Abstract
RNase P is a ribonucleoprotein involved in tRNA biosynthesis in all living organisms. Bacterial RNase P is comprised of a catalytic RNA subunit and a lone protein cofactor which plays a supporting, albeit essential, role in the tRNA processing reaction in vivo. In this study, we have searched various databases to identify homologs of the protein subunit of RNase P from diverse bacteria and used an alignment of their primary sequences to determine the most highly conserved residues, and thereby extend earlier predictions of which residues might play an important role in RNA recognition. By employing a genetic complementation assay, we have also gained insights into structure– function relationships in the protein subunit of bacterial RNase P.
INTRODUCTION
RNase P is responsible for cleavage of the 5′ leader sequence of precursor transfer RNAs (ptRNAs) to form mature tRNAs (1,2). On account of its indispensable role in tRNA biosynthesis, RNase P is absolutely essential for the viability of all living organisms and has been identified in Bacteria, Archaea and Eukarya (1,2). The bacterial RNase P holoenzyme is a ribonucleoprotein (RNP) heterodimer consisting of a catalytic RNA subunit (∼350–400 nucleotides) and a small protein cofactor (∼100–150 amino acid residues) (1–3). At high ionic strength in vitro, the RNA subunits of all bacterial and some archaeal RNase P are catalytically active in the absence of their cognate protein cofactor(s) (1–4). However, the protein subunit of bacterial RNase P is absolutely essential for RNase P function in vivo, presumably due to its pleiotropic effects on RNA catalysis (5–13). Results from in vitro experiments indicate that addition of the protein cofactor increases the catalytic efficiency of the RNase P holoenzyme, in large part due to enhanced affinity for the ptRNA substrate (7,11).
Since the discovery of the catalytic activity of M1 RNA, the RNA subunit of Escherichia coli RNase P, various biochemical and genetic studies have identified nucleotides in this RNA moiety as well as amino acid residues in its cognate protein cofactor (C5 protein) which are involved in substrate binding and catalysis (1,2,14–19). Taken together with structural perspectives on the individual RNA and protein subunits, obtained using computer-assisted molecular modeling and high-resolution approaches, respectively, these data are providing the first glimpses into the functioning of an ancient ribozyme (20–25). These results are the subject of recent reviews (1,2,26) and will not be elaborated here. Despite these advances in relating long-standing functional data and sequence information, our current knowledge of the mechanism of action of RNase P is incomplete owing to the lack of knowledge of the tertiary structures of the catalytic RNA moiety and the RNase P holoenzyme. In this study, we have used database mining in conjunction with a genetic complementation assay to elucidate the role of conserved residues in the protein subunit of E.coli RNase P and thereby furthered our understanding of the bacterial RNase P holoenzyme.
MATERIALS AND METHODS
Enzymes and reagents
Restriction and modifying enzymes were obtained from New England Biolabs, Gibco Life Technologies or USB. Oligonucleotides used for PCR and site-directed mutagenesis (Table 1) were synthesized by the Keck Facility at Yale University or IDT. The Hybond ECL nitrocellulose membrane and the ECL Western Blotting Analysis kit were purchased from Amersham Biosciences.
Table 1. Oligonucleotides used for site-directed mutagenesis.
| Name of primer | Sequence of oligonucleotide | Mutation(s) introduced |
|---|---|---|
| SDMO7 | 5′-A ACT CCC AGT CAA GCT ACA TTC GTC TTC CAG CAG CC-3′ | F18A |
| SDMO16 | 5′-A ACT CCC AGT CAA TTC ACA TTC GTC GCT CAG CAG CCA CAA CGG GC-3′ | F22A |
| SDMO716 | 5′-A ACT CCC AGT CAA GCT ACA TTC GTC GCT CAG CAG CCA CAA CGG GC-3′ | F18A/F22A |
| SDMOA49 | 5′-CTG AAT TCG CTG GGG CAT CCC CAT ATC GGT CTT ACA GTC GCC-3′ | R46H |
| SDMO32a | 5′-GCG TTC ATG GGC GCG CGC AAC GTT TTT CTT GGC G-3′ | R57A |
| SDMO33a | 5′-G ACG GAA GCT TTC AGC CGT CAG ACG TTT AAT CCG-3′ | R70A |
| SDMO925a | 5′-CCG GAA GCT TTC ACG CGT CAG AGC TTT GAT CCG ATT AGC TTC ATG G-3′ | R62A/R67A |
| SDMO933a | 5′-G ACG GAA GCT TTC AGC CGT CAG ACG TTT AAT CCG ATT AGC-3′ | R62A/R70A |
| SDMO2533a | 5′-G ACG GAA GCT TTC AGC CGT CAG AGC TTT AAT CCG ATT GC-3′ | R67A/R70A |
| fEcoRI | 5′-GC CTG AAT TCG CTG GG-3′ | – |
| rHindIII | 5′-G ACG GAA GCT TTC ACG CGT C-3′ | – |
aThese oligonucleotides are in the antisense orientation relative to the coding sequence for C5 protein. The bold letters indicate the codon that was altered.
Construction of pFHC1008′
The low copy-number plasmid pFHC1008, derived from pBR322, contains the entire rpmH operon, encoding both the ribosomal protein L34 and C5 protein under the control of their endogenous promoter (27). Therefore, expression of C5 protein from this plasmid is expected to mimic the regulation of gene expression observed with the chromosomal copy. To facilitate cloning of various mutant derivatives of C5 protein for our studies, we constructed pFHC1008′, a derivative of pFHC1008 which differs from the parental construct in that the EcoRI and HindIII sites within the gene encoding C5 protein have been rendered unique. We accomplished this goal using the following approach. First, we digested pBR322 with EcoRI and HindIII, filled in recessed ends using Klenow polymerase, and ligated the resulting blunt ends with T4 DNA ligase. Subsequently, this pBR322 derivative and pFHC1008 were both digested with NheI and AvaI. The NheI–AvaI fragment derived from pFHC1008 and containing the rpmH operon was then cloned into the NheI and AvaI sites in the pBR322 derivative to yield pFHC1008′.
Cloning of various mutant derivatives of C5 protein in pFHC1008′
Different strategies were employed for generating the 23 mutant constructs used in this genetic complementation study (Table 2).
Table 2. Summary of reagents used for obtaining constructs of mutant derivatives of C5 protein in pFHC1008′.
| Name of construct | Mutations introduced | Primers used for PCR | PCR template | Enzymes used for cloning |
|---|---|---|---|---|
| Group Ia | ||||
| pFHC1008′-Sn49 | R46H | - | - | EcoRI and HindIII |
| pFHC1008′-Sn32 | R57A | - | - | EcoRI and HindIII |
| pFHC1008′-Sn9 | R62A | - | - | EcoRI and HindIII |
| pFHC1008′-Sn24 | K66A | - | - | EcoRI and HindIII |
| pFHC1008′-Sn25 | R67A | - | - | EcoRI and HindIII |
| Group II | ||||
| pFHC1008′-Sn7 | F18A | SDMO7 and rHindIII | pFHC1008′ | HpaI and HindIII |
| pFHC1008′-Sn16 | F22A | SDMO16 and rHindIII | pFHC1008′ | HpaI and HindIII |
| pFHC1008′-Sn716 | F18A/F22A | SDMO716 and rHindIII | pFHC1008′ | HpaI and HindIII |
| Group III | ||||
| pFHC1008′-Sn33 | R70A | SDMO33 and fEcoRI | pFHC1008′ | EcoRI and HindIII |
| pFHC1008′-Sn925 | R62A/R67A | SDMO925 and fEcoRI | pFHC1008′ | EcoRI and HindIII |
| pFHC1008′-Sn933 | R62A/R70A | SDMO933 and fEcoRI | pFHC1008′ | EcoRI and HindIII |
| pFHC1008′-Sn2533 | R67A/R70A | SDMO2533 and fEcoRI | pFHC1008′ | EcoRI and HindIII |
| pFHC1008′-Sn4932 | R46H/R57A | SDMOA49 and rHindIII | pFHC1008′-Sn32 | EcoRI and HindIII |
| pFHC1008′-Sn499 | R46H/R62A | SDMOA49 and rHindIII | pFHC1008′-Sn9 | EcoRI and HindIII |
| pFHC1008′-Sn4924 | R46H/K66A | SDMOA49 and rHindIII | pFHC1008′-Sn24 | EcoRI and HindIII |
| pFHC1008′-Sn4925 | R46H/R67A | SDMOA49 and rHindIII | pFHC1008′-Sn25 | EcoRI and HindIII |
| pFHC1008′-Sn4933 | R46H/R70A | SDMOA49 and rHindIII | pFHC1008′-Sn33 | EcoRI and HindIII |
| Group IV | ||||
| pFHC1008′-Sn79 | F18A/R62A | SDMO7 and rHindIII | pFHC1008′-Sn9 | HpaI and HindIII |
| pFHC1008′-Sn724 | F18A/K66A | SDMO7 and rHindIII | pFHC1008′-Sn24 | HpaI and HindIII |
| pFHC1008′-Sn725 | F18A/R67A | SDMO7 and rHindIII | pFHC1008′-Sn25 | HpaI and HindIII |
| pFHC1008′-Sn169 | F22A/R62A | SDMO16 and rHindIII | pFHC1008′-Sn9 | HpaI and HindIII |
| pFHC1008′-Sn1624 | F22A/K66A | SDMO16 and rHindIII | pFHC1008′-Sn24 | HpaI and HindIII |
| pFHC1008′-Sn1625 | F22A/R67A | SDMO16 and rHindIII | pFHC1008′-Sn25 | HpaI and HindIII |
aThe different strategies used for obtaining the various groups of mutant derivatives are described in the Materials and Methods.
Group I constitutes a set of mutants that were constructed by replacing the EcoRI–HindIII fragment (within the gene encoding C5 protein) in pFHC1008′ with a corresponding fragment that harbors nucleotide substitutions to introduce a single amino acid change. This EcoRI–HindIII fragment was obtained from pMBA49 (28), pBSC5Sn32, pBSC5Sn9 (19), pBSC5Sn24 (19) or pBSC5Sn25 (19) to introduce R46H, R57A, R62A, K66A or R67A, respectively, in the pFHC1008′ backbone. Note that although pBSC5Sn32 was not described in our earlier study, it was constructed by employing the SDMO32 oligonucleotide (Table 1) and the Kunkel approach as already described for pBSC5Sn9, pBSC5Sn24 etc. (19).
The remaining constructs (groups II, III and IV) were all constructed by employing a PCR-based mutagenesis approach. Either the primers, the template, or both contained the mutations that were to be engineered. By incorporating in the PCR primers recognition sequences for different restriction enzymes, which have unique cleavage sites in pFHC1008′, we could digest both pFHC1008′ and the various PCR products with the appropriate pair of restriction enzymes and thereby replace the wild type fragment in pFHC1008′ with the mutant counterpart. All ligation reactions were transformed into E.coli DH5α cells. Plasmid DNA was isolated from the transformants and DNA sequencing was performed to ascertain the presence of the desired mutation and the absence of unwanted alterations.
Genetic complementation assay
NHY322 cells containing the rnpA mutation (i.e., C5 R46H) (29) in their chromosomes were transformed with pBR322 or plasmids encoding either wild type C5 protein or its mutant derivatives, and grown overnight at 30°C on LB media with 15 µg/ml tetracycline (rnpA chromosomal selection) and 100 µg/ml carbenicillin (plasmid selection). The permissive and non-permissive temperatures for this strain are 30 and 43°C, respectively. Colonies of the transformants obtained at 30°C and bearing the various plasmids were sequentially streaked on two different agar plates supplemented with the appropriate antibiotics and pre-warmed to 43 and 30°C. The genetic complementation ability of the various mutant derivatives of C5 protein was assessed by examining growth of colonies after an overnight incubation at the non-permissive temperature (28). Cells transformed with pBR322 (vector alone) and pFHC1008′ (i.e., wild type C5 protein) served as the negative and positive controls, respectively, for complementation of the NHY322 ts phenotype.
Western blot analysis
Samples for western analysis were obtained by employing the following procedure. First, NHY322 cells carrying the various plasmids were grown overnight in liquid LB media supplemented with 15 µg/ml tetracycline and 100 µg/ml carbenicillin at 30°C. The overnight cultures were diluted to OD600 ∼0.05 in fresh LB media supplemented with the appropriate antibiotics and grown at 30°C until OD600 ∼0.15 wherein one-half of the culture was shifted to 43°C and grown for 2 h with agitation. Since the mutant derivatives support growth to different densities at the non-permissive temperature (43°C), the amount of culture harvested was normalized on the basis of their respective OD600 values. The cell pellets were first resuspended in 80 µl water prior to addition of 20 µl 1 M DTT and 100 µl 2× Laemmli loading buffer. The samples were boiled for 5 min and a 10 µl aliquot was withdrawn for electrophoresis on a SDS–15% (w/v) polyacrylamide gel. Twenty nanograms of purified wild type C5 protein (19) and 2 µl Magic Markers (Invitrogen) were included in the gel to serve as positive control and size standards, respectively. After electrophoresis at 160 V for 1 h, the proteins in the gel were transferred to an ECL Hybond nitrocellulose membrane (Amersham Biosciences) at 175 mA for 1 h.
Note that all the following steps for immunodetection were performed at room temperature with continuous shaking on an orbital shaker. The membrane was pre-incubated for 1 h in blocking buffer [10 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.05% (w/v) Tween-20, and 3% (w/v) non-fat dry milk powder] prior to a 1 h incubation with a 1:1000 dilution of affinity-purified C5 antibody (in blocking buffer). The membrane was then washed three times for 7 min each in wash buffer [10 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.05% (w/v) Tween-20]. Subsequently, the membrane was incubated with a 1:5000 dilution in blocking buffer of goat anti-rabbit horseradish peroxidase-conjugated anti-sera (ECL kit, Amersham Biosciences). After three washes each sequentially in wash buffer with and without Tween-20, the membrane was incubated in the ECL detection solution for 1 min and exposed to X-ray film for an appropriate amount of time to obtain a good quality image.
RESULTS
Database mining
The BLAST (Basic Local Alignment Search Tool) algorithm (30) was used to search for RNase P protein subunit homologs in publicly available sequence databases, including partial genome sequences, of different bacteria. Prior to the initiation of our database mining efforts, 39 amino acid sequences of different bacterial RNase P protein subunits were already known and documented in the RNase P database (http://jwbrown.mbio.ncsu.edu/RNaseP/home.html; 31). We have now identified 73 new amino acid sequences corresponding to the protein subunits of RNase P from various bacteria. To analyze conservation of amino acid identity, we used ClustalW to obtain a sequence alignment of the amino acid sequences of 112 bacterial RNase P protein subunits and then manually edited this alignment at certain positions to maximize identity/homology (Fig. 1) (32). The 73 new sequences as well as the entire ClustalW alignment will be deposited in the RNase P database (Table S1 and Fig. S1, Supplementary Material).
Figure 1.
Sequence alignment of the protein subunit of RNase P from various bacteria. Although a ClustalW-based alignment of 112 homologs from 10 different phylogenetic groups of Bacteria was performed, only 14 representative sequences (E.coli, B.subtilis, Corynebacterium diphtheriae, Caulobacter crescentus, Campylobacter jejuni, Neisseria gonorrhoeae, Synechocystis sp. PCC 6803, Treponema denticola, Chlamydophila psittaci, Thermotoga maritima, Porphyromonas gingivalis, Deinococcus radiodurans, Chlorobium tepidum and Dehalococcoides ethenogenes) are shown in this illustration. Two sets of conserved residues at positions which exhibit at least 67% identity in the complete alignment are highlighted in blue and brown. †The demarcation of secondary structure elements is based on the tertiary structure of the protein subunit of B.subtilis RNase P (22).
Although we arbitrarily chose an identity cut-off of 67% while highlighting the conserved residues (Fig. 1), we appreciate that this minimal set of amino acid residues will underestimate conservation of similarity at other sites. We are currently conducting an extensive phylogenetic analysis of the sequences of RNase P from 112 bacteria to examine pre servation of size and polarity of amino acid residues at various positions, evolutionary relationships, lateral gene transfer etc. (E.Braun, M.Jovanovic and V.Gopalan, unpublished data). Nevertheless, we emphasize that the ClustalW alignment pinpoints the residues most likely to be important for function.
Location and possible role of conserved residues in the protein subunit of bacterial RNase P
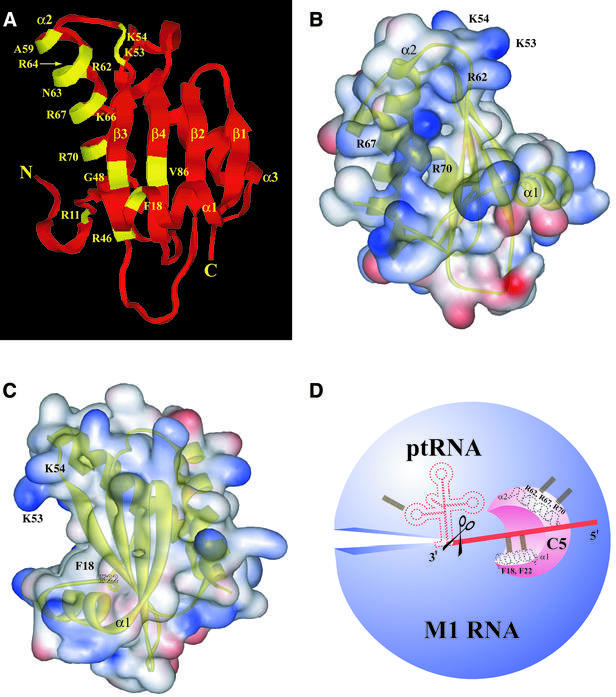
The tertiary structures of the RNase P protein subunit of Bacillus subtilis and Staphylococcus aureus RNase P provide a framework to understand the basis for conservation of amino acid identity at certain positions in the protein subunit of bacterial RNase P (Figs 1 and 2) (22,23). These two structures reveal an overall (identical) αβββαβα topology that includes an uncommon βαβ left-handed crossover connection from β3 to α2 to β4 (Fig. 2A). The positions and putative roles of two sets of conserved residues in RNase P catalysis are discussed below.
Figure 2.
(A) Location of conserved residues in the tertiary structure of the protein subunit of bacterial RNase P. Although the illustrations are based on the tertiary structure of the protein subunit of B.subtilis RNase P, the numbering is based on the E.coli counterpart. (B and C) Web Lab Viewer-generated views of the electrostatic surface potential map of the protein subunit of B.subtilis RNase P in which the RNR motif (B) and the putative ptRNA substrate-binding cleft (C) are depicted. Regions containing basic and acidic residues are depicted in blue and red, respectively. Labels for the residues coincide with the location of the side chains. (D) A schematic depicting RNA recognition sites in the bacterial RNase P holoenzyme. Potential interactions between (i) Arg62, Arg67 and Arg70 in helix α2 of C5 protein and M1 RNA; and (ii) Phe18 and Phe22 in helix α1 of C5 protein and the ptRNA leader sequence, are illustrated. The scissors indicate the site of cleavage on the ptRNA molecule by bacterial RNase P.
The RNR motif in helix 2. The RNR (Arg-Asn-Arg) motif, which refers to the stretch of residues A59xxRN(R/K)xKRxx(R/K)70 in α2, is conserved in nearly all the bacterial RNase P protein subunits (residues in blue in Figure 1; numbering is based on the sequence of the protein subunit of E.coli RNase P). It is noteworthy that Arg62 and Arg67 are invariant, and Arg70 is conserved in 93 of the 112 sequences that we examined. The high conservation of the RNR motif is consistent with various observations that suggest a role for this motif in binding the catalytic RNA moiety and generating the active site in the bacterial RNase P holoenzyme. First, Arg-rich sequences in some RNA-binding proteins do mediate specific RNA recognition (33–37). For example, the human immunodeficiency virus type-1 (HIV-1) Rev peptide utilizes several Arg side chains to make base-specific contacts with its cognate RNA ligand, the Rev response element (RRE) (37). Second, alteration of conserved residues in the RNR motif of C5 protein (the protein subunit of E.coli RNase P) compromised holoenzyme activity in in vitro assays (19). Third, OH·-mediated footprinting studies revealed that some of these conserved residues in C5 protein are proximal to certain conserved nucleotides that are part of the active site in M1 RNA (24,25). Lastly, a DEAD-box protein called Hera (heat-resistant RNA-dependent ATPase) from Thermus thermophilus has the RNR motif (38). As expected from the presence of this motif, Hera hydrolyzes ATP in the presence of M1 RNA. Moreover, deletion of this motif leads to a decrease in M1 RNA-dependent ATPase activity, thus providing indirect evidence that M1 RNA interacts with the RNR motif in Hera (38).
The large central cleft formed by packing of α1 against the β-sheet. Recent studies have provided evidence for the 20 Å-long and 10 Å-wide central cleft in ptRNA substrate binding (Fig. 2C) (12,23). Photo-crosslinking experiments performed with the ptRNAAsp substrate and the B.subtilis RNase P holoenzyme revealed that nucleotides –4 to –7 in the ptRNA leader sequence are proximal to Phe16 (α1), Phe20 (α1), Val32 (β2), Tyr34 (β2), Ser49 (β3) and Ile86 (β4) in the cleft (12) (note that these residues correspond to Phe18, Phe22, Ile33, Ile35 and Thr50, respectively, in the protein subunit of E.coli RNase P; see Fig. 1). We have also recently established that cleft residues in the protein subunit of E.coli RNase P crosslink to the ptRNA leader sequence at identical positions (25). Interestingly, residues in the RNR motif did not crosslink to the ptRNA leader (12).
Rationale for site-directed mutagenesis
Most of the conserved residues identified by the sequence alignment are present in two regions, i.e. the RNR motif in helix α2 and the central cleft of the αβ-sandwich structure of the protein subunit of bacterial RNase P (Fig. 2B and C). Based on the hypothesis that residues in these two regions contribute independently to interactions with M1 RNA and ptRNA, respectively (Fig. 2D), substitution mutants of C5 protein were constructed and the consequent effects on RNase P catalysis in vivo assessed using a genetic approach in E.coli.
A genetic complementation assay to examine the role of conserved residues in the protein subunit of bacterial RNase P
An R46H mutation in C5 protein results in the temperature sensitive (ts) phenotype of E.coli NHY322 cells (28,29). An accumulation of ptRNAs ensues when NHY322 cells are shifted from 30°C (permissive temperature) to 43°C (non-permissive temperature) and, therefore, growth is arrested at 43°C. The observation that increasing the concentration of either M1 RNA or the C5 R46H variant (from plasmid-encoded copies) can rescue the ts phenotype suggests that assembly of the R46H mutant holoenzyme is defective in NHY322 cells at the non-permissive temperature (28). Because transformation of NHY322 with a plasmid bearing the wild type C5 gene (or a functional derivative) can rescue this mutation and abolish the ts phenotype, it is easy to score which mutant derivatives of C5 protein support RNase P catalysis in vivo.
Since our recent studies have indicated that over-expression of C5 protein is toxic to the cell (data not shown), we consider copy number and promoter strength as critical parameters in the design of a complementation assay. We chose pFHC1008′ as the vector for genetic complementation based on the following rationale. The rnpA gene encoding C5 protein has been mapped to the dnaA region of the E.coli genome and is the second of five genes in the rpmH operon (27). Since the natural promoter and translational signals for rnpA (from the rpmH operon) are present in the low copy number plasmid pFHC1008′, it is an ideal vector for the complementation assay. We therefore cloned 23 mutant derivatives in pFHC1008′ and performed genetic complementation analysis to determine which residues in C5 protein are essential for the activity of RNase P in vivo. Note that one-fourth of these derivatives were tested in an earlier study using constructs in which protein expression was driven by the powerful T7 RNA polymerase (19).
The results obtained with the various mutant derivatives of C5 protein are summarized in Figure 3 and Table 3. All the mutants bearing a single substitution at conserved positions in the E.coli RNase P protein subunit (e.g., C5 F18A, R62A and R67A) rescued the NHY322 ts phenotype. These results indicate that individual alterations of conserved amino acid residues do not diminish the ability of C5 protein to support RNase P catalysis in vivo (at least under rich-media growth conditions).
Figure 3.
Results of the genetic complementation assay. The growth phenotypes observed at 30 and 43°C for various mutant derivatives of C5 protein are indicated. The key for the various mutant derivatives is provided in Table 3.
Table 3. Results of genetic complementation assays.
| Construct | Growth at 43°Ca | |
|---|---|---|
| 1 | Wild type | ++ |
| 2 | F18A | ++ |
| 3 | F22A | ++ |
| 4 | R46H | ++ |
| 5 | R57A | ++ |
| 6 | R62A | ++ |
| 7 | K66A | ++ |
| 8 | R67A | ++ |
| 9 | R70A | ++ |
| 10 | F18A/F22A | + |
| 11 | R62A/R67A | – |
| 12 | R62A/R70A | – |
| 13 | R67A/R70A | – |
| 14 | F18A/R62A | + |
| 15 | F18A/K66A | + |
| 16 | F18A/R67A | – |
| 17 | F22A/R62A | + |
| 18 | F22A/K66A | ++ |
| 19 | F22A/R67A | + |
| 20 | R46H/R57A | ++ |
| 21 | R46H/R62A | – |
| 22 | R46H/K66A | – |
| 23 | R46H/R67A | – |
| 24 | R46H/R70A | – |
aIf the complementation of NHY322 (i.e., growth at 43°C) observed with a mutant derivative was comparable to that of wild type C5 protein, the mutant was classified as ++; if the mutant derivative behaved like NHY322 cells transformed with pBR322, it was categorized as –. Mutant derivatives which exhibited intermediate behavior in terms of complementation were defined as +.
Since the E.coli RNase P protein cofactor could tolerate single substitutions of conserved residues, we investigated the possibility that functional redundancy and cooperativity between various conserved residues in the protein subunit of RNase P are required for its function in vivo. We postulated that the simultaneous mutation of two different conserved residues involved in (i) interacting with the ptRNA substrate; (ii) binding to the RNase P RNA subunit; or (iii) stabilizing the protein, might result in a non-functional RNase P holoenzyme complex. To accomplish this goal, we prepared four categories of mutants all of which bear two substitutions (i) both in helix α1 (e.g., C5 F18A/F22A); (ii) both in helix α2 (e.g., C5 R62A/R67A); (iii) one each in helices α1 and α2 (C5 F18A/R67A); and (iv) one in helix α2 and the other always being R46H (e.g., C5 R46H/R62A).
The C5 F18A/F22A mutant was able to weakly rescue the ts phenotype of the NHY322 cells. However, helix α2 double mutants (C5 R62A/R67A, C5 R62A/R70A and C5 R67A/R70), which were expected to bind the RNase P RNA subunit rather weakly, were unable to rescue the ts phenotype. Since Arg62, Arg67 and Arg70 are nearly invariant in the 112 sequences that we examined, this result is perhaps to be expected. Interestingly, the C5 F18A/R67A mutant was also unable to rescue the ts phenotype, indicating that a single insult in substrate binding (F18A, α1) coupled with another defect in RNase P RNA binding (R67A, α2) renders the protein defective in formation of a functional holoenzyme. However, there are some variations to this theme. For instance, while the C5 F18A/K66A rescues the ts phenotype weakly, the C5 F22A/K66A mutant is able to fully rescue the ts phenotype (see Discussion).
Based on the tertiary structures of the protein subunit of B.subtilis and S.aureus RNase P, it appears that Arg46 might be involved in a salt bridge with Asp84 in C5 protein and thus impart structural stability to the protein (23). This postulate helps rationalize the ts phenotype rendered by the C5 R46H mutation in NHY322 cells. Since a stable structural scaffold is vital for the correct positioning of the RNA-binding domains in C5 protein, we inquired into the effects of mutations introduced in conjunction with R46H. When the C5 R46H mutation is coupled with R62A, K66A, R67A or R70A, no complementation is observed at the non-permissive temperature (Fig. 3 and Table 3). Therefore, the combination of a mutation of a conserved residue in α2 (e.g., R62A, which potentially weakens holoenzyme assembly), with another tertiary fold-destabilizing mutation (e.g., R46H, which might weaken the structural core and indirectly affect holoenzyme formation) results in a non-functional protein. As a control for these experiments, we constructed C5 R46H/R57A since Arg57 is not a conserved residue and yet proximal to the RNR motif that we had mutated. We therefore postulated that C5 R46H/R57A will be functional in vivo since mutating Arg57 was not expected to adversely affect M1 RNA binding. Indeed, we observed robust genetic complementation with C5 R46H/R57A (Fig. 3).
Western blot analysis to examine levels of expression in vivo
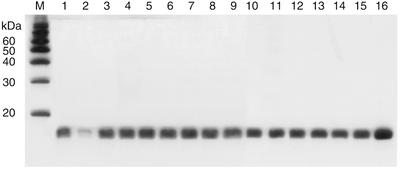
To ensure that differences in the complementation ability of the mutant derivatives of C5 protein did not arise from unforeseen variations in their levels of expression, we performed western blot analysis with crude extracts of the various transformants expressing the weakly- or null-complementing mutant derivatives. By using affinity-purified polyclonal antisera raised against C5 protein, we were able to confirm nearly identical levels of expression for the wild type as well as various mutant derivatives (Fig. 4). Although there is a faint signal from the chromosome-encoded C5 R46H protein in NHY322 cells, our data unambiguously establish that the signal in the western analysis is largely due to protein expressed from the plasmid-encoded copies (Fig. 4, compare lanes 1 and 2).
Figure 4.
Western blot analysis to examine the levels of expression of various mutant derivatives of C5 protein in E.coli NHY322 cells. Lane M, magic markers/size standards (Invitrogen); lane 1, wild type C5 protein; lane 2, pBR322 (vector control); lane 3, C5 R46H/R62A; lane 4, C5 R46H/K66A; lane 5, C5 R46H/R67A; lane 6, C5 R46H/R70A; lane 7, C5 F18A/F22A; lane 8, C5 F22A/R62A; lane 9, C5 F22A/R67A; lane 10, C5 F18A/R62A; lane 11, C5 F18A/K66A; lane 12, C5 F18A/R67A; lane 13, C5 R62A/R67A; lane 14, C5 R62A/R70A; lane 15, C5 R67A/R70A; and lane 16, purified C5 protein (20 ng).
DISCUSSION
Cooperativity among conserved residues is essential for the function of C5 protein
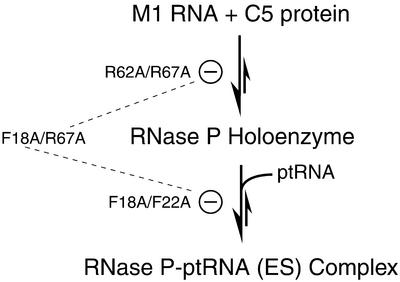
Our genetic complementation data support the notion that cooperativity and functional redundancy among various conserved residues in the protein subunit of RNase P are essential for its function in vivo. Our findings are easily interpreted on the basis of a working model for the functioning of the protein subunit of bacterial RNase P (Fig. 2D and Scheme 1).
Scheme 1.
Although Arg62, Arg67 and Arg70 are clearly some of the most invariant residues in all 112 bacterial RNase P protein subunits that we examined, their individual mutation to Ala was not detrimental for the function of C5 protein in vivo (Fig. 3). However, when two mutations were introduced in helix α2 (C5 R62A/R67A or R62A/R70A or R67A/R70A), it resulted in a complete loss of activity in vivo (Fig. 3). It is possible that the interaction of the bacterial RNase P RNA with its protein cofactor is mediated by multiple RNA–protein contacts between basic residues in helix α2 and conserved nucleotides in the RNA subunit. Moreover, a minimal threshold of contacts is likely required for assembling a stable holoenzyme complex. Violation of this simple requisite might explain the inability of double mutants of C5 protein, bearing substitutions in any two of the three conserved Arg residues in helix α2, to participate in assembly of a functional RNase P holoenzyme.
Although the specific binding of single-stranded (ss) RNA to the surface of β-sheets is a recurring theme in the structures of some RNP complexes (34–36,39), the absence of sequence homology in the leader sequences of ptRNAs warrants comment on how these sequences might bind specifically to the cleft in the protein subunit of RNase P where there is a paucity of phylogenetically conserved residues in the cleft. Spitzfaden et al. (23) examined perturbations in the 1H and 15N chemical shifts upon addition of small, ss-oligonucleotides (e.g., A10) to the protein subunit of S.aureus RNase P (23). Even though these small homo-oligomers are not akin to the ss-leader sequences that are heterogeneous in length and composition in the numerous ptRNA substrates of RNase P, the NMR data provide valuable insights (12,23). For instance, the titration of specific oligoribonucleotides led to chemical shift differences in residues in β1, β2 and α1 that were already proven to be proximal to the substrate by crosslinking (23). Modest chemical shift perturbations were also observed in the highly conserved residues Lys53 and Lys54, which are present in the loop connecting β3 to α2 at the upper boundary of the ptRNA-binding site (Fig. 2B). Taken together, Spitzfaden et al. (23) postulated that recognition of leader sequences might involve the solvent-exposed surface of the β-sheet. While there are few conserved amino acid residues in the cleft, there is considerable potential for exploiting multiple weak interactions (e.g., hydrogen bonding, van der Waals and ionic/hydrophobic interactions) with the unpaired bases that are fully exposed in the ptRNA leader sequence. Although the role of the cleft in binding ptRNA leader sequences through an ensemble of non-covalent interactions is likely to be conserved in the protein subunits of all bacterial RNase P, the types of interactions are likely to be diverse. For instance, in C5 protein, both Phe18 and Phe22 are possibly involved in stacking interactions with the ptRNA leader sequence (although the former, a highly conserved residue, is likely to play a more vital role in this regard). Consistent with this expectation, C5 F18A/F22A mutant is able to only weakly rescue the ts phenotype of the NHY322 cells in contrast to the wild type-like complementation ability of C5 F18A and C5 F22A (Fig. 3). The presence of residues such as Lys53 and Lys54, which could facilitate electrostatic interactions with the substrate, in C5 F18A/F22A might explain why this mutant is not entirely defective in substrate binding. Its turnover rate, despite poor substrate binding, must suffice to weakly rescue the ts phenotype.
The weak complementation exhibited by C5 F18A/F22A is also somewhat intriguing because of an earlier study in which purified C5 F18A/F22A when reconstituted with M1 RNA was reported to exhibit merely 3% of the activity of the wild type RNase P holoenzyme at 43°C (19). What might account for this apparent discrepancy between in vitro and in vivo results? First, alterations in Phe18 and Phe22 might weaken the hydrophobic interface between helix α1 and the central β-sheet and cause structural defects at high temperatures in vitro but such defects in protein folding (and therefore holoenzyme assembly) are alleviated by unknown mechanisms in vivo. Second, a fraction of the wild type holoenzyme activity might be sufficient to support minimal cell viability. Further experimentation is required to shed light in this regard.
The synergism between two distinct regions of C5 protein that is required to facilitate E.coli RNase P catalysis is illustrated by our results with mutant derivatives in which one residue has been simultaneously mutated in helices α1 and α2. Loss of function results even when two separate mutations are introduced in seemingly disparate regions. While C5 F18A, F22A, R62A, K66A, R67A and R70A are able to support robust complementation, the double mutants such as C5 F18A/K66A and C5 F18A/R67A are moderately functional and non-functional, respectively. We rationalize this additive effect on the basis that single mutations in helix α2 (either K66A or R67A) which weaken M1 RNA binding would result in loss of activity but perhaps not be lethal in vivo since the assembled holoenzyme, albeit in diminished levels compared to wild type, is able to bind the ptRNA substrate as effectively as the wild type and support a high catalytic turnover. However, when an α2 mutation (e.g., R67A) is coupled with an additional defect in substrate binding induced by an α1 mutation (e.g., F18A), the mutant (C5 F18A/R67A) holoenzyme becomes non-functional due to two independent phenotypic alterations (Scheme 1). Not only is there likely to be a defect in holoenzyme assembly but the mutant (C5 F18A/R67A) holoenzyme assembled is unable to bind the ptRNA substrate as tightly as its wild type counterpart. This type of reasoning permits us to interpret the results observed with the various double mutants bearing a substitution in both α1 and α2 (Fig. 3, Table 3). However, there are some exceptions to this interpretation likely reflective of the dispensable or indispensable nature of the mutated residues for RNase P catalysis. For example, C5 F22A/K66A is able to rescue the ts phenotype of NHY322 cells to near wild type levels presumably due to its mutations being in positions that are not vital to protein function. Such a speculation is strengthened by the fact that Phe22 (48%) and Lys66 (78%) are only semi-conserved in contrast to the near absolute invariance of Phe18 (88%), Arg62 (100%) and Arg67 (100%).
Dual RNA-binding specificity in the protein subunit of bacterial RNase P
RNA-binding proteins have minimally been classified as either groove binders or proteins that use a β-sheet to create large surfaces for RNA recognition (33–37). These two mechanisms underscore the need to accomplish recognition of highly structured RNA ligands as well as the sequence-dependent discrimination required to specifically bind ssRNAs. The groove binders position a secondary structure element (e.g., α-helix, β-ribbon) in the cognate RNA ligand, particularly in widened major grooves that deviate from the typical A-form helix due to the presence of internal loops or bulges. The proteins that employ a β-sheet surface can simultaneously accomplish multiple RNA–protein contacts and thereby permit recognition of various functional groups on the bases present in hairpin loops or single-stranded regions (34,35). Could both these mechanisms be employed concomitantly by an RNA-binding protein to recognize different structural features in two distinct RNA ligands?
During RNase P catalysis, the bacterial RNase P protein subunit probably employs different amino acid residues to mediate concomitant (but distinct) interactions with the catalytic RNA moiety and the ptRNA substrate (Fig. 2D). By binding the distal part of the leader sequence (positions –4 to –7) of a ptRNA substrate and simultaneously interacting with conserved nucleotides in the catalytic RNA subunit that are essential for ptRNA processing, the protein could both enhance the affinity for a ptRNA and modestly increase the rate of chemical cleavage (10,11,25,26). Specifically, the conserved basic residues in helix α2 (i.e., the RNR motif) could bind a tertiary structural motif in M1 RNA, while the cleft (formed by the β-sheet and α1) engages in multiple weak interactions with the unstructured ss-leaders of ptRNA substrates.
Although there is growing support for the dual roles of C5 protein in RNase P catalysis, there is a definite need for experimental evidence such as a direct demonstration with RNA-binding assays that alterations in the RNR motif weaken binding to the RNA moiety. This approach will also help decipher the molecular determinants in the RNR motif that contribute to the favorable binding energy for assembly of the holoenzyme (–13.3 kcal/mol for E.coli RNase P) (40). Also, the participation of the cleft residues in substrate binding needs to be further delineated using crosslinking and kinetic studies. Such experiments are currently in progress.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Cecilia Guerrier-Takada for guidance during the initial stages of this study. This research was supported by grants from the National Science Foundation (MCB 0091081), American Heart Association (Southern Ohio Valley affiliate) and Message Pharmaceuticals, Inc., to V.G. and from the National Institutes of Health (GM19422) to S.A.
REFERENCES
- 1.Altman S. and Kirsebom,L.A. (1999) Ribonuclease P. In Gesteland,R.F., Cech,T. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 351–380.
- 2.Harris M.E., Frank,D. and Pace,N.R. (1998) Structure and catalytic function of the bacterial ribonuclease P ribozyme. In Simons,R.W. and Grunberg-Manago,M. (eds), RNA structure and function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 309–337.
- 3.Guerrier-Takada C., Gardiner,K., Marsh,T., Pace,N.R. and Altman,S. (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell, 35, 849–857. [DOI] [PubMed] [Google Scholar]
- 4.Pannucci J.A., Haas,E.S., Hall,T. and Brown,J.W. (1999) RNase P RNAs from Archaea are catalytically active. Proc. Natl Acad. Sci. USA, 96, 7803–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schedl P. and Primakoff,P. (1973) Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc. Natl Acad. Sci. USA, 70, 2091–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakano H., Yamada,S., Ikemura,T., Shimura,Y. and Ozaki,H. (1974) Temperature-sensitive mutants of Escherichia coli for tRNA biosynthesis. Nucleic Acids Res., 1, 355–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peck-Miller K.A. and Altman,S. (1991) Kinetics of the processing of the precursor to 4.5 S RNA, a naturally occurring substrate for RNase P from Escherichia coli. J. Mol. Biol., 221, 1–5. [DOI] [PubMed] [Google Scholar]
- 8.Tallsjo A. and Kirsebom,L.A. (1993) Product release is a rate-limiting step during cleavage by the catalytic RNA subunit of Escherichia coli RNase P. Nucleic Acids Res., 21, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J.J., Kilani,A.F., Zhan,X., Altman,S. and Liu,F. (1997) The protein cofactor allows the sequence of an RNase P ribozyme to diversify by maintaining the catalytically active structure of the enzyme. RNA, 3, 613–623. [PMC free article] [PubMed] [Google Scholar]
- 10.Kurz J.C., Niranjanakumari,S. and Fierke,C.A. (1998) Protein component of B. subtilis RNase P specifically enhances the affinity for precursor tRNAAsp. Biochemistry, 37, 2393–2400. [DOI] [PubMed] [Google Scholar]
- 11.Crary S.M., Niranjanakumari,S. and Fierke,C.A. (1998) The protein component of Bacillus subtilis ribonuclease P increases catalytic efficiency by enhancing interactions with the 5′ leader sequence of pre-tRNAAsp. Biochemistry, 37, 9409–9416. [DOI] [PubMed] [Google Scholar]
- 12.Niranjanakumari S., Stams,T., Crary,S., Christianson,D.W. and Fierke,C.A. (1998) Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc. Natl Acad. Sci. USA, 95, 15212–15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurz J.C. and Fierke,C.A. (2002) The affinity of magnesium binding sites in the Bacillus subtilis RNase P-pre-tRNA complex is enhanced by the protein subunit. Biochemistry, 41, 9545–9558. [DOI] [PubMed] [Google Scholar]
- 14.Hardt W.D., Warnecke,J.M., Erdmann,V.A. and Hartmann,R.K. (1995) Rp-phosphoro-thioate modifications in RNase P RNA that interfere with tRNA binding. EMBO J., 12, 2935–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris M.E. and Pace,N.R. (1995) Identification of phosphates involved in catalysis by the ribozyme RNase P RNA. RNA, 1, 210–220. [PMC free article] [PubMed] [Google Scholar]
- 16.Siew D., Zahler,N.H., Cassano,A.G., Strobel,S.A. and Harris,M.E. (1999) Identification of adenosine functional groups involved in substrate binding by the ribonuclease P ribozyme. Biochemistry, 38, 1873–1883. [DOI] [PubMed] [Google Scholar]
- 17.Kufel J. and Kirsebom,L.A. (1998) The P-15 loop of Escherichia coli RNase P RNA is an autonomous divalent metal ion binding domain. RNA, 4, 777–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopalan V., Golbik,R., Schreiber,G., Fersht,A.R. and Altman,S. (1997) Fluorescence properties of a tryptophan residue in an aromatic core of the protein subunit of ribonuclease P from Escherichia coli. J. Mol. Biol., 267, 765–769. [DOI] [PubMed] [Google Scholar]
- 19.Gopalan V., Baxevanis,A., Landsman,D. and Altman,S. (1997) Functional analysis of conserved amino acid residues in the protein subunit of ribonuclease P from Escherichia coli. J. Mol. Biol., 267, 818–829. [DOI] [PubMed] [Google Scholar]
- 20.Harris M.E., Kazantsev,A., Chen,J.L. and Pace,N.R. (1997) Analysis of the tertiary structure of the ribonuclease P ribozyme–substrate complex by site-specific photoaffinity crosslinking. RNA, 3, 561–574. [PMC free article] [PubMed] [Google Scholar]
- 21.Massire C., Jaeger,L. and Westhof,E. (1998) Derivation of the three-dimensional architecture of bacterial RNase P RNAs from comparative sequence analysis. J. Mol. Biol., 279, 773–793. [DOI] [PubMed] [Google Scholar]
- 22.Stams T., Niranjanakumari,S., Fierke,C. and Christianson,D.W. (1998) Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science, 280, 752–755. [DOI] [PubMed] [Google Scholar]
- 23.Spitzfaden C., Nicholson,N., Jones,J.J., Guth,S., Lehr,R., Prescott,C.D., Hegg,L.A. and Eggleston,D.S. (2000) The structure of ribonuclease P protein from Staphylococcus aureus reveals a unique binding site for single-stranded RNA. J. Mol. Biol., 295, 105–115. [DOI] [PubMed] [Google Scholar]
- 24.Biswas R., Ledman,D., Fox,R.O., Altman,S. and Gopalan,V. (2000) Mapping RNA–protein interactions in ribonuclease P from Escherichia coli using disulfide-linked EDTA-Fe. J. Mol. Biol., 296, 19–31. [DOI] [PubMed] [Google Scholar]
- 25.Tsai H-.Y., Masquida,B., Biswas,R., Westhof,E. and Gopalan,V. (2003) Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenyzme. J. Mol Biol., in press. [DOI] [PubMed] [Google Scholar]
- 26.Kurz J.C. and Fierke,C.A. (2000) Ribonuclease P: a ribonucleoprotein enzyme. Curr. Opin. Chem. Biol., 4, 553–558. [DOI] [PubMed] [Google Scholar]
- 27.Hansen F.G., Hansen,E.B. and Atlung,T. (1985) Physical mapping and nucleotide sequence of the rnpA gene that encodes the protein component of ribonuclease P in E. coli. Gene, 38, 85–93. [DOI] [PubMed] [Google Scholar]
- 28.Baer M.F., Wesolowski,D. and Altman,S. (1989) Characterization in vitro of the defect in a temperature-sensitive mutant of the protein subunit of RNase P from Escherichia coli. J. Bacteriol., 171, 6862–6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirsebom L.A., Baer,M.F. and Altman,S. (1988) Differential effects of mutations in the RNA and protein moieties of RNase P on the efficiency of suppression by various tRNA suppressors. J. Mol. Biol., 204, 879–888. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown J.W. (1999) The Ribonuclease P Database. Nucleic Acids Res., 27, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- 34.Nagai K. (1996) RNA–protein complexes. Curr. Opin. Struct. Biol., 6, 53–61. [DOI] [PubMed] [Google Scholar]
- 35.Draper D.E. (1999) Themes in RNA–protein recognition. J. Mol. Biol., 293, 255–275. [DOI] [PubMed] [Google Scholar]
- 36.Brodersen D.E., Clemons,W.M.,Jr, Carter,A.P., Wimberly,B.T. and Ramakrishnan,V. (2002) Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol., 316, 725–768. [DOI] [PubMed] [Google Scholar]
- 37.Battiste J.L., Mao,H., Rao,N.S., Tan,R., Muhandiram,D.R., Kay,L.E., Frankel,A.D. and Williamson,J.R. (1996) Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science, 273, 1547–1551. [DOI] [PubMed] [Google Scholar]
- 38.Morlang S., Weglohner,W. and Franceschi,F. (1999) Hera from Thermus thermophilus: the first thermostable DEAD-box helicase with an RNase P protein motif. J. Mol. Biol., 294, 795–805. [DOI] [PubMed] [Google Scholar]
- 39.Handa N., Nureki,O., Kurimoto,K., Kim,I., Sakamoto,H., Shimura,Y., Muto,Y. and Yokoyama,S. (1999) Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature, 398, 579–585. [DOI] [PubMed] [Google Scholar]
- 40.Talbot S.J. and Altman,S. (1993) Kinetic and thermodynamic analysis of RNA–protein interactions in the RNase holoenzyme from Escherichia coli. Biochemistry, 33, 1406–1411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.