this pause and the frameshift efficiency (19). It has been suggested that a specific interaction between the frameshift stimulatory signal and the ribosome is required for the shift to occur (20,21). The slippery sequence is U UUU UUA for HIV-1, and the frameshift stimulatory signal is commonly assumed to be a simple stem–loop, with an 11-bp stem, separated from the slippery sequence by an eight-base spacer. This contrasts with the majority of known frameshift events, where the stimulatory signal is a pseudoknot, although there are other well documented cases of stem– loop-containing frameshift signals (6). The structure of the HIV-1 frameshift stimulatory signal was first proposed from a computer structure analysis program (22) and later demonstrated by structural probing experiments, using a short RNA transcript encompassing the HIV-1 classic frameshift region and ending immediately after the stem–loop (23).
In this study, we investigated whether the sequence following the stem–loop could influence the frameshift efficiency in HIV-1. We inserted the HIV-1 frameshift region in the beginning of the coding sequence of a luciferase reporter gene, such that its expression depends on a –1 frameshift, and assessed the expression of luciferase in COS cells and in vitro, in a rabbit reticulocyte lysate (RRL). Based on site-directed mutagenesis experiments, we propose that the frameshift stimulatory signal of HIV-1 is larger than assumed, and consists of a two-stem helix, where the upper stem–loop corresponds to the classic stem–loop and the lower stem is made by base pairing the spacer sequence to a segment following the classic stem–loop. While this study was underway, a paper was published by Dinman et al. (24), supporting our findings that the frameshift region of HIV-1 is longer than believed but suggesting a different structure for the frameshift stimulatory signal.
MATERIALS AND METHODS
Construction of plasmids
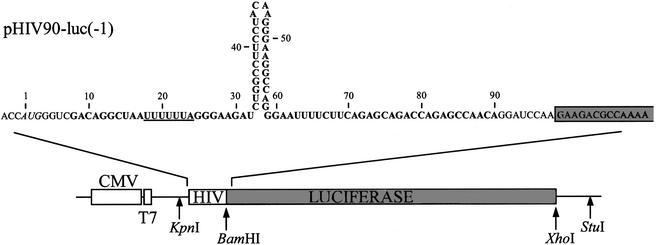
Plasmid pHIV90-luc is derived from pcDNA3.1/Hygro(+) (Invitrogen). A portion of the firefly luciferase gene encompassing the coding sequence and part of the 3′ untranslated region, contained between the BamHI and XhoI sites from pGEM®-luc (Promega) was first introduced between the same restriction sites of pcDNA3.1/Hygro(+), and the AUG initiation codon of luciferase was removed by standard mutagenesis, generating pcDNA3.1-luc. The HIV-1 gag/pol frameshift region encompassing bases 2076 to 2158 of HIV-1 HXB2 molecular clone (GenBank accession no. K03455) was amplified from plasmid pHC(–1) (25), where it had been inserted in the beginning of the chloramphenicol acetyltransferase gene. The amplification used the following forward and reverse primers, respectively: 5′-TAATACGACTCACTATAGGG-3′ and 5′-GCATGCTGGGGATCCTGTTGGC-3′. The resulting PCR fragment was inserted before the second codon of luciferase, between KpnI and BamHI restriction sites of pcDNA3.1-luc, generating pHIV90-luc(–1) (see details in Fig. 1). In this construct, the gag/pol frameshift region corresponds to bases 8 to 90. A short region containing an AUG initiation codon precedes the HIV portion, and the HIV-luc fusion is under the control of a cytomegalovirus (CMV) and a T7 promoter. This construction is such that the luciferase coding sequence is in the –1 frame relative to the initiator codon, so that only ribosomes that make a –1 frameshift produce luciferase. An in-frame plasmid control, pHIV90-luc(0), was created by inserting an additional adenine at position 81 in pHIV90-luc(–1), using PCR amplification. Derivatives of pHIV90-luc (deletion and substitution mutants) were created by PCR, by first amplifying mutated DNA fragments from pHIV90-luc for the (0) and (–1) constructs with two primers for deletion mutants or mutants with substitutions in the 3′ strand of the lower stem of the stimulatory signal, and with four primers for mutants with substitutions in the slippery sequence or in the 5′ strand of the lower stem, according to the procedure of Ho et al. (26). The amplified DNA fragments were then subcloned between the KpnI and BamHI sites of pcDNA3.1-luc, and all the constructs were verified by sequencing the entire insert.
Figure 1.
Description of pHIV90-luc, a frameshift reporter construct. Plasmid pHIV90-luc is a derivative of pcDNA3.1/Hygro(+) (Invitrogen) in which a BamH1/XhoI fragment encoding the firefly luciferase gene sequence has been added, with an insertion at the beginning of the coding sequence corresponding to the HIV-1 gag/pol frameshift region (see Materials and Methods). The KpnI and BamHI sites were used for subcloning the HIV-1 sequence between the vector and the luciferase sequences. The AUG initiator codon is italicized. Bases from positions 8–90 (in bold) originate from HIV-1. The slippery site is UUUUUUA (underlined). It is followed by a spacer, the classic frameshift stimulatory signal (a stem–loop) and an additional 30 nt sequence. The figure presents construct (–1), which produces luciferase only when ribosomes make a –1 frameshift. Addition of an adenine at position 81 corresponds to construct (0), which produces luciferase when ribosomes do not shift the reading frame.
Transient transfections and luciferase assays
COS1 cells were cultured in Dulbecco’s modification of Eagle’s medium supplemented with 10% (v/v) fetal bovine serum (BioMedia, Canada). Transfections were carried out in duplicate, using a standard calcium-phosphate precipitation method (27). Cells (1.5 × 105) were seeded in 35 mm dishes the day before transfection, 90 µl of the calcium-phosphate transfection precipitate containing 5 µg of a pHIV-luc construct and 2 µg of pcDNA3.1/Hygro/lacZ coding for β-galactosidase were overlayed over the cells. Cells were harvested 48 h post-transfection, washed twice with 2 ml of PBS, and 600 µl of the Cell Culture Lysis Reagent (Promega) were added. Following an incubation of 15 min at room temperature, cells were harvested and the supernatants assayed immediately for luciferase and β-galactosidase activity. For luciferase assays, 1.5 µl of cell extracts was added to 100 µl of the Luciferase Assay Reagent (Promega) and the light output was measured in relative light units with a Berthold Lumat LB 9507 luminometer. The β-galactosidase activity was measured with the chlorophenolred-β-galactopyranoside substrate (Calbiochem), as described (28), with aliquots of 10 µl of cell extracts, and used to normalize luciferase activities for variations in transfection efficiency.
In vitro transcription and translation
In vitro transcriptions were carried out essentially as previously described (29), using StuI-linearized pHIV-luc constructs. The RNA transcript was extracted twice with 1 vol of phenol/choloroform [50:50 (v/v)], once with 1 vol of chloroform/isoamyl alcohol [24:1 (v/v)], followed by precipitation with 1/10 vol of 3 M sodium acetate (pH 6.0) and 2.5 vol of 100% ethanol. The RNA pellet was dissolved in water, and unincorporated nucleotide triphosphates removed by G-50 Sephadex chromatography (MicroSpin G50 column, Amersham Pharmacia Biotech). RNA was quantified by spectrophotometry and checked for integrity by electrophoresis on 1.5% (w/v) agarose gels containing 0.1% (w/v) sodium dodecyl sulfate. For translation assays, RNA transcripts were heated at 65°C for 10 min and then briefly kept on ice prior to use, 0.2 µg of these transcripts were translated in 25 µl of RRL (Promega) at 30°C for 15 minutes, and the reaction was stopped by addition of EDTA at a final concentration of 6 mM. Luciferase activity was monitored as mentioned above, with 2.5 µl of the translation mixture.
Enzymatic probing of RNA structure
Enzymatic probing of the structure of an RNA fragment encompassing the frameshift region of HIV-1 was performed as described with minor modifications (30). An oligonucleotide cassette containing a T7 promoter followed by the HIV-1 gag/pol frameshift region (bases 16 to 70 according to the numbering of the construct depicted in Fig. 1) was cloned between the NaeI and Bsp119I sites of pGEM®-7Zf(–) (Promega), generating the recombinant plasmid pGEM-HIV. The RNA transcript, produced by in vitro transcription of the Bsp119I-linearized plasmid, was 5′-end labeled with [γ-32P], using a standard dephosphorylation–rephosphorylation method (31), purified from a 10% acrylamide–7 M urea gel and dissolved in 500 mM NH4OAc, 10 mM Mg(OAc)2, 1 mM EDTA and 0.1% SDS. Probing with RNase V1 (in a total volume of 10 µl containing 105 c.p.m. of 5′-end labeled RNA supplemented with 1 µg of yeast tRNA) was carried out at 25°C for 15 min in 10 mM Tris–HCl (pH 8), 10 mM MgCl2, 100 mM KCl and 0–0.01 U of enzyme (Ambion). The reaction was stopped by adding an equal volume of formamide gel loading buffer, the sample heated for 2 min at 95°C and immediately loaded for analysis on a 20% polyacrylamide– 7 M urea gel.
RESULTS
Length of the HIV-1 frameshift region
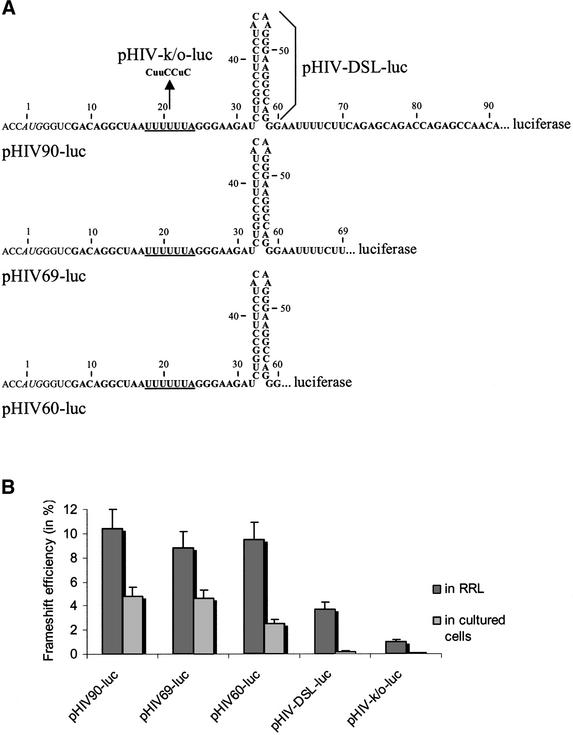
Several studies on HIV-1 frameshift have been performed where the frameshift region, encompassing the slippery site, the spacer and the classic stem–loop, was inserted in the beginning of the coding sequence of a reporter gene, such that its expression requires a translational frameshift (32,33). We decided to investigate whether the frameshift region that was previously studied contains all the signals required for maximal frameshifting and examined whether the sequence following the classic stem–loop could influence the frameshift. To this end, we made a HIV-1 frameshift reporter construct, using as a vector the pcDNA3.1/Hygro(+) plasmid, which contains a CMV and a T7 promoter, allowing us to assess the expression of a reporter gene in cultured cells and in a RRL, respectively. The firefly luciferase (luc) reporter gene was first introduced into this plasmid, and the HIV-1 frameshift region, longer by 30 bases than the classic frameshift region, was inserted in the beginning of the luc coding sequence. This generated plasmid pHIV90-luc(–1) (see Fig. 1 and Materials and Methods). This HIV-1 insertion is such that expression of the LUC protein requires a –1 frameshift. To quantify the frameshift efficiency, an ‘in-frame’ version [pHIV90-luc(0)] was derived from pHIV90-luc(–1), in which an additional adenine base was inserted upstream of the luciferase coding sequence so that expression of the reporter does not require a frameshift event. Linearization of the plasmid with StuI and transcription with T7 RNA polymerase generate a transcript of ∼1.9 kb, which was translated in a RRL. For assays in cultured cells, constructs were transfected into COS cells and luciferase levels were measured 48 h post-transfection. In both types of experiments, the frameshift efficiency was calculated by determining the ratio of the luciferase activity with the (–1) construct to the luciferase activity of the (0) construct plus that of the (–1) construct, assuming that the same level of frameshift should take place in the in-frame and in the (–1) construct. Different deletions were made in the frameshift region, generating pHIV60-luc(–1), which corresponds to the insertion of the classic frameshift region, ending after the stem–loop, and pHIV69-luc(–1), which contains the classic frameshift region plus 10 bases after the stem–loop (see Fig. 2A). For each of these constructs, an in-frame (0) control was also created. Results from experiments made in cultured cells and in vitro are presented in Figure 2B. As a control, mutations that altered the slippery sequence were made in pHIV90-luc to generate pHIV-k/o-luc. In cultured COS cells, these mutations almost abolished the frameshift (0.1%). The level of frameshifting in cultured cells was ∼2.5% in the presence of the classic stem–loop (pHIV60-luc), a value in full agreement with previous studies using reporters where the classic frameshift region of HIV-1 was inserted in the beginning of the coding sequence and whose expression depends upon a –1 frameshift (32,33). It was reduced to 0.2% in the absence of the stem–loop (pHIV-DSL-luc), stressing the importance of the classic stem–loop for the frameshift event. When frameshifting was assessed with the construct containing a longer frameshift region (pHIV90-luc), the frameshift efficiency was increased to 4.8% in cultured cells, 2-fold higher than with the classic frameshift region. With construct pHIV69-luc, where the classic frameshift region was extended by a small pyrimidine-rich 10-base segment, the frameshift efficiency (4.6%) was identical to the frameshifting level observed with construct pHIV90-luc. These results indicate that a segment of 10 bases adjacent to the classic stem–loop contributes to increase the level of frameshift, showing that the frameshift region is longer than believed. Similar values for frameshift efficiencies were obtained when transfecting another cell line, 293T (data not shown). Moreover, the frameshift efficiencies were independent of the reporter gene, since replacing the luciferase reporter with the green fluorescent protein reporter provided identical results (data not shown). Our findings that the frameshift region of HIV-1 is longer than believed are, thus, in perfect agreement with the recent study of Dinman et al. (24). The same constructs that were used in cultured cells were also assayed in vitro in a RRL. With the construct containing the conventional frameshift region, it was found that the classic stem–loop influences the frameshift efficiency but to a lesser extent than in cultured cells. Indeed, the frameshift level was 9.5% in its presence and decreased to 3.7% in its absence. With mutations that alter the slippery sequence, the frameshift efficiency was 1%, suggesting the possibility of aberrant initiation events. Lengthening the frameshift region resulted in a frameshift efficiency of 8.8 and 10.4% (constructs pHIV69-luc and pHIV90-luc, respectively), showing no significant variation, compared with the value of 9.5% obtained with the classic frameshift region. It has already been observed that the slippery sequence of HIV-1 is very efficient on its own in vitro (4–5%) compared with other natural slippery sequences, for which the frameshift efficiency is 1% or less (reviewed in 34). Previous reports have also shown that in vitro the frameshift of HIV-1 is more efficient and less sensitive to the presence of the stimulatory signal, compared with the situation in cultured cells (33,35). It has been proposed that the lower in vitro rate of translation accounts for the higher efficiency of the frameshift promoted by the slippery sequence of HIV-1 on its own (35), and this higher frameshift efficiency probably minimizes the effect of the frameshift stimulator. The weak effect of the classic stimulator on the frameshift efficiency, as shown by our results and in agreement with previous reports, led us to conclude that an in vitro system is not sensitive enough to investigate the length of the frameshift region and the structure of the stimulatory signal of HIV-1. Therefore, the following experiments were performed only in cultured cells.
Figure 2.
Effect on frameshifting of mutations in the HIV-1 frameshift region. (A) A series of mutations were made within the frameshift region of pHIV90-luc (the dots correspond to the BamHI linker connecting the frameshift region to the luciferase coding sequence): deletion mutants pHIV69-luc and pHIV60-luc, where the region 3′ to the classic stem–loop is shortened; a slippery site mutant, pHIV-k/o-luc, where the slippery sequence (underlined) is mutated (bases that are changed are in uppercase letters); a deletion mutant, pHIV-DSL-luc, where the classic stem–loop is eliminated (deletion of bases 31–60). (B) Frameshift efficiency in vitro and in cultured cells with the pHIV-luc constructs described above. In vitro translation experiments were made in 25 µl of RRL with 0.2 µg of mRNAs transcribed from StuI-digested pHIV90-luc and mutant constructs. Assays in cultured cells were made by co-transfecting COS1 cells with 5 µg of pHIV-luc and 2 µg of pcDNA3.1/Hygro/lacZ. Frameshift efficiencies were determined as described in the text. Each value represents the mean ± standard deviation of five to six independent experiments. The bars indicate the standard deviation on the means.
Proposed structure for the frameshift stimulatory signal of HIV-1
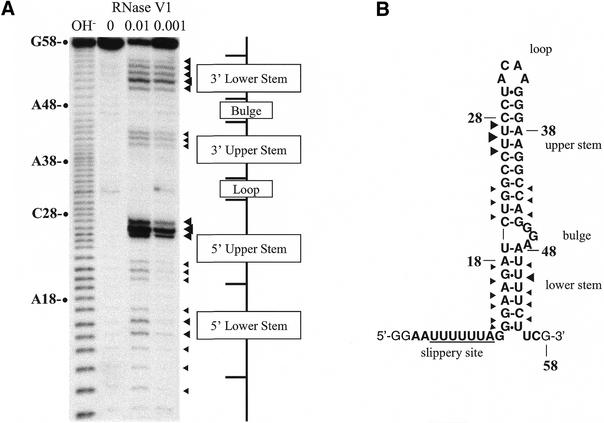
Our results reveal that, in cultured cells, a pyrimidine-rich segment downstream of the classic stem–loop enhances the frameshift efficiency for constructs containing an extended frameshift region of HIV-1. To account for the contribution of the downstream segment, we investigated the structure of an RNA fragment encompassing the gag/pol frameshift region of HIV-1. Enzymatic probing with RNase V1, an enzyme that cleaves RNA in helical conformation, showed that the purine-rich spacer region following the slippery sequence and preceding this classic stem–loop and the pyrimidine-rich downstream segment were attacked by the enzyme (Fig. 3). Interestingly, a similar observation had been made by Aupeix et al. (36). Based on these observations, we suggested that the frameshift region folds into a two-stem helix, made of an upper stem–loop, corresponding to the classic stem–loop, and a lower stem, formed by pairing the purine-rich spacer region to the pyrimidine-rich segment downstream of the classic stem–loop. A three-purine bulge interrupts these two stems (Fig. 3B). The thermodynamic stability of the novel frameshift stimulatory signal is –24.6 kcal/mol, as predicted by the MFOLD program at Michael Zucker’s MFOLD server at http://bioinfo.math.rpi.edu/∼mfold/rna/form1.cgi (37), whereas the classic frameshift signal has a thermodynamic stability of –21.3 kcal/mol. We then reasoned that if the lower stem contributes to stimulate frameshifting, mutations that prevent the formation of this stem should decrease the frameshift efficiency.
Figure 3.
Novel structure proposed for the frameshift stimulatory signal of HIV-1. (A) Structure probing of the frameshift stimulatory signal by RNase V1 attack. An RNA transcript encompassing the HIV-1 gag/pol frameshift region was 5′ end-labeled with [γ-32P] and digested with RNase V1. Digestion products were analyzed on a 20% acrylamide–7 M urea gel. The sites of cleavage were identified by comparison with a ladder of bands created by limited alkaline hydrolysis of the RNA (OH–) and by the position of RNase T1 cuts (not shown). Uniquely cleaved nucleotides were identified by their absence in the untreated control lane (0). The amount of units of enzyme added to each reaction is also indicated. (B) Description of the novel two-stem model for the frameshift stimulatory signal as suggested by structure probing. The upper stem corresponds to the classic stem–loop and the lower stem is formed by pairing the spacer to a segment downstream of this stem–loop. The sensitivity of nucleotides in the HIV-1 frameshift region to RNaseV1 is shown. The size of the arrows is approximately proportional to the intensity of the cleavage at that site. Bases in bold originate from HIV-1.
Mutagenesis studies
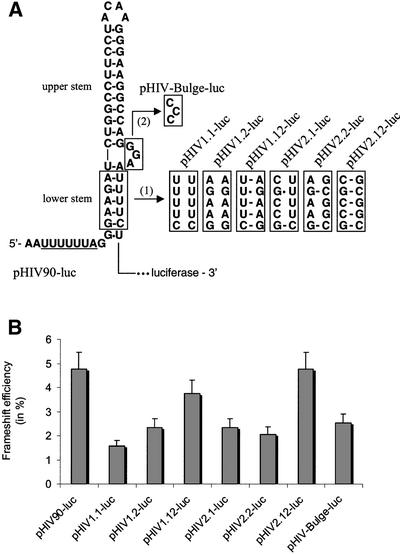
To further investigate the structure that we propose for the frameshift stimulatory signal of HIV-1, we introduced mutations in the two strands of the putative lower stem, which individually disrupt this stem, but when present simultaneously allow its re-formation (Fig. 4A). Frameshift efficiencies with the different constructs are shown in Figure 4B. We first replaced five bases of the 5′ strand of the putative lower stem of the HIV-1 frameshift stimulator with five bases of the 3′ strand of this stem, generating construct pHIV1.1-luc. This mutation was found to reduce frameshifting ∼3-fold in cultured cells (1.6% frameshifting), near the level obtained with the construct containing the classic frameshift region. Similarly, the frameshift efficiency was decreased 2-fold when replacing five bases in the 3′ strand of the lower stem with five bases of the 5′ strand (pHIV1.2-luc; 2.3% frameshifting). However, when both strands were mutated simultaneously, allowing re-formation of the lower stem, the frameshift was enhanced, although still inferior to the wild-type level (pHIV1.12-luc; 3.8% frameshifting). A second series of complementary and compensatory base-pair change mutations were also made in the putative lower stem. These mutations are such that they can create four new G-C pairs in the lower stem, so as to increase its stability (Fig. 4A). In the compensatory mutants, while pHIV1.12-luc has the same thermodynamic stability as pHIV90-luc (ΔG° = –24.6 kcal/mol), this stability is significantly increased in pHIV2.12-luc (ΔG° = –32.9 kcal/mol). Assays with this second series of mutations in the lower stem confirmed the results obtained with the first series of mutations, showing that changes in either the 5′ strand or the 3′ strand of the lower stem reduced the frameshifting level ∼2-fold (pHIV2.1-luc and pHIV2.2-luc; 2.3 and 2.1%, respectively), and that the frameshift was restored to a wild-type value when both strands were mutated so as to re-form a lower stem (pHIV2.12-luc; 4.8%). It can be observed that the frameshift efficiency was not higher with the more stable lower stem. We also investigated whether the three-purine bulge separating the two stems participates in the frameshift event. To address this question, mutations were made that replaced the three purines of the bulge with pyrimidines (pHIV-Bulge-luc). We found that this mutation reduced the frameshifting level ∼2-fold (2.5% compared with 4.8% for pHIV90-luc), bringing it to the level observed when only the classic stem–loop is present and showing that the bulge contributes to the frameshift.
Figure 4.
Effect on frameshifting of mutations in the lower stem of the proposed frameshift stimulatory signal. (A) Description of mutations made within the gag/pol frameshift region of pHIV90-luc. (1) Two series of mutants (the pHIV1 and pHIV2 series) were made in the lower stem. For each series, mutations were introduced either in the 5′ or 3′ strand of the lower stem, impairing the formation of this stem (pHIV1.1-luc and pHIV1.2-luc; pHIV2.1-luc and pHIV2.2-luc), or allowing re-formation of this stem (pHIV1.12-luc and HIV2.12-luc). (2) The three purines forming the bulge separating the two stems were substituted with pyrimidines (pHIV-Bulge-luc). (B) Frameshift efficiency in cultured cells with the pHIV-luc constructs described above. Assays were as described in the legend to Figure 2.
Conservation of the two-stem helix among HIV-1 isolates
Since the frameshift efficiency is critical for maintenance of the Gag-Pol to Gag ratio that allows viral replication, the structure of the frameshift stimulatory signal should be highly conserved in HIV-1 natural isolates. To verify the capacity of the spacer following the slippery sequence to base pair with the pyrimidine-rich region downstream of the classic stem– loop, we analyzed 139 complete pol sequences retrieved from the Los Alamos National Laboratory HIV Sequence Database (38) (Table 1). This analysis shows that the proposed duplex structure can be formed in 96.4% of the sequences, with 58.3% of them conserving the capacity to form the proposed structure with only Watson–Crick or G-U base-pairs and 38.1% containing non-Watson–Crick A-C base-pairs (35.2 and 2.9% with one and two A-C base pairs, respectively), which are known not to distort significantly the geometry of an RNA helix (39). Only 3.6% of the sequences analyzed contain mismatches able to distort the RNA helix. Therefore, the analysis of sequences strongly supports our suggestion that the capacity to form the lower stem in the structure that we proposed for the frameshift stimulatory signal of HIV-1 is evolutionarily conserved. As mentioned in the Introduction, a different structure was recently proposed for the frameshift stimulatory signal of HIV-1 by Dinman et al. (24). In their model, the pyrimidine-rich sequence downstream of the classic stem–loop interacts with three bases in the loop capping this stem through Watson–Crick interactions and forms a triple helix with 4 bp on top of this stem. Analysis of the pol sequences retrieved from the Los Alamos National Laboratory HIV Sequence Database does not contradict their model, as shown in Table 1, if one assumes that a U can replace a C in the third strand of the triplex and interact with the G of a C-G pair without destabilizing the triplex. This, however, is not the case (39–41), especially in a short triplex formed with only 4 bp where three U*G-C triple interactions are proposed. If, nevertheless, we accept the assumption of Dinman et al. (24), 92.8% of the sequences could form the triple helix, with 41% of them conserving the capacity to form the triplex structure and 51.8% containing changes that minimally affect the capacity to form this structure. Only 7.2% of the sequences could not form this structure. However, a major criticism that we address to the model of Dinman et al. (24) is that the pyrimidine-rich third strand that forms the triplex is in an antiparallel orientation relative to the purine-rich strand of the duplex with which it interacts, an orientation that is not sterically favored (see 40–42), making this structure highly improbable. Moreover, Dinman et al. (24) claim that structural probing supports their model but this probing was done with a short fragment of the frameshift region of HIV-1 encompassing only the classic stem–loop and the downstream sequence. When a longer fragment encompassing, in addition, the slippery sequence and the spacer region was probed, this spacer region and the downstream segment to which we propose that it base pairs were found to be sensitive to RNase V1, an endonuclease that cleaves RNA in helical conformation (Fig. 3) (36). This probing fully supports the lower stem of the structure that we propose for the frameshift stimulatory signal of HIV-1. Finally, it can be observed that our mutants pHIV1.12-luc and pHIV2.12-luc have frameshift efficiencies near wild-type level although they cannot participate in the structure proposed by Dinman et al. (24).
Table 1. Conservation of the proposed structure for the frameshift stimulatory signal among HIV-1 isolates.
| Description of the sequence | Proposed structure | |
|---|---|---|
| Two-stem helixa | Triplex helixb | |
| Total of sequences that do not affect the capacity to form the structure | 81 (58.3%) | 57 (41.0%) |
| Consensus sequence | 19 (13.7%) | 13 (9.3%) |
| Changes that do not affect the capacity to form the structurec | 62 (44.6%) | 44 (31.7%) |
| Changes that have a minimal impact on the capacity to form the structured | 53 (38.1%) | 72 (51.8%) |
| Total changes that do not or minimally affect the capacity to form the structure | 134 of 139 (96.4%) | 129 of 139 (92.8%) |
| Mismatches that disrupt the capacity to form the structuree | 5 of 139 (3.6%) | 10 of 139 (7.2%) |
Complete pol sequences (139) from HIV-1 were retrieved from the Los Alamos National Laboratory HIV Sequence Database (38), and analyzed manually for the capacity to form the proposed frameshift stimulatory signals. The 139 sequences include natural isolates of HIV-1 group M (main), the most widespread group in the world (38). Sequences belonging to members of divergent lineages such as group O (outlier) (four sequences) and N (non-M, non-O) (two sequences) have not been included in the analysis.
aTwo-stem helix proposed in this study.
bTriplex helix proposed by Dinman et al. (24).
cSequential changes allowing the formation of Watson–Crick and G-U base pairs and, in the model of Dinman et al. (24), conserving the capacity to form the triplex structure.
dSequential changes allowing the formation of A-C base pairs that minimally distort RNA double helices.
eMismatches (U-U, A-A, C-C, G-A, U-C, C-U and C-A base pairs) that disrupt double-helical structures and, in the model of Dinman et al. (24), impair the capacity to form the triplex structure.
DISCUSSION
Our results show that the HIV-1 frameshift stimulatory signal folds into a more complex structure than commonly assumed. A two-stem structure was proposed where the sequences flanking the classic stem–loop base-pairs, thus forming the lower stem of an extended helix. This structure was proposed from structure probing of the frameshift stimulatory signal and supported by mutagenesis studies. Indeed, mutations that disrupted base pairing between the sequences forming the lower stem of our proposed structure (pHIV1.1-luc and pHIV1.2-luc; pHIV2.1-luc and pHIV2.2-luc) reduced frameshift efficiencies. It could be argued that sequences flanking the stem–loop influence frameshifting by modulating stem– loop unfolding kinetics (43). However, compensatory mutations that enable re-formation of the lower stem, by exchanging the sequence of each strand, restored frameshifting near the wild-type level (pHIV1.12-luc and pHIV2.12-luc), and this could not result from a simple effect of mutations in the flanking sequences on the unfolding kinetics of the classic stem–loop. As mentioned in the Introduction, the function of the stimulatory signal is not completely understood, and it is thought that specific interactions between the ribosome and the frameshift stimulatory signal are required to achieve efficient frameshifting. The upper stem of the stimulator obviously plays a major role in the interaction with the ribosome since its presence strongly increases frameshifting, compared with what is observed when only the slippery sequence is present, whereas the lower stem only contributes to enhance frameshifting ∼2-fold. However, as indicated above, the frameshift efficiency controls the Gag-Pol to Gag ratio, and small variations in frameshifting can dramatically affect the virus propagation. Indeed, studies with the yeast L-A double-stranded RNA virus, for which the synthesis of Gag-Pol also requires a programmed –1 frameshift, showed that either increasing or decreasing the frameshift efficiency by >2-fold, by altering the slippery sequence, disrupted viral propagation (8). Also, a 3-fold increase in the Gag-Pol to Gag ratio in HIV-1 producing cells, achieved by co-transfecting HIV-1 proviral DNA with an HIV-1 Gag-Pol expression vector, resulted in a 10-fold decrease in virion infectivity (4). The novel frameshift stimulatory structure is only slightly more stable than the classic signal, and it is unlikely that its enhancing effect results from this increased stability. Indeed, it was even observed that increasing the stability of the classic stem–loop by lengthening the stem could reduce frameshifting (44). Our data show the importance of the three-purine bulge interrupting the two stems in the HIV-1 frameshift stimulatory signal. The formation of this bulge requires the presence of the lower stem and substitution of the purines of the bulge with pyrimidines decreased frameshifting to the level observed in the absence of the lower stem. Although purely speculative, we suggest that this bulge, by interacting with the ribosome through purine-mediated interactions, could contribute to enhance the interaction of the upper stem with the ribosome.
A feature that differentiates the frameshift stimulatory signal in HIV-1 from other signals is that it is distant from the slippery site by one base only. In most cases where frameshifting occurs, there is a spacer sequence of five to eight bases between the slippery sequence and the stimulatory signal (5). However, in the Rous sarcoma virus (RSV) RNA, which uses a programmed –1 frameshift, the frameshift stimulator, which is a pseudoknot, is also located one base after the slippery sequence (30). Since frameshift takes place when the slippery sequence occupies the A and P sites of the ribosome, either the RSV frameshift stimulatory signal or the stimulatory signal that we propose for HIV-1 would have to evade the unwinding activity of the translating ribosome or re-fold once inside the ribosome. Interestingly, the possibility that a signal stimulating a recoding event can evade the ribosome unwinding activity or re-fold within the ribosome has also been proposed for the ribosomal hopping that occurs during translation of the T4 gene 60 (45). In this recoding event, a stem–loop in close proximity to the decoding site stimulates tRNA slippage, implying that this structure is present within the ribosome.
In conclusion, we propose a refinement of the structure of the HIV-1 frameshift stimulatory signal, which consists of increasing the size and complexity of this signal. A complete characterization of the signal that promotes frameshifting in HIV and of its interaction with the ribosome will be extremely useful in providing valuable information for the development of antiviral agents that target programmed –1 ribosomal frameshift.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Guy Lemay for his interest in this work. We are most grateful to Sergey Steinberg for stimulating discussions and comments. We are also grateful to Pascal Chartrand, Luc Desgroseillers and Francis Robert for critical reading of this manuscript. This study was supported by a grant from the Canadian Institutes for Health Research.
REFERENCES
- 1.Jacks T., Power,M.D., Masiarz,F.R., Luciw,P.A., Barr,P.J. and Varmus,H.E. (1988) Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature, 331, 280–283. [DOI] [PubMed] [Google Scholar]
- 2.Park J. and Morrow,C.D. (1991) Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J. Virol., 65, 5111–5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karacostas V., Wolffe,E.J., Nagashima,K., Gonda,M.A. and Moss,B. (1993) Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology, 193, 661–671. [DOI] [PubMed] [Google Scholar]
- 4.Shehu-Xhilaga M., Crowe,S.M. and Mak,J. (2001) Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol., 75, 1834–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brierley I. (1995) Ribosomal frameshifting on viral RNAs. J. Gen. Virol., 76, 1885–1892. [DOI] [PubMed] [Google Scholar]
- 6.Brierley I. and Pennell,S. (2001) Structure and function of the stimulatory RNAs involved in programmed eukaryotic –1 ribosomal frameshifting. In Stillman,B. (ed.), The Ribosome, 66th Cold Spring Harbor Symposium on Quantitative Biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 233–248. [DOI] [PubMed]
- 7.Tzeng T.H., Tu,C.L. and Bruenn,J.A. (1992) Ribosomal frameshifting requires a pseudoknot in the Saccharomyces cerevisiae double-stranded RNA virus. J. Virol., 66, 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinman J.D. and Wickner,R.B. (1992) Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol., 66, 3669–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuchihashi Z. and Brown,P.O. (1992) Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. Genes Dev., 6, 511–519. [DOI] [PubMed] [Google Scholar]
- 10.Chandler M. and Fayet,O. (1993) Translational frameshifting in the control of transposition in bacteria. Mol. Microbiol., 7, 497–503. [DOI] [PubMed] [Google Scholar]
- 11.Larsen B., Wills,N.M., Gesteland,R.F. and Atkins,J.F. (1994) rRNA-mRNA base pairing stimulates a programmed –1 ribosomal frameshift. J. Bacteriol., 176, 6842–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gesteland R.F. and Atkins,J.F. (1996) Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem., 65, 741–768. [DOI] [PubMed] [Google Scholar]
- 13.Shigemoto K., Brennan,J., Walls,E., Watson,C.J., Stott,D., Rigby,P.W. and Reith,A.D. (2001) Identification and characterisation of a developmentally regulated mammalian gene that utilises –1 programmed ribosomal frameshifting. Nucleic Acids Res., 29, 4079–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacks T., Madhani,H.D., Masiarz,F.R. and Varmus,H.E. (1988) Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell, 55, 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farabaugh P.J. (2000) Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog. Nucleic Acid Res. Mol. Biol., 64, 131–170. [DOI] [PubMed] [Google Scholar]
- 16.Somogyi P., Jenner,A.J., Brierley,I. and Inglis,S.C. (1993) Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol., 13, 6931–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu C., Tzeng,T.H. and Bruenn,J.A. (1992) Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl Acad. Sci. USA, 89, 8636–8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopinski J.D., Dinman,J.D. and Bruenn,J.A. (2000) Kinetics of ribosomal pausing during programmed –1 translational frameshifting. Mol. Cell. Biol., 20, 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kontos H., Napthine,S. and Brierley,I. (2001) Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol., 21, 8657–8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen L.X. and Tinoco,I.,Jr (1995) The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J. Mol. Biol., 247, 963–978. [DOI] [PubMed] [Google Scholar]
- 21.Kang H. and Tinoco,I.,Jr (1997) A mutant RNA pseudoknot that promotes ribosomal frameshifting in mouse mammary tumor virus. Nucleic Acids Res., 25, 1943–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le S.Y., Chen,J.H. and Maizel,J.V. (1989) Thermodynamic stability and statistical significance of potential stem–loop structures situated at the frameshift sites of retroviruses. Nucleic Acids Res., 17, 6143–6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang H. (1998) Direct structural evidence for formation of a stem–loop structure involved in ribosomal frameshifting in human immunodeficiency virus type 1. Biochim. Biophys. Acta, 1397, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dinman J.D., Richter,S., Plant,E.P., Taylor,R.C., Hammell,A.B. and Rana,T.M. (2002) The frameshift signal of HIV-1 involves a potential intramolecular triplex RNA structure. Proc. Natl Acad. Sci. USA, 99, 5331–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doyon L., Payant,C., Brakier-Gingras,L. and Lamarre,D. (1998) Novel Gag-Pol frameshift site in human immunodeficiency virus type 1 variants resistant to protease inhibitors. J. Virol., 72, 6146–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.K. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- 27.Jordan M., Schallhorn,A. and Wurm,F.M. (1996) Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res., 24, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eustice D.C., Feldman,P.A., Colberg-Poley,A.M., Buckery,R.M. and Neubauer,R.H. (1991) A sensitive method for the detection of beta-galactosidase in transfected mammalian cells. Biotechniques, 11, 739–743. [PubMed] [Google Scholar]
- 29.Dragon F., Payant,C. and Brakier-Gingras,L. (1994) Mutational and structural analysis of the RNA binding site for Escherichia coli ribosomal protein S7. J. Mol. Biol., 244, 74–85. [DOI] [PubMed] [Google Scholar]
- 30.Marczinke B., Fisher,R., Vidakovic,M., Bloys,A.J. and Brierley,I. (1998) Secondary structure and mutational analysis of the ribosomal frameshift signal of Rous sarcoma virus. J. Mol. Biol., 284, 205–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knapp G. (1989) Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol., 180, 192–212. [DOI] [PubMed] [Google Scholar]
- 32.Cassan M., Delaunay,N., Vaquero,C. and Rousset,J.P. (1994) Translational frameshifting at the gag-pol junction of human immunodeficiency virus type 1 is not increased in infected T-lymphoid cells. J. Virol., 68, 1501–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reil H., Kollmus,H., Weidle,U.H. and Hauser,H. (1993) A heptanucleotide sequence mediates ribosomal frameshifting in mammalian cells. J. Virol., 67, 5579–5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brierley I., Jenner,A.J. and Inglis,S.C. (1992) Mutational analysis of the ‘‘slippery-sequence’’ component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol., 227, 463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkin N.T., Chamorro,M. and Varmus,H.E. (1992) Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J. Virol., 66, 5147–5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aupeix K., Le Tinevez,R. and Toulmé,J.J. (1999) Binding of oligopyrimidines to the RNA hairpin responsible for the ribosome gag-pol frameshift in HIV-1. FEBS Lett., 449, 169–174. [DOI] [PubMed] [Google Scholar]
- 37.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 38.Human Retroviruses and AIDS (2002) A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences. Los Alamos National Laboratory, Los Alamos, NM, http://hiv-web.lanl.gov.
- 39.Nowakowski J. and Tinoco,I.,Jr (1999) RNA structure in solution. In Neidle,S. (ed.), Oxford Handbook of Nucleic Acid Structure. Oxford University Press, New York, NY, pp. 567–602.
- 40.Han H. and Dervan,P.B. (1993) Sequence-specific recognition of double helical RNA and RNA·DNA by triple helix formation. Proc. Natl Acad. Sci. USA, 90, 3806–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frank-Kamenetskii M.D. and Mirkin,S.M. (1995) Triplex DNA structures. Annu. Rev. Biochem., 64, 65–95. [DOI] [PubMed] [Google Scholar]
- 42.Lim A.C. and Barton,J.K. (1998) Rh(Phen)2Phi3+ as a shape-selective probe of triple helices. Biochemistry, 37, 9138–9146. [DOI] [PubMed] [Google Scholar]
- 43.Kim Y.-G., Mass,S. and Rich,A. (2001) Comparative mutational analysis of cis-acting RNA signals for translational frameshifting in HIV-1 and HTLV-2. Nucleic Acids Res., 29, 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bidou L., Stahl,G., Grima,B., Liu,H., Cassan,M. and Rousset,J.P. (1997) In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem–loop stimulatory signal. RNA, 3, 1153–1158. [PMC free article] [PubMed] [Google Scholar]
- 45.Herr A.J., Gesteland,R.F. and Atkins,J.F. (2000) One protein from two open reading frames: mechanism of a 50 nt translational bypass. EMBO J., 19, 2671–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]