Abstract
Objective: Health information retrieval (HIR) on the Internet has become an important practice for millions of people, many of whom have problems forming effective queries. We have developed and evaluated a tool to assist people in health-related query formation.
Design: We developed the Health Information Query Assistant (HIQuA) system. The system suggests alternative/additional query terms related to the user's initial query that can be used as building blocks to construct a better, more specific query. The recommended terms are selected according to their semantic distance from the original query, which is calculated on the basis of concept co-occurrences in medical literature and log data as well as semantic relations in medical vocabularies.
Measurements: An evaluation of the HIQuA system was conducted and a total of 213 subjects participated in the study. The subjects were randomized into 2 groups. One group was given query recommendations and the other was not. Each subject performed HIR for both a predefined and a self-defined task.
Results: The study showed that providing HIQuA recommendations resulted in statistically significantly higher rates of successful queries (odds ratio = 1.66, 95% confidence interval = 1.16–2.38), although no statistically significant impact on user satisfaction or the users' ability to accomplish the predefined retrieval task was found.
Conclusion: Providing semantic-distance-based query recommendations can help consumers with query formation during HIR.
Introduction
Health information retrieval (HIR) on the Internet has become a common and important practice for millions of people.1 Health consumers of varying backgrounds perform HIR for themselves as well as for friends and family, and to merely satisfy their own curiosity, as well as to make medical decisions. Because of the vast amount of information available and the ad hoc nature of information gathering by consumers, HIR is not always efficient and effective.
Query formation is a major aspect of consumer HIR that is in need of improvement. One observation study has shown that consumers' HIR queries tend to be too short and general.2 Although current search engines are fairly good at retrieving appropriate information, they still depend on the queries to set the correct retrieval goal. If queries do not reflect users' specific information needs, they will lead to results that do not address those information needs. For instance, we once interviewed a user who wanted to know “Are there natural substitutes for the hormone replacement therapy Prempro?” One of the queries this person typed in was “natural hrt.” It was not surprising that the query failed to yield the correct answer.
Internet queries tend to be short regardless of the search domain: users do not type more than 2 or 3 query terms on average.3,4 When searching for health information though, many consumers' limited knowledge of medical vocabulary contributes to the construction of simplistic queries. For instance, when a consumer we interviewed could not remember the exact name of a drug, he had to use the more general query term “antidepressant.”
To help consumers better articulate their health information needs, we have developed and evaluated a novel system, the Health Information Query Assistant (HIQuA), to recommend alternative/additional query terms. The recommended terms are deemed to be closely related to the initial query and can be used as building blocks to construct more accurate and specific queries. By relying on user recognition instead of recall, our tool attempts to make query formation easier.
Background and Significance
Consumer Health Queries
We have interviewed consumers and analyzed log data of health-related consumer queries in some of our previous work. Three findings from our previous studies are: (1) consumer queries are short (usually no more than 1 to 2 words on average),5 (2) most terms in consumer queries can be mapped to concepts in medical vocabularies,5 and (3) the terms and concepts consumers use often do not accurately reflect their information needs and do not form effective queries.2,6 The problem of overly general queries and ineffective search strategies in consumer HIR has also been reported by Eysenbach et al.7
In HIR, many queries are not only short, but also not specific enough to describe the information needs. One reason is that consumers, unlike clinicians or research scientists, have limited knowledge of medicine. As a result, they require more assistance in query construction.
The work by Fredin et al. also sheds some light on this issue.8 They suggest that Internet information retrieval is an iterative process, during which the information retrieval goals are constantly refined and revised. Consequently, queries need to be refined and revised. Our system, if successful, can make the process of refinement and revision more convenient for users.
Query Modification
Researchers have developed many techniques to improve information retrieval performance, one of which is query expansion, i.e., adding additional terms to the original query.9 Typical sources of additional terms are thesauri or the retrieved documents themselves. A thesaurus may offer synonyms, antonyms, descendents, or other related terms. Retrieval feedback methods analyze the “best” returned documents, as determined by the user or by some ranking algorithm. Co-occurrence data of the query and other terms in certain data sets, for instance, log data that records the search behavior of previous users, have also become sources for expansion terms.10,11 Not all methods automatically add the related terms to the original query. In interactive systems, related terms are suggested to a user.12 The user may ignore or use the suggested terms to expand or replace the original queries.
In recent literature, variations of the basic query expansion techniques have been reported. Some techniques combine different expansion methods, for example, combining retrieval feedback with co-occurrence information13 or combining several thesauri.14 Some have explored the fuzzy nature of relatedness between terms or concepts.15,16,17 The fact that Web users are often not good at constructing queries has led to more studies on interactive methods, while the availability of large query logs from Web sites has provided a rich source for mining term and concept relations.18,19,20,21
In biomedical informatics, there have been a number of applications that have used query expansion techniques for searching literature.22,23,24,25,26,27 The sources of expansion terms have been medical vocabularies, retrieval feedback, and co-occurrence data. A set of methods has also been developed to transform natural language questions or queries into computer-friendly representations such as Boolean expressions or conceptual graphs.28,29,30,31 (Similar studies have been carried out on searching patient medical records,32,33 which we will not elaborate on here.)
For retrieving consumer health content, previous studies have explored query expansion, reformulation, and suggestion. For example, the study of Gobel et al.34 added broader and narrower concepts automatically to user queries according to entries in the MeSH Thesaurus. McCray and colleagues' study35 utilized a variety of strategies such as synonym expansion, spelling correction, and suggesting more general queries when no results are found, among others. Finally, we have conducted experiments,36 as have Patrick et al.,37 that examined the impact of reformulating consumer queries with professional synonyms. Of all the studies mentioned, however, none explored concept relations beyond synonymy or hierarchy.
When dealing with consumer HIR, the main query expansion approaches have pros and cons. Automated expansion that is based on a thesaurus or on co-occurrence data does not put any extra burden on the user; however, it can end up being even less effective than the original query if the original query does not represent the user's search goal well. Retrieval feedback methods suffer from the same problem even though they may rely on some user participation before automatically generating the new query. The strength of the retrieval feedback method is its ability to learn from examples. Query suggestion methods, on the other hand, require greater user participation, which can be viewed as extra work for users, but the benefit of these methods is that even if the initial query is poorly constructed, the user is empowered to articulate his or her needs and refine his or her queries. This article describes a query suggestion method.
For identifying related terms to suggest to users, we considered several sources that have been exploited by previous studies:
Usage patterns of consumers: Forming recommendations from consumers' usage patterns has the advantage of reflecting the semantic distance among concepts in the consumers' mental models. The downside is that it also relies on the extent of the consumers' knowledge and their recall abilities, which could be quite limited.
Controlled medical vocabularies: In medicine there is a great wealth of available semantic knowledge embedded in controlled vocabularies, so making thesaurus-based suggestions is feasible and common. The disadvantage of relying on a thesaurus is that it may sometimes lead to recommendations that are unrecognizable to a consumer. Thesaurus knowledge also typically focuses on definitional and hierarchical relations. For instance, “pneumonia” is an “infectious disease” and is a “lung disorder.” These are important fundamental relations; however, other types of relations (e.g., the relations between medications and diseases) are not extensively included in medical vocabularies.
Concept co-occurrence in medical literature: This provides another source to estimate the semantic relatedness of concepts. Medical literature reflects up-to-date knowledge in the health domain. Past research has shown that a high frequency of concepts co-occurring in literature is a decent indicator of a close semantic distance between them.38,39 Generally speaking, its coverage of relations is more comprehensive than manually constructed vocabularies, but less reliable.
To provide HIR users with recommendations that reflect their mental models while avoiding being limited by users' recall abilities, we decided to combine these 3 sources. As some other research has done, our method treats semantic distance between concepts as a fuzzy concept.15,16,17 Our method for estimating semantic distance and combining sources was designed specifically for the consumer HIR context and consequently differs from other published methods.
Design
Overview
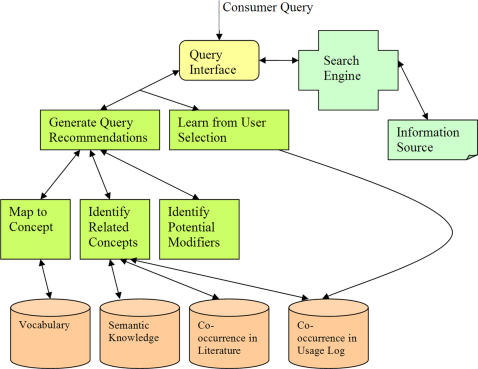
The main function of the system is to identify medical concepts that are semantically related to a user's initial query and recommend them to the user. The semantic distance among concepts is calculated based on their co-occurrence frequency in query log data and in medical literature, and on known semantic relationships in the medical domain. Topic-specific modifiers are also recommended for concepts of several common semantic types. In addition, the system continuously learns from user selections in order to improve future performance. ▶ shows the overall design of the system.
Figure 1.
Overall design of HIQuA. The system suggests alternative/additional query terms related to the user's initial query, which can be used as building blocks to construct a better query.
Distance-Based Query Recommendations
To provide a query recommendation, the system first maps the query into 1 or more concepts and then identifies concepts that are related to those concepts.
Mapping to Concepts
In HIQuA, semantic relations and semantic distances exist among concepts, not character strings. Query strings are thus first mapped to concepts, which are defined by the Unified Medical Language System (UMLS).40 Each initial query may be mapped to 1 or more concepts.
If the entire query string cannot be mapped to one UMLS concept, HIQuA attempts to find concepts with names that match the longest possible substrings of submitted search terms. The search string “thrombosis attack coronary,” for instance, will return two concepts named “Thrombosis,” and “Heart Attack.” (“Heart Attack” is the preferred name in the UMLS for the concept to which the string “attack coronary” maps.)
On the other hand, a single string may sometimes not only match a concept without being broken into substrings, but can even match more than one concept. The word “cancer,” for instance, maps to two UMLS concepts: “Malignant Neoplasms” and “Cancer Genus.” (In biological taxonomy, “cancer” is a genus of rock crabs.) Of these two, “Malignant Neoplasms” is clearly the more appropriate concept to match to in our application. Because queries are short and provide little context for disambiguation, we are only able to disambiguate between concepts based on the following factors: (1) whether the matched term is considered a suppressible name for the concept by the UMLS (according to the UMLA FAQ, certain names are “suppressible” if they have “invalid face meanings or are otherwise problematic” [from http://umlsinfo.nlm.nih.gov/synonym3.html]); (2) the editing distance (i.e., the number of editing operations—deletions, insertions, and substitutions—necessary to make two strings identical) between the term and the preferred name of the concept (the shorter the better); (3) the number of vocabulary sources containing the concept (suggesting a common rather than a rare semantic); and (4) whether the term is marked in UMLS with “ < 1>” (indicating that it is the primary meaning of a term). The limitations of our mapping technique are discussed in the Limitations section.
Identifying Related Concepts
The recommended concepts should be related to the initial query concept(s); in other words, they should have a short semantic distance from the initial concept(s). For estimating semantic distance we used three sources: (1) the semantic relations of concepts in medical vocabularies, (2) co-occurrence of concepts in consumer HIR sessions, and (3) co-occurrence of concepts in medical literature.
Medical vocabularies are a reliable source of known semantic relations between concepts because they have been constructed and reviewed by domain experts. We used the UMLS Metathesaurus relationship (MRREL) table as our medical vocabulary source.
To complement the medical vocabularies, we used co-occurrence data of concepts in medical literature. The UMLS Metathesaurus co-occurrence (MRCOC) table was used as our literature co-occurrence source.
The third source of semantic distance is the co-occurrence of concepts in consumer HIR sessions. The underlying relations among these co-occurring concepts could be co-occurring symptoms of a disease, a symptom and its location, a medication and a disease for which it has been prescribed, or somewhat obscure connections, such as the relationship between “diet” and “food allergies.” Of course concept co-occurrence in search sessions could be incidental, but a high frequency of co-occurrence is unlikely to be due to chance. The query log of a consumer health information Web site, MedlinePlus,41 was obtained from the National Library of Medicine and used as the co-occurrence source. This log contained 12 million queries that we split into sessions based on the IP address and time of the queries: queries from the same IP address within 5 minutes of each other were considered to be in the same session. Co-occurrence was calculated based on concepts that appeared together in the same sessions.
For each concept mapped to the initial query, HIQuA extracts related concepts from the three sources described above. The first source, the MRREL table, lists semantic relationships between concept, such as “parent,” “child,” “synonymous,” and “similar to,” among others. The second source, the MRCOC table, lists pairs of concepts that have appeared together in medical literature, along with the frequency of their co-occurrence. The final source, the query log table (QRYLOG), lists pairs of concepts that users have often performed searches on in conjunction with each other. In addition, this table is continuously updated by HIQuA as it continues to gain information about users' search habits.
We set no limit on the number of related concepts. We did, however, establish the following exclusion criteria in order to eliminate inscrutable relations and accidental co-occurrences (see ▶).
Table 1.
Concept Exclusion Criteria for the Different Sources
| Source | Elimination Rule |
|---|---|
| MRREL | Relationship type = “can be qualified by”* |
| MRREL | Relationship type = “is an allowed qualifier for”* |
| MRCOC | Number of co-occurrence < 3 |
| MRCOC | Co-occurrence type = “qualification”* |
| QRYLOG | Number of co-occurrence < 3 |
| ALL Sources | Related concept name > 35 characters |
| ALL Sources | Related concept is a “stop concept”† |
The qualification relationship appears to be both overly broad and inscrutable.
We maintained a list of “stop concepts” that we have found to be unusually unhelpful as query concepts. Examples from this “stop concept” list include the concepts with the names “Preposition For,” “Of,” and “With.”
Because relevance is a fuzzy concept, we used fuzzy logic methods to represent the semantic distance among concepts based on each source and then combined the three distances. Instead of determining if two concepts are related, or what the chances are that they are related, the system calculates to what degree or how closely they can be viewed as related. The calculation of degree of relevance in our system is frequency based: the frequency of occurrence of a relation in various medical vocabularies, or the frequency of concept co-occurrence in literature or log data. In the case of medical vocabularies, consideration is also given to the particular type of the relation. For instance, “parent-child” relations are considered to be more important than others.
Once information on related concepts is retrieved from the three tables, their relevance to the query concept is calculated. The method of estimating distance differs slightly by information source.
A frequency score is assigned to each unique pair of concepts from a source: Score(Cx, Cy, s), Cx is the query concept, Cy is a related concepts and s is the source. For relations derived from MRCOC and QRYLOG, the frequency score of a relation is simply the frequency of co-occurrence of the 2 concepts in that relation. For these 2 sources, Score(Cx, Cy, s) = Score(Cy, Cx, s).
For relations derived from MRREL, the frequency score is the weighted co-occurrence of the two concepts in the table. Because relationships in UMLS are derived from many different sources, two concepts can appear as a pair several different times. The pair “heart attack” and “ischemic heart disease” appears 14 times, for instance, under four different relationships (parent, sibling, broader, and other). Because we consider the child relationship especially relevant, a weight of two is given each time a concept is identified as a child of the query concept. When a parent-child relationship is involved, Score(Cx, Cy, s)! = Score(Cy, Cx, s).
Then, for each related concept a fuzzy score (0 to 1) is computed, representing the degree of relatedness between that concept and the initial concept. The fuzzy membership, i, for each set of concepts from a source is defined as:
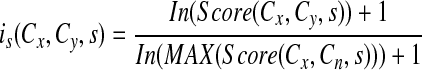 |
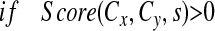 |
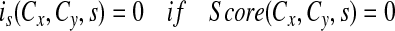 |
Cn is any concept that is found to be related to Cx based on one of the sources. The log transformation is a common technique used to normalize highly skewed data; we found the distribution of frequency scores to be highly skewed.
Using A, B, and C to represent the three sources, we combined the degrees of relevance following two fuzzy rules:
If two concepts are relevant in A and B and C, then they are relevant. (Rule 1)
If two concepts are relevant in A or B or C, then they are relevant. (Rule 2)
For Rule 1, fuzzy intersection of the three fuzzy sets is computed. For Rule 2, fuzzy union of the three fuzzy sets is computed.
The traditional definition of the fuzzy union has been the maximum function, and the traditional definition of the fuzzy intersection has been the minimum function. These functions yield rather crisp results, and when more than two fuzzy sets are involved they fail to take into account those membership values between the maximum and minimum.9 So we have used the smoother algebraic sum and algebraic product functions to compute the fuzzy union and fuzzy intersection, respectively. The membership of an element i in the intersection of three fuzzy sets, A, B, and C, is defined as the product of i's degree of membership in A and i's degree of membership in B and i's degree of membership in C:
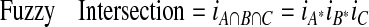 |
The fuzzy union is accordingly defined as the algebraic sum (i.e., the simple sum minus the algebraic products):
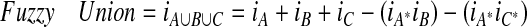 |
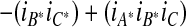 |
When translating the membership value into semantic distance, intersection is given more weight:
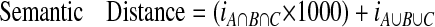 |
The top n concepts with the shortest semantic distance from a query concept are considered related to it.
To provide an example of the calculation of semantic distance, ▶ shows the top 10 related concepts for “shingles” from each source. Table 3 (Table 3 is available as a JAMIA online supplement at www.jamia.org) shows the top 10 concepts with the shortest semantic distances to “shingles” and how they were calculated to take into consideration information from each source. The list of query suggestions (i.e., Varicella, Herpes Zoster Ophthalmicus, Pneumonia, Pregnancy, Neuralgia, Chicken Pox, Ramsey-Hunt Syndrome, Herpes Simplex, Antiviral, Varicella Encephalitis) that would be displayed to the user is the list from Table 3. They are ordered according to the final score they achieved after computing their scores from each of the three sources. Concepts that either appear in all three lists or have an extremely high score in just one of the lists are likely to make it into the final list.
Table 2.
Top 10 Concepts Related to “Shingles” from 3 Sources
| Query Term = Shingles | |||||
|---|---|---|---|---|---|
| Weight of the semantic relationship with the top 10 concepts from the medical vocabularies (Methathesaurus Relationship table, MRREL) | Frequency of co-occurrence with the top 10 concepts from medical literature (Methathesaurus Co-occurrence table, MRCOC) | Frequency of co-occurrence of the top 10 concepts derived from query log data (QRYLOG table) | |||
| Herpes Zoster Ophthalmicus | 12 | Varicella | 237 | Pregnancy | 36 |
| Ramsey-Hunt Syndrome | 11 | Neuralgia | 222 | Itching | 15 |
| Herpes Simplex | 9 | Chicken Pox | 174 | Scabies | 15 |
| Varicella Encephalitis | 9 | Antiviral | 173 | Hives | 13 |
| Chicken Pox | 8 | Acyclovir | 122 | Integumentary System | 12 |
| Herpes Infection | 8 | Herpes Simplex | 60 | Small Pox | 12 |
| Disseminated Zoster | 7 | Pain | 51 | Viral | 12 |
| Herpes Zoster with Meningitis | 7 | Skin Diseases, Viral | 48 | Psoriasis | 11 |
| Zoster without Complications | 7 | Varicella Vaccine | 46 | Poison Ivy | 11 |
| Herpes Zoster Iridocyclitis | 6 | AIDS | 40 | Virus | 10 |
There are many more results in each of these lists, but only the top 10 are shown here for brevity's sake.
Semantic-Type Based Recommendations
Certain classes of topics, such as diseases, procedures, or medications, are common subjects of consumer queries. We found from our previous studies that people are often only interested in a certain aspect of the topics of interest to them, but are not always explicit about this in the query. For instance, one person may be interested in the risk factors for a disease but another may be interested in the prognosis. To encourage consumers to specify these aspects (which we refer to as query modifiers), the system first classifies the concepts based on their semantic types. For a few major semantic types (e.g., disease and procedure), we identified type-specific modifiers based on published literature of consumer HIR needs.1,2 The system suggests some of these modifiers, which have been hard-coded into the system, if a concept of 1 of these few types appears in a query. For instance, based on the semantic type “disease,” the system will suggest the concepts “Symptoms,” “Risk Factors,” “Causes,” “Outlook,” “Diagnosis,” “Treatment,” and “Morbidity.” If the semantic type is “procedure,” however, the system will suggest “Risks,” “Benefits,” “Success Rate,” “Preparation,” “Indications,” “Complications,” and “Convalescence.” These are suggestions based not on the concept the user entered, but on the type of concept the user entered.
Learning from User Selection
The related concepts identified through fuzzy rules are only an informed guess of what consumers may find useful in constructing a query. The relevance and value of a recommendation will ultimately be confirmed by usage, which provides a means for us to improve the quality of the recommendations. Our system learns from usage by continuously updating the QRYLOG table: when a suggested concept is selected by a user, its occurrence with the query concept is increased by one. The original QRYLOG table only contains concepts that consumers can recall; the new co-occurrence indicates what can be recognized. Assuming that “psoriasis” and “eczema” are both in the suggestion list for a query of “skin” with similar ranking, if users consistently click on “psoriasis” but never “eczema,” the co-occurrence of “psoriasis” and “skin” will become higher over time, which in turn will boost its ranking over “eczema.”
Implementation
HIQuA is implemented using a 3-tier client-server architecture. The client is a Java applet that runs in a Web page, the middle tier is an Apache Tomcat Web server, and the back end is a MySQL database server containing millions of medical concepts and relations derived from the Unified Medical Language System (UMLS), which is provided by the National Library of Medicine, and the query log data. Queries are submitted to 1 of several major search engines, with Google™ being the default.
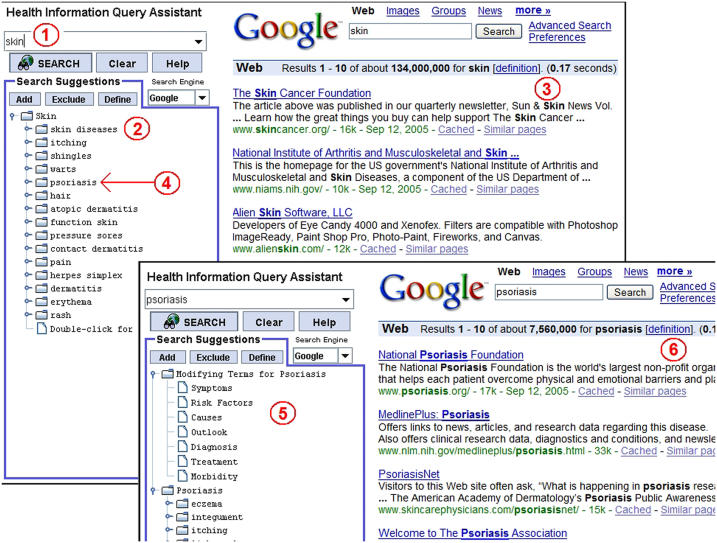
For a given query, HIQuA suggests a list of modifiers and related concepts. Users may look up definitions of the suggestions, add them to the query, exclude them from the query, or replace the initial query with the suggested terms. The modified query is then submitted to the search engine for free-text search. Users may also further explore the related concepts of any recommended concept; these further related concepts are identified by HIQuA in the same fashion as for the original query concept. A screen shot of HIQuA is shown in ▶.
Figure 2.
Screen shots of HIQuA based on the actual search behavior of one of the study subjects. (1) The user submitted a query on the word “skin.” (2) HIQuA displayed a list of query suggestions. (3) The query was sent to Google™. (4) The user recognized “psoriasis” as the condition he was interested in, so he clicked on that suggestion. (5) HIQuA displayed suggestions related to psoriasis. (6) Google™ displayed search results related to psoriasis.
Usability tests were performed to ensure that the user interface was clearly understood by consumers. We recruited 25 consumers from the Brigham and Women's Hospital in Boston to test the system, and iteratively improved the system according their feedback. Users sometimes discovered bugs due to using the system in unexpected ways. They also pointed out what they found confusing and made some useful suggestions regarding features they would like to see, one of which was a navigable history list of searches (▶).
Evaluation
Data Collection
We performed a formal evaluation of the system (BWH IRB Protocol #2003P000710). Consumers who were not health care professionals were recruited from the Bunker Hill Community College (BHCC). The eligibility criteria for the study were some experience with the Web, age 18 or above, not a physician or nurse, and able to read and write English. Flyers and posters advertising the study were posted in the BHCC lobbies. A BHCC computer laboratory room was borrowed to be the study site. Two discount movie tickets (approximate $10 value) were given to each subject who completed the study, which usually required 20 to 30 minutes. The recruitment and testing took place during June and July of 2004.
A total of 213 subjects participated in the study. All subjects were asked to use the HIQuA system (in conjunction with Google™) to search the Web for health information. Query recommendations were blocked out for half of the subjects by randomization. Each study subject was asked to first fill out a brief demographic questionnaire and then perform 1 of 2 predefined health-retrieval tasks—finding five factors that increase one's chance of having a heart attack or finding three methods to treat baldness. We used two different questions for the predefined task and randomized half of the subjects to each question because it would reduce the chance of many subjects unexpectedly having prior knowledge of the given question. The task was described to the participants as follows:
Task #1
Version A: Please find five things that increase a person's chances of having a heart attack.
Version B: Some people are concerned about going bald. Please list three ways to potentially treat their condition.
Each study subject was also asked to perform a self-defined health-retrieval task. For the self-defined task, subjects were given verbal instruction to elaborate on their information needs and retrieval goals in writing prior to the search, so that we could later evaluate their queries in the context of the goals. We did not ask for the search results of the self-defined task to be written down due to practical time concerns (our system did, however, record their queries, allowing us to reconstruct their results later). The self-defined task is described to the participants as follows:
Task #2
Please search for any health-related question that you are curious about.
The subjects were also asked to rate their own overall satisfaction of the search experience on a scale of 1 to 5 at the end of the searches:
- Please rank your satisfaction with your search experience (circle one:)
- 1 = Extremely dissatisfied
- 2 = Not satisfied
- 3 = Neutral
- 4 = Satisfied
- 5 = Extremely satisfied
All queries typed by study subjects and the recommendations selected by the recommendation group were automatically recorded in a log file.
Data Analysis
To evaluate the impact of HIQuA recommendations, we measured and compared three main outcomes in the recommendation group and the nonrecommendation group: (1) User satisfaction; (2) Query success rate; (3) Score of the predefined task.
Univariate analysis was first carried out to look at the unadjusted association between the groups (recommendation vs. nonrecommendation) and potential demographic factors including age, race, sex, years of Internet experience, health-related Web experience, and health status. Only health-related Web experience and health status were found to be statistically significant. These two confounders were later used in the multivariable logistic regression models and the general linear model to obtain the effect of query recommendations on the three outcomes.
User Satisfaction
To analyze the first outcome, user satisfaction, the 5-point user satisfaction ratings were categorized into two categories: satisfied (including extremely satisfied and satisfied) and not satisfied (including extremely unsatisfied, unsatisfied, and neutral). A multivariable logistic regression model was set up to look at the effect of being in the group receiving query recommendations on user satisfaction, while controlling for the confounders. The odds ratios and 95% confidence intervals were computed.
Query Success Rate
To analyze the second outcome, query success rate, a query that resulted in one or more relevant documents in the top 10 search results was considered successful. A multivariable logistic regression model was set up to look at the effect of being in the group receiving query recommendations on the percent of successful queries, while controlling for the confounders. The odds ratios and 95% confidence intervals were computed.
In this analysis, we only considered the top 10 results because too many results were generated by queries for us to assess recall and precision more comprehensively, and, in any case, consumers typically only examine the top few search results.9 The success of the queries was evaluated by three human reviewers: each query (including recommendations that were clicked on) was submitted to the search engine and the top 10 returned pages were examined for relevance based on the pre- or self-defined retrieval goal.
We were able to assess the results for self-defined retrieval goals because most participants followed the study instruction and wrote down clear descriptions of their information needs (and we were able to reconstruct their search results from the queries recorded in HIQuA's logs). Every query and search result was examined by at least two reviewers and differences between reviewers were resolved through discussion. For a page to be judged relevant, it needed to contain at least some information that met the search goal stated by the participant, and the information could not be misleading or in the form of commercial advertisement. For example, for a subject's question “How can I prevent sexually transmitted diseases?,” a page on sexually transmitted disease treatments, a page denouncing abstinence as a government conspiracy, and a page advertising a particular brand of condom were all judged to be irrelevant. A total of 280 self-defined tasks were analyzed.
Score of the Predefined Task
To analyze the third outcome, the score of the predefined task, the answers given by subjects for the predefined retrieval task were graded according to a gold standard that was established based on literature review. When grading, a correct answer was given a score of 1, incorrect answers were given a score of −1, and the absence of an answer was graded as 0. Incorrect answers were graded lower than the absence of an answer because being misinformed can be more harmful than being uninformed. Because we asked subjects to find and report 5 risk factors for heart disease or 3 treatments for baldness, all answers to a question were summed up and divided by 5 or 3, respectively, to generate a normalized score. Analysis of variance (General Linear Model) was used to compare the predefined task score of the group receiving query recommendations versus the group that did not receive query recommendations. We adjusted for the 2 confounders in the analysis.
Results
A total of 213 subjects participated in the study. We had a fairly diverse population of subjects in terms of race and ethnicity. On average, the subjects were young, reasonably well educated, and healthy (Table 4 is available as a JAMIA online supplement at www.jamia.org). Please note that the education level indicates the highest level started, not finished. Many of the subjects were attending the community college where we conducted our study. Over 40% did not speak English as their first language, and the command of English varied significantly among these non-native speakers. The subjects were generally familiar with the Web, though not all had had Web HIR experience.
User Satisfaction
Of those in the group receiving query recommendations, 85.2% of the subjects were satisfied with their search experience—a result that was a little higher than for the nonrecommendation group (80.6%). However, the difference was not statistically significant (p = 0.136). According to the odds ratio calculated using logistic regression, the odds of being satisfied increased by 79% if the participant was in the recommendation group. The confidence interval for the odds ratio, however, is wide and crosses 1.0; thus the association between groups and user satisfaction is not statistically significant (▶).
Table 5.
Comparison of User Satisfaction, Query Success, Predefined Task Score of the Recommendation Group and Nonrecommendation Group
| Recommendation Group | Nonrecommendation Group | p Value | Confidence Interval and Odds Ratio* | |
|---|---|---|---|---|
| User satisfaction | 85.2% | 80.6% | 0.136 | 1.79 (0.83–3.83) |
| Query success rate | 76.0% | 65.7% | 0.006 | 1.66 (1.16–2.38) |
| Predefined task score† | 0.440 ± 0.702 | 0.577 ± 0.653 | 0.233 | −0.137 (−0.320 to 0.049) |
| 1.0 | 1.0 |
95% confidence interval for estimated odds ratios and mean difference from logistic regression and linear model.
Mean ± standard deviation, and median.
Query Success Rate
There was a statistically significant difference (p = 0.006) in the percentage of successful queries between the recommendation group (76.0%) and the nonrecommendation group (65.7%) (▶). According to the odds ratio calculated using logistic regression, being in the recommendation group increased the odds of submitting a successful query by 66% (with a 95% confidence interval of between 16% and 138%).
Because a statistically significant difference was found for the query success rate between the recommendation group versus the nonrecommendation group, we further examined the source of the difference. The queries manually typed in by both groups of subjects did not have a statistical difference in success rate, as one would expect. The suggested queries that were selected by subjects in the recommendation group did have a higher success rate (p < 0.0001) than the typed-in queries (Figure 3 is available as a JAMIA online supplement at www.jamia.org). (This comparison was also adjusted for the two confounders—health-related Web experience and health status.)
Score of Predefined Task
The normalized mean score of the predefined task was higher for the nonrecommendation group (0.577) than for the recommendation group (0.440), although not statistically significant (p = 0.233). In ▶, we report both mean and median for the third outcome because the distribution of scores was asymmetric.
To summarize, the use of query recommendations led to a higher rate of successful queries. The impact (positive or negative) of query recommendations on satisfaction or accomplishing a predefined retrieval task was not clear.
Limitations
There are known limitations to our development and evaluation methodology. First, the target users are consumers, which is a very diverse group. It can be argued that each consumer has a different mental model; however, even a diverse group shares common terms, concepts, and concept relations. Take the term “anorexia,” for instance—in the professional setting it usually refers to the symptom “loss of appetite” while in the lay setting it usually refers to the disease anorexia nervosa. We use the adjective usually here because there are always exceptions. Yet if there did not exist some common mental model among consumers, and between consumers and physicians, it would be impossible for physicians to communicate with consumers and for consumers to communicate with each other. Nevertheless, the diversity of the consumer population makes measurement of semantic distance between concepts inherently less precise.
For query expansion, consumer queries are mapped to UMLS concepts by string matching. Accurate mapping is not always feasible because the UMLS concepts and concept names primarily represent the language of health care professionals. We are currently involved in a collaborative effort (www.consumerhealthvocab.org) to address this very issue. Disambiguation also remains challenging. We may disambiguate incorrectly and thus present the user with unhelpful suggestions. In these cases, the user is free to ignore the suggestions. Note that without being privy to the internal thoughts of the user, we often cannot know whether we have disambiguated incorrectly, e.g., perhaps a particular user actually desires to find information on a genus of crabs when entering the query “cancer.”
Our and others' previous analyses of consumer health queries and online postings showed that about 50% to 80% of consumer query terms can be mapped to UMLS.5,42,43 Although this mapping rate is not ideal, it provides a starting point for our concept-based query expansion. In our identification of related concepts, three sources are involved which have a small common overlap (less than 5% by our observation) while being largely complementary to each other. Having a related concept declared relevant by more than one source or having one source rank a related concept extremely high suggests a shorter semantic distance. The rules we used are a fuzzy representation of this basic logic, which is not equivalent to an algebraic mean of rankings from each source. We acknowledge that this might not be the optimal solution, but rather, a solution which reflects the intuitive ways in which people combine information from multiple sources. (There is no universal solution to the general problem of combining semantic-distance information from multiple sources—different approaches apply to different domains.)
Regarding the second outcome measurement (query success rate), the query success was determined by the researchers instead of study participants. Researchers judged the relevance of a page based on whether it met the retrieval goals stated by participants, or the predefined retrieval goal. A potential problem of this approach is that researchers could make mistakes in interpreting the retrieval goals written by participants, although most goals were relatively straightforward, e.g., “How can I prevent sexually transmitted diseases?” On the other hand, researchers tend to be more consistent and better equipped to review the relevance of a page of health information than study participants.
Time spent by participants conducting the searches was recorded, but not reported as an outcome. One reason is that we found that there could be different causes for spending more time at a task: it sometimes resulted from finding interesting material to read and explore and sometimes from not being able to find the desired information.
Finally, we did not distinguish officially published literature from unpublished literature (“grey literature”) in this study, and neither did we distinguish high-quality from low-quality material. The quality and credibility of content are important issues, but it was beyond the scope of HIQuA development.
Discussion
We have designed, implemented, and evaluated a tool to help consumers with query construction during HIR. The resulting system, HIQuA, recommends medical concepts and modifiers related to an initial user query as building blocks to form more specific or complex queries. The HIQuA system uses fuzzy logic to combine semantic distance information from three sources (concept co-occurrence in query log and medical literature, and semantic relationships in medical vocabularies), for the purpose of identifying relevant concepts. It also learns from user selection to continuously refine the recommendations. The evaluation showed that the availability of recommendations led to a significantly higher rate of successful queries, although there was not any significant impact on user satisfaction or on accomplishing a predefined retrieval task.
Because HIQuA can be used to explore the semantic neighborhood of tens of thousands of medical concepts, consumers may first browse the concept space to find the right term(s) to describe their needs and then look in the content space for the relevant information. There exist Web directories that consumers can browse for health information, but these directories mostly reflect hierarchical or classification knowledge regarding medical concepts. HIQuA constructs a concept neighborhood based on a much broader scope of medical knowledge and takes consumer usage patterns and consumer mental models into account.
In presenting the related terms to users, we did not simply use the UMLS preferred name because many preferred names are not the most user-friendly among all the synonyms. We have identified a set of consumer-preferred names for tens of thousands of UMLS concepts primarily based on how often a name is used by lay people. These names are used whenever available as the display names for concepts. They are also naturally free of the “NOS”-type postfixes present in some vocabularies, because no consumer ever adds a “ < 1>” or “NOS” behind a term. (“NOS” stands for not otherwise specified; “ < 1>” is sometimes added by a vocabulary to indicate that a certain string is preferred for one concept over another).
Without knowing the context of a query, HIQuA makes recommendations on the basis of two postulations: (1) a user may want to refine or replace the search term with other related terms; (2) the relatedness of terms can be derived from co-occurrences in usage and from known semantic relations. HIQuA is limited in its capacity to understand the real information needs underlying a query, especially a short one. It thus can only make best guesses about which other terms might be of use to a consumer.
The evaluation showed that HIQuA recommendations helped consumers to generate more successful queries, which helped to validate the design and implementation of the system. Several factors contributed to our failure to show a statistically significant impact of the system on overall user satisfaction or on the score of the predefined task. First, not every consumer needs the help of recommendations when performing every single task. Some subjects in the nonrecommendation group can accomplish the given or self-defined retrieval tasks successfully on their own. Second, not all subjects made use of the recommendations. Six people in the recommendation group did not click on any recommendations. On the other hand, there were also some curiosity clicks: at least one subject clicked on every query term suggested by HIQuA, many of which did not help with the retrieval tasks. Third, query recommendations would not be of help to people with very poor health literacy and very poor general literacy levels. Several study subjects misinterpreted the predefined question or the information they had found: a few subjects wrote down causes for baldness although the question was how to treat the condition. Some subjects clearly were unable to discern the promotional or misleading information from “good” information and thus gave wrong answers. Fourth, satisfaction is a very subjective measurement. Some people answered the predefined question completely incorrectly, yet reported satisfaction with the search experience. Because of these factors and the sample size, it was understandable that a statistically significant difference on the user satisfaction score or on the predefined task score was not found between the recommendation and nonrecommendation groups. A larger sample size might have resulted in statistically significant findings.
The innovation of the HIQuA system is that it estimates semantic distance based on three types of information sources (i.e., query log, literature corpus, and manually constructed thesaurus) and uses fuzzy logic to do so. Previous information science research has explored each of these types individually for query expansion purposes. As discussed in the Background section, there have also been studies that used multiple information sources and utilized fuzzy logic in query expansion. No prior study, however, has used fuzzy rules to combine multiple co-occurrence data with relations from vocabularies.
In the specific area of consumer HIR, research on query expansion or suggestion has depended on medical vocabularies as the main knowledge source. However, the HIQuA system explores other sources and makes use of semantic relations beyond synonymy and hierarchical relationships. The use of the query log is especially important because it is a record of consumer language and consumer search behaviors.
Query suggestions will not be needed by every user for every search; however, the evaluation has shown that our system could be a helpful tool for query formation when a user does need it. Because there are millions of consumers conducting HIR, even a fraction of the entire user population comprises a great number of users. As a general purpose application in the health care domain, HIQuA could potentially benefit many users conducting HIR.
To help consumers obtain better satisfaction and retrieval performance when querying, we will continue to work on the refinement of this system as well as some other HIR issues such as content annotation and quality assessment.
Conclusion
We have developed a query suggestion tool to help consumers search for health information online. Our approach is designed to address the problems of user query construction by providing frequency- and knowledge-based query recommendations. Our trial showed that providing HIQuA recommendations resulted in statistically significantly more successful consumer queries over not providing the recommendations, although no statistically significant impact on user satisfaction or ability to accomplish a predefined retrieval task was found. Although query expansion has been studied extensively, using fuzzy logic to combine information derived from usage logs, literature co-occurrence, and vocabulary information is novel. While prior research in query expansion or query recommendations for consumer HIR has been mainly thesaurus-based, our study tested the feasibility of (and showed promising results for) employing more diverse sources to find related terms or concepts. Because query formation is a challenging task for many HIR users, we believe that our system, or a similar system, could have a positive impact on HIR for consumers by providing meaningful and consumer-centered suggestions.
Supplementary Material
Supported by National Institutes of Health grant R01 LM07222.
The authors thank Lowell Ling and Chantal Friedman for assisting with the evaluation study.
References
- 1.Fox S, Fallows D. Health searches and email have become more commonplace, but there is room for improvement in searches and overall Internet access: Pew Internet & American Life Project; 16July2003.
- 2.Zeng QT, Kogan S, Plovnick RM, Crowell J, Lacroix EM, Greenes RA. Positive attitudes and failed queries: an exploration of the conundrums of consumer health information retrieval. Int J Med Inform. 2004;73:45–55. [DOI] [PubMed] [Google Scholar]
- 3.Silverstein C, Marais H, Henzinger M, Moricz M. Analysis of a very large web search engine query log. ACM SIGIR Forum. 1999;33:6–12. [Google Scholar]
- 4.Spink A, Wolfram D, Jansen MBJ, Saracevic T. Searching the Web: the public and their queries. J Am Soc Inform Sci Technol. 2001;52:226–34. [Google Scholar]
- 5.Zeng Q, Kogan S, Ash N, Greenes RA, Boxwala AA. Characteristics of consumer terminology for health information retrieval. Methods Inf Med. 2002;41:289–98. [PubMed] [Google Scholar]
- 6.Kogan S, Zeng Q, Ash N, Greenes RA. Problems and challenges in patient information retrieval: a descriptive study. Proc AMIA Symp. 2001:329–33. [PMC free article] [PubMed]
- 7.Eysenbach G, Kohler C. How do consumers search for and appraise health information on the World Wide Web? Qualitative study using focus groups, usability tests, and in-depth interviews. BMJ. 2002;324:573–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fredin ES, David P. Browsing and the hypermedia interaction cycle: a model of self-efficacy and goal dynamics. Journal Mass Commun Q. 1998;75:35–55. [Google Scholar]
- 9.Baeza-Yates R, Ribeiro-Neto B. Modern information retrieval. New York, NY: ACM Press; 1999.
- 10.Cui H, Wen J-R, Nie J-Y, Ma W-Y. Query expansion by mining user logs. IEEE Trans Knowledge Data Eng. 2003:829–39.
- 11.Stenmark D. Query expansion on a corporate intranet: using LSI to increase precision in explorative search. In: Proceedings of the 38th Annual Hawaii International Conference on System Sciences; 03-06 Jan. 2005; Page 101c.
- 12.Harman D. Towards interactive query expansion. In: 11th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, May 1988. Grenoble, France: ACM Press, 1988, 321–31.
- 13.Xu J, Croft WB. Query expansion using local and global document analysis. In: The Nineteenth Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, 1996:4–11.
- 14.Mandala R, Tokunaga T, Tanaka H. Combining multiple evidence from different types of thesaurus for q uery expansion. In: The 22nd Annual International ACM SIGIR Conference on Research and development in information retrieval; 1999; Berkeley, CA, 1999:191–7.
- 15.Lee H-M, Lin S-K, Huang C-W. Interactive query expansion based on fuzzy association thesaurus for Web information retrieval. In: Fuzzy systems, 2001. The 10th IEEE International Conference, 2001:724–7, vol 3.
- 16.Takagi T, Tajima M. Query expansion using conceptual fuzzy sets for search engine. In: Fuzzy systems, 2001. The 10th IEEE International Conference, 2001:1303-8.
- 17.Akrivas G, Wallace M, Andreou G, Stamou G, Kollias S. Context-sensitive semantic query expansion. In: Artificial Intelligence Systems, 2002. (ICAIS 2002). 2002 IEEE International Conference, 2002:109–14.
- 18.Anick P. Using terminological feedback for web search refinement: a log-based study. In: The 26th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, Toronto, Canada, 2003:88–95.
- 19.Muramatsu J, Pratt W. Transparent queries: investigating users' mental models of search engines. In: The 24th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, New Orleans, LA, 2001:217–24.
- 20.Ruthven I. Re-examining the potential effectiveness of interactive query expansion. In: The 26th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, Toronto, Canada, 2003:213–20.
- 21.Billerbeck B, Scholer F, Williams HE, Zobel J. Query expansion using associated queries. In: The Twelfth International Conference on Information and Knowledge Management. New Orleans, LA: ACM Press, 2003, 2–9.
- 22.Doszkocs TE. AID, an associative interactive dictionary for online searching. Online Rev. 1978;2:163–72. [Google Scholar]
- 23.Pollitt S. CANSEARCH: an expert systems approach to document retrieval. Inform Process Manage. 1987;23:119–38. [Google Scholar]
- 24.Srinivasan P. Retrieval feedback in MEDLINE. J Am Med Inform Assoc. 1996;3:157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hersh W, Price S, Donohoe L. Assessing thesaurus-based query expansion using the UMLS Metathesaurus. Proc AMIA Symp. 2000:344–8. [PMC free article] [PubMed]
- 26.Aronson AR, Rindflesch TC. Query expansion using the UMLS Metathesaurus. Proc AMIA Annu Fall Symp. 1997:485–9. [PMC free article] [PubMed]
- 27.Nenadic G, Mima H, Spasic I, Ananiadou S, Tsujii J. Terminology-driven literature mining and knowledge acquisition in biomedicine. Int J Med Inform. 2002;67:33–48. [DOI] [PubMed] [Google Scholar]
- 28.Joubert M, Fieschi M, Robert JJ, Volot F, Fieschi D. UMLS-based conceptual queries to biomedical information databases: an overview of the project ARIANE. Unified Medical Language System. J Am Med Inform Assoc. 1998;5:52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson SB, Aguirre A, Peng P, Cimino J. Interpreting natural language queries using the UMLS. Proc Annu Symp Comput Appl Med Care. 1993:294–8. [PMC free article] [PubMed]
- 30.Merz RB, Cimino C, Barnett GO, Blewett DR, Gnassi JA, Grundmeier R, et al. Q & A: a query formulation assistant. Proc Annu Symp Comput Appl Med Care. 1992:498–502. [PMC free article] [PubMed]
- 31.Salton G, Buckley C, Fox EA. Automatic query formulations in information retrieval. J Am Soc Inf Sci. 1983;34:262–80. [DOI] [PubMed] [Google Scholar]
- 32.Hripcsak G, Allen B, Cimino JJ, Lee R. Access to data: comparing AccessMed with Query by Review. J Am Med Inform Assoc. 1996;3:288–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fisk JM, Mutalik P, Levin FW, Erdos J, Taylor C, Nadkarni P. Integrating query of relational and textual data in clinical databases: a case study. J Am Med Inform Assoc. 2003;10:21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gobel G, Andreatta S, Masser J, Pfeiffer KP. A MeSH based intelligent search intermediary for Consumer Health Information Systems. Int J Med Inform. 2001;64:241–51. [DOI] [PubMed] [Google Scholar]
- 35.McCray AT, Ide NC, Loane RR, Tse T. Strategies for supporting consumer health information seeking. Medinfo. 2004;11(Pt 2):1152–6. [PubMed] [Google Scholar]
- 36.Plovnick RM, Zeng QT. Reformulation of consumer health queries with professional terminology: a pilot study. J Med Internet Res. 2004;6(3):e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patrick TB, Monga HK, Sievert ME, Houston Hall J, Longo DR. Evaluation of controlled vocabulary resources for development of a consumer entry vocabulary for diabetes. J Med Internet Res. 2001;3(3):E24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendonca EA, Cimino JJ. Automated knowledge extraction from MEDLINE citations. Proc AMIA Symp. 2000:575–9. [PMC free article] [PubMed]
- 39.Hristovski D, Stare J, Peterlin B, Dzeroski S. Supporting discovery in medicine by association rule mining in Medline and UMLS. Medinfo. 2001;10(Pt 2):1344–8. [PubMed] [Google Scholar]
- 40.Humphreys BL, Lindberg DA, Schoolman HM, Barnett GO. The Unified Medical Language System: an informatics research collaboration. J Am Med Inform Assoc. 1998;5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller N, Lacroix EM, Backus JE. MEDLINEplus: building and maintaining the National Library of Medicine's consumer health Web service. Bull Med Libr Asnsoc. 2000;88:11–7. [PMC free article] [PubMed] [Google Scholar]
- 42.Tse T. Identifying and characterizing a “consumer medical vocabulary.”. College Park, MD: University of Maryland, 2003.
- 43.Brennan PF, Aronson AR. Towards linking patients and clinical information: detecting UMLS concepts in e-mail. J Biomed Inform. 2003;36:334–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.