Abstract
Spinal muscular atrophy is a neurodegenerative disorder caused by the deletion or mutation of the survival-of-motor-neuron gene, SMN1. An SMN1 paralog, SMN2, differs by a C→T transition in exon 7 that causes substantial skipping of this exon, such that SMN2 expresses only low levels of functional protein. A better understanding of SMN splicing mechanisms should facilitate the development of drugs that increase survival motor neuron (SMN) protein levels by improving SMN2 exon 7 inclusion. In addition, exonic mutations that cause defective splicing give rise to many genetic diseases, and the SMN1/2 system is a useful paradigm for understanding exon-identity determinants and alternative-splicing mechanisms. Skipping of SMN2 exon 7 was previously attributed either to the loss of an SF2/ASF–dependent exonic splicing enhancer or to the creation of an hnRNP A/B–dependent exonic splicing silencer, as a result of the C→T transition. We report the extensive testing of the enhancer-loss and silencer-gain models by mutagenesis, RNA interference, overexpression, RNA splicing, and RNA-protein interaction experiments. Our results support the enhancer-loss model but also demonstrate that hnRNP A/B proteins antagonize SF2/ASF–dependent ESE activity and promote exon 7 skipping by a mechanism that is independent of the C→T transition and is, therefore, common to both SMN1 and SMN2. Our findings explain the basis of defective SMN2 splicing, illustrate the fine balance between positive and negative determinants of exon identity and alternative splicing, and underscore the importance of antagonistic splicing factors and exonic elements in a disease context.
Spinal muscular atrophy (SMA types I, II, and III [MIMs 253300, 253550, and 253400]) is an autosomal recessive neurodegenerative disease characterized by the selective destruction of spinal cord α-motor neurons. SMA is a leading genetic cause of infant mortality, with an estimated incidence of 1 in 6,000–10,000 live births, and is caused by the homozygous loss or mutation of the SMN1 gene (Entrez Gene ID number 6606), which codes for the survival motor neuron (SMN) protein (UniProt accession number Q16637-1) (Lefebvre et al. 1995; reviewed by Frugier et al. [2002]). SMN is a ubiquitously expressed, essential protein necessary for the efficient assembly of ribonucleoprotein complexes (reviewed by Meister et al. [2002] and Gubitz et al. [2004]). SMN2 (Entrez Gene ID number 6607), a nearly identical paralog of SMN1, is present in the human genome as part of an inverted duplication on 5q13. SMN2 expresses limited amounts of functional, full-length SMN protein, which is apparently sufficient for normal activity in most cell types but not in motor neurons. SMN2 is efficiently transcribed but cannot fully compensate for the loss of SMN1, because a translationally silent, single-nucleotide transition in SMN2 at position 6 of exon 7 (c6t) causes predominant exon 7 skipping (Lefebvre et al. 1995; Lorson et al. 1999; Monani et al. 1999) and results in an unstable protein (SMNΔ7) (UniProt accession number Q16637-3) with a different C-terminus (Lorson and Androphy 2000). Increased expression from SMN2 can reduce the severity of SMA, as seen when multiple copies of SMN2 are present, both in patients and mouse models (reviewed by Monani et al. [2000] and Frugier et al. [2002]). Current efforts to develop therapeutics for SMA are largely directed at increasing SMN2-derived full-length SMN protein abundance or activity at the transcriptional and posttranscriptional levels (Jarecki et al. 2005 and references therein), including approaches aimed specifically at changing the ratio of the two main splicing isoforms in favor of the full-length active one (Lim and Hertel 2001; Miyajima et al. 2002; Cartegni and Krainer 2003; Skordis et al. 2003).
Signals located within an exon can have positive or negative effects on the recognition of that exon during splicing. Exonic splicing enhancers (ESEs) stimulate splicing and are often required for efficient intron removal, whereas exonic splicing silencers (ESSs) inhibit splicing. ESSs and ESEs frequently antagonize each other—for example, in the context of alternative splicing regulation (reviewed by Hastings and Krainer [2001] and Cáceres and Kornblihtt [2002]). The specific sequence and location of these cis-elements determine the proteins that mediate the splicing activity. Well-characterized ESEs include those recognized by SR proteins, a family of RNA-binding proteins with distinctive serine/arginine–rich domains (reviewed by Cáceres and Kornblihtt [2002] and Cartegni et al. [2002]). Some ESSs are recognized by hnRNP A1 (UniProt accession number P09651-2), an abundant RNA-binding protein that plays an important role in splicing repression (reviewed by Cáceres and Kornblihtt [2002] and Dreyfuss et al. [2002]). As exemplified by SMA, point mutations that alter splicing signals can cause a loss of normal gene expression as a result of aberrant splicing. Indeed, aberrant splicing is a prevalent cause of genetic diseases (reviewed by Cáceres and Kornblihtt [2002], Faustino and Cooper [2003], and Pagani and Baralle [2004]).
Two different models have been proposed to explain the observed difference in splicing between SMN1 and SMN2: loss of an enhancer or gain of a silencer. The first model proposes the existence in SMN1 of an ESE recognized by the SR protein SF2/ASF (UniProt accession number Q07955) (Cartegni and Krainer 2002). The C→T transition in SMN2 disrupts this ESE and results in exon 7 skipping. This transition eliminates a high-score motif for SF2/ASF, according to ESEfinder, a program that predicts ESEs that are responsive to one of several SR proteins on the basis of experimental and statistical analyses (Cartegni and Krainer 2002; Cartegni et al. 2003). Mutational analysis, RNA-binding data, transfection experiments, and in vitro splicing assays using extracts and recombinant proteins all supported a direct role for SF2/ASF in SMN1 exon 7 inclusion that is not maintained in SMN2 (Cartegni and Krainer 2002), which is consistent with the notion that exon 7 skipping in SMN2 transcripts results from the loss of the SF2/ASF–dependent ESE. Furthermore, SMN2 splicing can be corrected by targeting exon 7 with specific synthetic effector molecules that mimic the function of SR proteins (Cartegni and Krainer 2003) or with bifunctional modified antisense oligonucleotides that comprise SF2/ASF–binding sites (Skordis et al. 2003). The second model asserts that, rather than eliminating an ESE, the presence of a T at position +6 of SMN2 exon 7 creates an ESS that is recognized by hnRNP A1 and leads to inefficient exon 7 inclusion (Kashima and Manley 2003). Increased exon 7 inclusion following small interfering RNA (siRNA)–mediated knockdown of hnRNP A1 and the related protein hnRNP A2 (UniProt accession number P22626-2), as well as UV cross-linking assays, suggested that hnRNP A1 inhibits SMN2 exon 7 inclusion, whereas genetic depletion of SF2/ASF in the chicken cell line DT-40 does not affect full-length SMN1 levels (Kashima and Manley 2003). These two models are not necessarily incompatible, since both the loss of an SF2/ASF–specific ESE as well as the simultaneous creation of an hnRNP A1–binding site could, in principle, be contributing factors to SMN2 exon 7 skipping. Indeed, these proteins are known to antagonize each other, a competition that appears to be based on their relative concentrations and RNA-binding properties (Mayeda and Krainer 1992; Cáceres et al. 1994; Eperon et al. 2000; Zhu et al. 2001; reviewed by Black [2003]).
Because of the obvious implications for SMA, a clearer understanding of the mechanism of SMN1/SMN2 exon 7 splicing would significantly impact the development of therapeutic approaches. We have now rigorously tested the enhancer-loss and silencer-gain models, using extensive mutational analysis, RNA interference (RNAi)–mediated knockdown, overexpression of hnRNP A1 and SF2/ASF, in vivo and in vitro splicing assays, and RNA-protein interaction analysis. We find that SMN2 exon 7 skipping primarily results from the loss of the SF2/ASF–dependent ESE. Although hnRNP A1 indeed has a strong inhibitory effect on exon 7 inclusion, this effect is independent of the C→T transition and, therefore, is not specific to SMN2. The observed antagonism between SF2/ASF and hnRNP A1 in the definition of SMN1/2 exon 7, together with the variable expression of these splicing regulators, may cause tissue-specific differences in the extent of exon 7 inclusion and thereby contribute to the motor neuron–restricted phenotype observed in SMA.
Material and Methods
RNA Affinity Chromatography
RNA affinity chromatography was performed with the modification of a published technique (Caputi et al. 1999). RNA oligonucleotides SMN.WT (5′-GGUUUCAGACAAAAUCA-3′) and SMN.C6T (5′-GGUUUUAGACAAAAUCA-3′) were obtained from Dharmacon. One hundred micrograms of each RNA was oxidized with sodium m-periodate in a 24-μl reaction. One hundred microliters (1:1 slurry) of adipic acid dihydrazide agarose beads (Sigma) was mixed with the SMN1 or SMN2 RNA and was rotated overnight at 4°C. A 250-μl in vitro splicing reaction mix (Cartegni et al. 2002) including 100 μl of HeLa nuclear extract was added to 50 μl of the SMN1 or SMN2 beads that were prewashed in buffer D with 0.1 M KCl, and the mixtures were incubated for 40 min at 30°C. The beads were divided into three aliquots, which were washed three times in buffer D with either 0.1 M KCl, 0.15 M KCl, or 0.2 M KCl. After the final wash, the beads were resuspended in 40 μl of protein sample buffer and were heated at 90°C for 10 min to elute the bound proteins. Ten microliters of each protein sample was loaded on a 12% SDS polyacrylamide gel, which was then electroblotted onto a nitrocellulose membrane and was probed with monoclonal antibodies against hnRNP A1 or SF2/ASF (Hanamura et al. 1998).
Minigene Construction, Transfections, and Expression Analysis
All SMN constructs were derived from pCI-SMNx-wt (Cartegni et al. 2002), which is a derivative of pCITel (Lorson and Androphy 2000). Mutations were introduced by site-directed mutagenesis, with the use of a QuikChange kit (Stratagene). Plasmids (1 μg) were transfected into 293-HEK cells by use of FuGENE (Roche). For SF2/ASF overexpression experiments, early-passage mouse embryo fibroblasts (MEF) cells were cotransfected with 2 μg of pCGT7-SF2/ASF and 400 ng of the SMN minigene plasmids. Total RNA was isolated after 48 h by use of Trizol (Life Technologies). Reverse transcription was performed using 1 μg of DNase-treated total RNA, oligo(dT), and Superscript II reverse transcriptase (Life Technologies). Semiquantitative PCR was performed with the cDNA equivalent of 10 ng of starting RNA, by use of AmpliTaq Gold (Roche) and 30 amplification cycles (94°C for 30 s, 58°C for 60 s, and 72°C for 60 s) in reactions containing [α-32P]dCTP. Primers were pCIFwdB (5′-GACTCACTATAGGCTAGCCTCG-3′) and SMN8-300+5′R (5′-AAGTACTTACCTGAAATCTAATCCACATTCAAATTTTCTCAACTG-3′). Products were separated on 6% native polyacrylamide or 1% agarose gels. The figures show either autoradiograms or ethidium bromide–stained gels, but, in all cases, the quantitation of splicing was based on phosphorimage analysis (Fujix BAS2000). The relative abundance of each product was calculated after adjustment for differences in base composition. For the statistical analysis in figure 1 and table 1, each mutant was tested in two to four separate transfections. Correlation coefficients were calculated with the use of nonparametric Spearman rank correlation and a bootstrap technique to determine the CIs. Additional experiments using fewer PCR amplification cycles gave similar results.
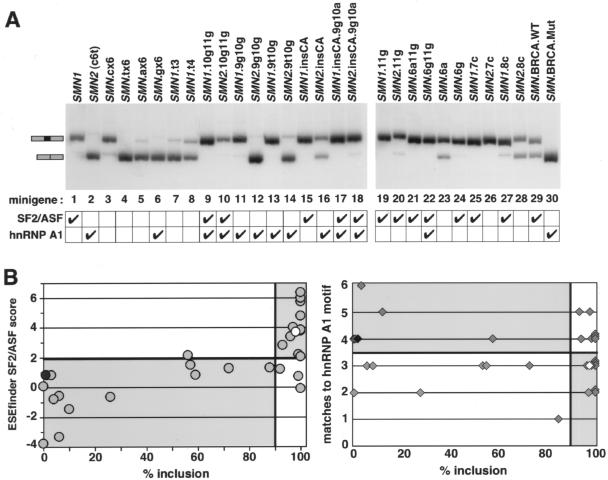
Figure 1.
Mutational analysis of SMN exon 7 splicing. A, Radioactive RT-PCR analysis of RNA from SMN mutant minigenes expressed in 293-HEK cells. Transcripts that either include or skip exon 7 are indicated. Check marks indicate the presence of a high-score SF2/ASF motif (⩾1.96), as predicted by ESEfinder 2.0 (Cartegni et al. 2003), or a ⩾4 of 6 match to the hnRNP A1 consensus motif (Burd and Dreyfuss 1994). Lane numbers correspond to the 30 minigene construct numbers listed in table 1. B, Results of multiple experiments (table 1) quantitated by phosphorimage analysis and presented as scatterplots. The percentage of exon 7 inclusion was plotted against SF2/ASF ESEfinder 2.0 scores (left) or against matches to the hnRNP A1 consensus binding motif (right). Thresholds are set at 1.96 for SF2/ASF, at between 3 and 4 matches for hnRNP A1, and at 90% for exon inclusion. The white and black symbols represent SMN1 and SMN2, respectively. Quadrants where the ESE-loss (left) and ESS-gain (right) models predict that the points should fall are shaded.
Table 1.
Summary of SMN Exon 7 Mutations and Levels of Exon Inclusion
| MinigeneNumber | Minigenea | Sequenceb | SF2/ASFScorec | hnRNP A1Scored | % (±SD)Includede |
| 1 | SMN1 | GGTTTCAGACAAAATCA |
3.77 | 3 | 98 ± 3 |
| 2 | SMN2 (c6t) | GGTTTTAGACAAAATCA |
.81 | 4 | 1 ± 2 |
| 3 | SMN.cX6 | GGTTTCcccCcAAATCA |
1.29 | 1 | 88 ± 5 |
| 4 | SMN.tX6 | GGTTTTtttttAAATCA |
−3.77 | 2 | 0 ± 0 |
| 5 | SMN.aX6 | GGTTTaAaAaAAAATCA |
−3.33 | 3 | 6 ± 7 |
| 6 | SMN.gX6 | GGTTTggGgggAAATCA |
.81 | 4 | 3 ± 3 |
| 7 | SMN1.t3 | GGTTTCAGtttAAATCA |
−.56 | 3 | 6 ± 3 |
| 8 | SMN1.t4 | GGTTTCAttCttAATCA |
−.65 | 2 | 26 ± 15 |
| 9 | SMN1.10g11g | GGTTTCAGAggAAATCA |
5.74 | 4 | 100 ± 0 |
| 10 | SMN2.10g11g | GGTTTTAGAggAAATCA |
2.79 | 4 | 93 ± 5 |
| 11 | SMN1.9g10g | GGTTTCAGggAAAATCA |
1.19 | 5 | 92 ± 2 |
| 12 | SMN2.9g10g | GGTTTTAGggAAAATCA |
−.78 | 6 | 4 ± 5 |
| 13 | SMN1.9t10g | GGTTTCAGtgAAAATCA |
.71 | 4 | 99 ± 1 |
| 14 | SMN2.9t10g | GGTTTTAGtgAAAATCA |
−1.46 | 5 | 10 ± 2 |
| 15 | SMN1.insCA | GGTTTcaCAGACAAAATCA |
3.77 | 3 | 100 ± 1 |
| 16 | SMN2.insCA | GGTTTcaTAGACAAAATCA |
.81 | 4 | 59 ± 5 |
| 17 | SMN1.insCA9g10a | GGTTTcaCAGgaAAAATCA |
5.98 | 4 | 100 ± 0 |
| 18 | SMN2.insCA9g10a | GGTTTcaTAGgaAAAATCA |
3.67 | 5 | 98 ± 2 |
| 19 | SMN1.11g | GGTTTCAGACgAAATCA |
6.34 | 2 | 100 ± 0 |
| 20 | SMN2.11g | GGTTTTAGACgAAATCA |
3.39 | 3 | 96 ± 2 |
| 21 | SMN.6a11g | GGTTTaAGACgAAATCA |
3.84 | 3 | 100 ± 0 |
| 22 | SMN.6g11g | GGTTTgAGACgAAATCA |
4.77 | 4 | 100 ± 1 |
| 23 | SMN.6a | GGTTTaAGACAAAATCA |
1.26 | 3 | 72 ± 2 |
| 24 | SMN.6g | GGTTTgAGACAAAATCA |
2.19 | 3 | 99 ± 1 |
| 25 | SMN1.7c | GGTTTCcGACAAAATCA |
2.04 | 2 | 100 ± 0 |
| 26 | SMN2.7c | GGTTTTcGACAAAATCA |
−.09 | 3 | 100 ± 0 |
| 27 | SMN1.8c | GGTTTCAcACAAAATCA |
4.01 | 2 | 97 ± 2 |
| 28 | SMN2.8c | GGTTTTAcACAAAATCA |
1.49 | 3 | 57 ± 3 |
| 29 | SMN.BRCA.WT | GGTTTCtGAgttAATCA |
2.14 | 3 | 56 ± 3 |
| 30 | SMN.BRCA.Mut | GGTTTCttAgttAATCA |
.08 | 4 | 0 ± 0 |
The 30 minigenes used in transfection experiments, in the same order as the lanes in figure 1A.
Nucleotide sequence around the ESE/ESS motifs. The ESEfinder 2.0 SF2/ASF top-scoring motif for each sequence is shown in bold italic type (if >0). The best fit to the hnRNP A1–binding consensus (TAGGGA/T) is underlined. SMN1 constructs have a T, and SMN2 constructs a C, at position +6. Mutations are indicated in lowercase.
Top SF2/ASF score. High-score motifs are shown in bold italic type (threshold = 1.96).
Number of nucleotide matches to the hnRNP A1 hexamer consensus, shown in bold italic type when >3.
Average inclusion percentages and SDs (n = 2–4) measured after radioactive RT-PCR in the linear range.
For analysis of endogenous SMN splicing, 293-HEK cells were transfected with 2 μg of pCGT7-A1 expression plasmid (Cáceres et al. 1994). RT-PCR of total RNA was performed as described above, with primers SMNex6FXho (5′-CGATCTCGAGATAATTCCCCCACCACCTCCC-3′) and SMNex8RNot (5′-ATATGCGGCCGCCACATACGCCTCACATACA-3′). Products were purified using PCR purification columns (Qiagen), were digested with DdeI, and were separated on 6% native polyacrylamide gels. Protein lysates were collected with Trizol and were analyzed by western blotting with an anti-T7-Tag antibody (Novagen) and mAb 4G3 (Habets et al. 1989) against human U2B′′ snRNP protein.
RNA Interference
Twenty-four hours before transfection of siRNAs with Oligofectamine (Invitrogen), 2.5×104 HeLa cells were seeded into 24-well plates. After 24 h, SMN minigenes were transfected using FuGENE, and the cells were incubated for another 24 h and then harvested for RNA isolation and western blotting. The following siRNAs were used: hnRNP A1, 5′-CAGCUGAGGAAGCUCUUCA-3′; hnRNP A2, 5′-GGAACAGUUCCGUAAGCUC-3′; SF2/ASF, 5′-ACGAUUGCCGCAUCUACGU-3′; and luciferase control, 5′-GCCAUUCUAUCCUCUAGAGGAUG-3′. RT-PCR was performed as described above. Western blotting was performed using antibodies specific for α-tubulin (Sigma B512) and for hnRNP A1 (4B10) and hnRNP A2 (DP3B3), which were kindly provided by Gideon Dreyfuss.
Nuclear Extract Depletion and in Vitro Splicing Analysis
HeLa cell nuclear extract was affinity depleted of hnRNP A/B proteins, as described elsewhere (Zhu et al. 2001). Three microliters of mock-depleted or hnRNP A/B–depleted extract was used in a standard reaction for SMN in vitro splicing (Cartegni and Krainer 2002). Extracts were complemented with recombinant hnRNP A1 protein, which was overexpressed in Escherichia coli and was purified as described elsewhere (Mayeda and Krainer 1992).
Results
SF2/ASF and hnRNP A1 Binding to SMN1 versus SMN2 RNA
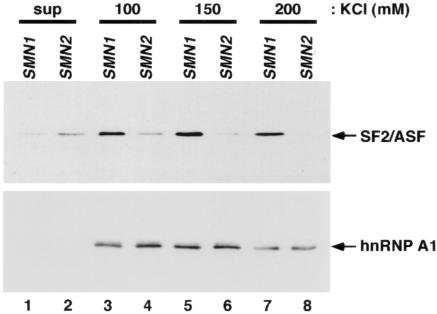
We previously used site-specific labeling to show the preferential binding of SF2/ASF to exon 7 RNA from SMN1 compared with SMN2 (Cartegni and Krainer 2002). Preferential binding of hnRNP A1 to SMN2 exon 7 RNA was shown by the uniform labeling of a 5′ fragment of the exon (Kashima and Manley 2003). These two observations were obtained using somewhat different methodologies and are, therefore, not directly comparable, but both results relied on immunoprecipitation after UV-induced cross-linking of radiolabeled RNAs to proteins in nuclear extract. Biases can arise because different nucleotides cross-link to proteins with significantly different efficiencies. In particular, rU generally cross-links more efficiently than does rC (Hockensmith et al. 1991). SMN2 exon 7 RNA has a U at position +6 and would, therefore, be expected to cross-link more efficiently to a bound protein than would SMN1, which has a C at the same position. This difference in cross-linking efficiency could account for the apparent difference in hnRNP A1 binding to SMN1 and SMN2 RNAs (Kashima and Manley 2003). Conversely, this difference in cross-linking efficiency could have resulted in an underestimate of the preferential binding of SF2/ASF to the region around position +9 of SMN1, as noted elsewhere (Cartegni and Krainer 2002).
To obtain a more direct and unbiased assessment of proteins that bind to the RNA encompassing the C→T transition, we performed RNA affinity chromatography. The first 17 nt of SMN1 or SMN2 exon 7 RNA were covalently linked to agarose beads via the 3′ end and were incubated with HeLa cell nuclear extract. Proteins that remained tightly bound to each RNA after washing at different salt concentrations were analyzed using monoclonal antibodies specific for hnRNP A1 or SF2/ASF. hnRNP A1 bound efficiently to SMN1 and SMN2 RNA fragments with approximately equal efficiency (fig. 2, bottom). In fact, incubation of nuclear extract with either RNA resulted in the efficient depletion of hnRNP A1 from the extract, as evident in the supernatant fractions. In contrast, SF2/ASF bound very specifically to the SMN1 RNA fragment (fig. 2, top), with minimal binding to the corresponding SMN2 RNA, which is consistent with the notion that the C→T transition results in the loss of an SF2/ASF–dependent ESE. Although this experimental approach does not reveal the stoichiometry or the precise location of the bound proteins along the immobilized RNA probes, it is unlikely that more than one or two molecules of SF2/ASF or hnRNP A1 can bind to a 17mer RNA, given the available information about their heptamer and hexamer recognition motifs (Burd and Dreyfuss 1994; Cartegni et al. 2003), as well as previous structural data (Ding et al. 1999; Maris et al. 2005).
Figure 2.
Binding specificity of SF2/ASF and hnRNP A1. Western blot of proteins recovered from agarose beads covalently linked to RNAs corresponding to the first 17 nt of SMN1 or SMN2. The RNA beads were incubated with identical aliquots of HeLa cell nuclear extract under splicing conditions; they were then divided into three aliquots for washing at the indicated salt concentrations, and the proteins that remained bound were recovered by heating in SDS-sample buffer. Blots were probed with monoclonal antibodies against SF2/ASF (top) or hnRNP A1 (bottom). Supernatant (sup) and bound samples shown correspond to 1/100th and 1/16th of the initial binding reactions, respectively.
Extensive Mutational Analysis of SMN1 Exon 7
To assess the relationship between hnRNP A1 motifs and exon skipping versus SF2/ASF motifs and exon inclusion, we designed a large panel of SMN minigenes with different mutations surrounding the +6 position of exon 7 (table 1). We used ESEfinder 2.0 (Cartegni et al. 2003) to calculate SF2/ASF–dependent ESE motif scores and used the consensus motif derived from SELEX (Burd and Dreyfuss 1994) to assign putative hnRNP A1–binding sites. We transfected the constructs into 293-HEK cells and measured the extent of exon 7 inclusion in the spliced transcripts. The minigene constructs (minigenes 1–30) are listed in table 1 in the same order as the 30 lanes in figure 1.
In the design of the first set of mutants, we reasoned that the gain of an element should be sequence specific, requiring particular nucleotides that match the consensus binding motif. In contrast, the loss of an element should not require highly specific sequence changes as long as the element is destroyed. We initially substituted 6 nt of the presumptive splicing elements with cytosines (minigene 3: SMN.cX6), thymidines (minigene 4: SMN.tX6), adenosines (minigene 5: SMN.aX6), or guanosines (minigene 6: SMN.gX6) (table 1). Three of these four mutations caused loss of exon 7 inclusion to levels similar to SMN2 (c6t) (fig. 1, lanes 4–6). The SMN.cX6 mutation was an exception (see below), since it only reduced inclusion by ∼15% (fig. 1, lane 3). Substitution with a more limited number of thymidines (minigene 7: SMN1.t3 and minigene 8: SMN1.t4) in the putative enhancer element, while maintaining the C at position +6, also led to very significant exon skipping (table 1 and fig. 1, lanes 7 and 8). These two mutants are equivalent to ones first used to identify the sequences surrounding position +6 as being important for efficient SMN1 exon 7 inclusion (Lorson and Androphy 2000). The ability to cause exon 7 skipping in such a nonspecific manner strongly suggests that the mutations disrupt a positively acting splicing element (i.e., an ESE) required for SMN1 exon 7 inclusion.
To assess the involvement of hnRNP A1 in exon 7 splicing, we next tested whether mutations that strengthen or weaken hnRNP A1–binding motifs correlate with the extent of exon 7 skipping. SMN1 and SMN2 have three and four nucleotide matches, respectively, to the optimal hnRNP A1–binding hexamer, TAGGGA/T (Burd and Dreyfuss 1994). We tested three pairs of SMN1/SMN2 minigenes with double mutations at positions +9 to +11 (10g11g, 9g10g, and 9t10g) combined with C or T at position +6 (minigenes 9–14, table 1 and fig. 1). Despite having at least the same number of matches to the consensus hnRNP A1 motif as SMN2, all but two of the mutations resulted in increased exon 7 inclusion, showing that the introduction of an hnRNP A1–binding motif does not consistently result in exon 7 skipping. In the remaining two cases, a perfect or near-perfect match at that site (i.e., minigene 12: SMN2.9g10g and minigene 14: SMN2.9t10g) was indeed associated with exon skipping, but because these mutations also simultaneously eliminate the SF2/ASF high-score motif, the cause of exon skipping cannot be unequivocally assigned (table 1 and fig. 1, lanes 12 and 14).
The presence of UAG at the 5′ end of the hnRNP A1 consensus motif appears to be important for hnRNP A1 binding and silencing activity in other contexts (Burd and Dreyfuss 1994; Caputi et al. 1999). To further evaluate whether the presence of an hnRNP A1–binding site correlates with SMN2 exon 7 skipping, we tested two sets of mutations (minigenes 25–28), 7c and 8c, that target the UAG/CAG motif in SMN1 and SMN2 and are equivalent to previously tested mutations (Kashima and Manley 2003). Both SMN1.7c and SMN1.8c maintain a high-score SF2/ASF motif and, as predicted by the ESE-loss model, gave high levels of exon 7 inclusion (fig. 1, lanes 25 and 27). SMN2.7c lacks both a high-score SF2/ASF motif and an hnRNP A1–binding motif but maintained a high level of exon 7 inclusion (lane 26). In SMN2.8c, the SF2/ASF motif was also eliminated, correlating with the ∼50% exon 7 skipping despite the absence of the UAG motif (lane 28). Indeed, when a UAG motif was present without the concomitant disruption of the ESE (lanes 10, 18, and 20), exon 7 inclusion was close to 100%, pointing to a marginal role of the UAG motif in this context.
Several mutants have good matches to both SF2/ASF and hnRNP A1 motifs (table 1, minigenes 9, 10, 17, 18, and 22) and gave high levels of exon 7 inclusion (fig. 1), suggesting that the SF2/ASF high-score motif is the functional or dominant element in determining exon 7 splicing. Two of these mutants are part of a set specifically designed to test this hypothesis, by inserting a CA dinucleotide upstream of the +6 C or T in SMN1 or SMN2, respectively (table 1, minigenes 15 and 16). The insertion, which leaves the preexisting SF2/ASF and hnRNP A1 putative sites unaltered, maintained the high level of exon 7 inclusion for SMN1 and partially improved it for SMN2 (fig. 1A, lanes 15 and 16). When combined with a 9g10a mutation in the context of SMN2 (table 1, minigene 18), the CA dinucleotide insertion restores a high-score SF2/ASF motif and simultaneously strengthens the hnRNP A1–binding motif. Despite the strengthened hnRNP A1 motif, exon 7 inclusion increased to nearly 100% (fig. 1, lane 18), indicating that the presence of an hnRNP A1 motif at that position contributes minimally to the levels of exon 7 skipping, whereas an SF2/ASF motif appears to be a strong determinant of inclusion.
Eight mutations had neither SF2/ASF motifs nor hnRNP A1 motifs (table 1, minigenes 3–5, 7, 8, 23, 26, and 28). Six of these eight mutations gave significant exon 7 skipping, consistent with the lack of the ESE motif (fig. 1, lanes 4, 5, 7, 8, 23, and 28). The other two mutations, SMN2.7c and, to a lesser degree, SMN.cX6, gave levels of exon 7 inclusion comparable to SMN1 (lanes 3 and 26). Given that exon 7 inclusion for the other mutations does not correlate with the lack of an hnRNP A1–binding site, we infer that 7c and cX6 may fortuitously introduce an unrelated protein-binding site and/or affect RNA secondary structure in a way that facilitates splicing.
We also tested an SF2/ASF–dependent ESE found in BRCA1 exon 18, which is abrogated by a point mutation that causes skipping of the exon (Liu et al. 2001). In the context of the SMN minigene, the heterologous BRCA1 ESE stimulated exon 7 inclusion above SMN2 levels, whereas complete exon skipping was observed with the mutant version (table 1, minigenes 29 and 30, and fig. 1). Together with the reciprocal finding that the SMN1 SF2/ASF ESE, but not the SMN2 version, can replace the BRCA1 ESE (Cartegni and Krainer 2002), this result shows that different SF2/ASF ESE heptamers are functionally interchangeable.
Our ESE-loss model takes into account mutations that were tested in previous studies (Lorson and Androphy 2000; Cartegni and Krainer 2002; Kashima and Manley 2003) and that were also included in the present study to confirm previous results and to comprehensively reassess the roles of SF2/ASF and hnRNP A1. For example, one set of mutants (table 1, minigenes 19–24) was originally designed to eliminate the SF2/ASF–dependent ESE by introducing changes at position +6 and, then, to restore it by means of a compensatory mutation, a11g (Cartegni and Krainer 2002). In agreement with our previous observations, exon 7 inclusion was rescued (fig. 1, lanes 19–24). Two constructs, however, gave significantly different results compared with a previous report (Kashima and Manley 2003). In that study, SMN2-TG and SMN2-8C rescued SMN2 exon 7 inclusion to levels similar to SMN1. Our equivalent constructs, SMN2.9t10g and SMN2.8c, gave only 13% and 56% exon inclusion, respectively (table 1, minigenes 14 and 28, and fig. 1A), consistent with the reduction in the SF2/ASF motif scores (see the “Discussion” section).
It is apparent from the above results that not every mutant analyzed is consistent with the presence or absence of either an SF2/ASF or an hnRNP A1 motif. This is to be expected, because any point mutation can potentially create or destroy additional regulatory elements. However, the contribution of a particular motif to the recognition of exon 7 can be assessed by the extent of correlation between splicing and motif scores when the entire panel of mutants is analyzed. We, therefore, compiled the results from all of the mutations (table 1) into a data set presented as scatterplots (fig. 1B). The graphs show the relationship between exon 7 inclusion and either ESEfinder SF2/ASF motif scores (Cartegni et al. 2003) or matches to the hnRNP A1–binding site consensus (Burd and Dreyfuss 1994). The threshold for the hnRNP A1–binding motifs is set at more than three matches, because the transition from three matches in SMN1 to four matches in SMN2 was proposed to be the determinant of SMN2 exon 7 skipping (Kashima and Manley 2003). To set a high threshold for ESE activity, the extent of exon 7 inclusion considered indicative of high inclusion was set at 90%. According to the ESE-loss model, the data points should be distributed in the lower left and upper right quadrants, which was indeed the case for 26 of the 30 experimental points (fig. 1B). Moreover, there is a strong correlation between SF2/ASF scores and exon 7 inclusion (Spearman rank correlation coefficient 0.75; 95% CI 0.53–0.88; perfect correlation = 1) but not between hnRNP A1–binding motifs and exon 7 skipping (correlation coefficient −0.14; 95% CI −0.48 to 0.25; perfect correlation = −1). Indeed, the ESS-gain model predicts that the data points should be distributed in the lower right and upper left quadrants, but, instead, the distribution appears to be random (fig. 1B).
One caveat of this statistical analysis is that the SF2/ASF and hnRNP A1 motifs were originally derived by different methods, and they have different extents of degeneracy. To allow an unbiased comparison of the correlation between exon 7 inclusion and the two motifs, we derived a position weight matrix for hnRNP A1 from the original SELEX data (Burd and Dreyfuss 1994), using the same methodology as originally employed for the derivation of the SF2/ASF motif (Liu et al. 1998). The new hnRNP A1 matrix and the statistical analysis of the data are presented in appendix A and gave results consistent with the match/mismatch approach. Once again, there was no correlation between hnRNP A1 motif scores and the extent of exon 7 skipping (figs. A1 and A2). Taken together, these results strongly suggest that the presence of an SF2/ASF motif is important for exon 7 inclusion, whereas an hnRNP A1 motif is neither necessary nor sufficient to promote exon 7 skipping.
Effects of Depletion and Overexpression on SMN Splicing in Vivo
We have confirmed and extended previous work that demonstrated that simultaneous siRNA-mediated knockdown of hnRNP A1 and a closely related hnRNP A/B family member, hnRNP A2, significantly increases SMN2 exon 7 inclusion in HeLa cells (Kashima and Manley 2003) (fig. 3A and 3C). The original interpretation of these results was that hnRNP A1/A2 inhibits splicing of SMN2 exon 7 via an ESS introduced into SMN2 by the C→T transition. Although this is a formal possibility, we wished to address the specificity of the hnRNP A1/A2 effect and an alternative interpretation—namely, that nonspecific binding by hnRNP A1/A2 (or specific binding elsewhere) becomes inhibitory in the absence of the SF2/ASF–dependent ESE.
Figure 3.
Analysis showing that depletion of hnRNP A1/A2 in vivo relieves exon 7 skipping, irrespective of the C→T transition. A, HeLa cells were treated with siRNAs against hnRNP A1, A2, or both and then were transfected with SMN minigene plasmids. Endogenous levels of hnRNP A1 and A2 and tubulin control were measured by western blotting 24 h after transfection (upper images). Radioactive RT-PCR analysis was performed, and spliced products were visualized by ethidium-bromide staining (lower images). B, Sequences of the relevant portions of intron 6 (lowercase) and exon 7 (uppercase) in the minigenes. Mutations are underlined, and the −8 and +6 positions are boxed. C, RT-PCR products shown in panel A were quantitated on a phosphorimager. Note that the SMN1.t3 and SMN1.t4 mutants gave higher levels of inclusion in HeLa cells than in 293-HEK cells (compare with fig. 1A and table 1). D, Overexpression of SF2/ASF promotes exon 7 inclusion in SMN1.PyD. MEF cells were cotransfected with an SF2/ASF expression plasmid or vector alone (C, lane 1) and the indicated SMN minigenes. RT-PCR analysis and quantitation were performed as in panels A and C.
If the observed hnRNP A1/A2 inhibitory activity is general, rather than specific, it should repress exon 7 inclusion irrespective of the C→T transition. We, therefore, designed a mutation that weakens exon 7 inclusion in SMN1 without affecting the region of the SF2/ASF–dependent ESE. The mutation is a C→G transversion within the polypyrimidine tract of intron 6 (SMN1.PyD) (fig. 3B). This mutation weakens the 3′ splice site and decreases exon 7 inclusion by >50% (fig. 3A). If hnRNP A1–mediated repression is indeed dependent on the T at position +6, there should be no effect of hnRNP A1/A2 depletion on splicing of this minigene, because the C was retained and no ESS was introduced. However, hnRNP A1/A2 depletion resulted in an approximately twofold increase in exon 7 splicing with this mutant (SMN1.PyD) (fig. 3A, lanes 1–4, and fig. 3C).
We next analyzed two additional mutations, SMN1.t3 and SMN1.t4, which have a C at position +6 of exon 7 but have neither SF2/ASF nor hnRNP A1 motifs (table 1). As predicted by the ESE-loss model, exon 7 inclusion levels were low with both mutants. Upon hnRNP A1 and A2 depletion, however, inclusion increased approximately twofold (fig. 3A and 3C), supporting the conclusion that the hnRNP A/B–mediated inhibition of exon 7 inclusion is an indirect effect, independent of the C→T transition in SMN2 and consistent with the general inhibitory properties of hnRNP A/B proteins (Mayeda et al. 1993; Cartegni et al. 2002; Black 2003).
The SMN1.PyD mutant splices at suboptimal levels but, at the same time, maintains an intact, responsive ESE and, therefore, can be used to test whether SF2/ASF can promote exon 7 inclusion in cells. We overexpressed SF2/ASF in MEF cells, which have low basal levels of endogenous SF2/ASF, compared with transformed cell lines. As expected, there was no significant improvement in splicing with the SMN1 and SMN2 minigenes, since SMN1 exon 7 inclusion is already very efficient and SMN2 lacks the SF2/ASF motif (fig. 3D). In contrast, the level of exon 7 inclusion from the SMN1.PyD mutant increased about twofold, indicating that the ESE is responsive to varying levels of SF2/ASF in vivo.
If hnRNP A1 specifically inhibits exon 7 inclusion in SMN2, then hnRNP A1 overexpression would be expected to inhibit inclusion even further, whereas SMN1 should not be affected. In contrast, if hnRNP A1–mediated inhibition is a global and/or nonspecific effect, its overexpression should also cause an increase in SMN1 exon 7 skipping. To determine which prediction is correct, we transiently overexpressed hnRNP A1 in 293-HEK cells and analyzed the splicing of RNA transcripts from the endogenous SMN1 and SMN2 genes. The endogenous SMN2 gene can be distinguished from SMN1 by a G→A transition in the 3′ UTR in exon 8 that creates a DdeI site (Gennarelli et al. 1995; Parsons et al. 1996). Analysis of DdeI-digested RT-PCR reactions showed that the SMN1 cDNAs from control cells correspond to essentially 100% inclusion of exon 7 (fig. 4, lane 1). In contrast, overexpression of hnRNP A1 resulted in the appearance of SMN1 transcripts lacking exon 7 (lane 2), indicating that hnRNP A1 can inhibit endogenous SMN1 exon 7 splicing. An increase in exon 7 skipping was also observed for the SMN2 transcripts.
Figure 4.
Analysis showing that overexpression of hnRNP A1 in vivo inhibits SMN1 and SMN2 exon 7 inclusion. Top, Radioactive RT-PCR analysis was performed with RNA from control mock-transfected 293-HEK cells (C, lane 1) or cells expressing T7-tagged hnRNP A1 (lane 2). To distinguish between SMN1 and SMN2 endogenous mRNAs, the cDNAs were digested with DdeI to specifically cleave SMN2 in exon 8. Products were separated on a 6% native polyacrylamide gel. The white box represents the 3′ end of exon 8 that was cleaved from SMN2 spliced products. The proportion of SMN1 and SMN2 transcripts lacking exon 7 (% skip) is shown below the autoradiogram. Bottom, Western blot of lysates from mock-transfected cells or cells expressing T7-tagged hnRNP A1. Blots were probed with anti-T7 antibody and an antibody against U2B′′ as a control.
Inhibition of SMN1 and SMN2 Exon 7 Splicing by hnRNP A1 in Vitro
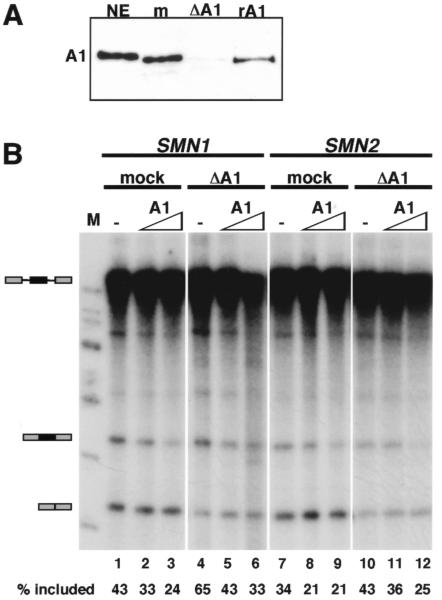
The role of hnRNP A1 in SMN1 and SMN2 exon 7 splicing can be tested directly with the use of an in vitro splicing assay (Cartegni and Krainer 2002). SMN1 exon 7 inclusion is less efficient in vitro than in vivo, facilitating the detection of changes that lead to increased exon inclusion. Indeed, depletion of hnRNP A/B proteins from nuclear extracts (fig. 5A) resulted in a significant increase in SMN1 exon 7 inclusion, relative to mock-depleted extracts (fig. 5B, lanes 1 and 4). This effect was even greater than that on SMN2 (fig. 5B, lanes 7 and 10). The addition of recombinant hnRNP A1 resulted in dose-dependent inhibition of both SMN1 and SMN2 exon 7 inclusion (fig. 5B, lanes 4–6 and 10–12). Because SMN1 exon 7 inclusion was also inhibited, these results further demonstrate that the hnRNP A1–mediated effect is independent of the C→T transition in SMN2.
Figure 5.
Analysis showing that hnRNP A1 inhibits SMN1 and SMN2 exon 7 splicing in vitro. A, Western blot of hnRNP A1 in 1 μl of HeLa nuclear extract (NE), mock-depleted extract (m), extract depleted of hnRNP A/B proteins (ΔA1), and 50 ng of recombinant hnRNP A1 (rA1). B, In vitro splicing of SMN1 and SMN2 transcripts in 3 μl of mock- or hnRNP A/B–depleted extract supplemented with 0 (−), 170, or 340 ng of recombinant hnRNP A1. Marker lane is indicated (M).
Discussion
The SMN2 gene can potentially express sufficient functional SMN protein to fully compensate for the loss of SMN1 in all patients with SMA, which makes it an attractive target for drug therapy. The development of therapeutic approaches would be greatly facilitated by precise knowledge of the mechanism by which SMN2 exon 7 fails to be efficiently incorporated into mature transcripts. The identification of the cis-elements and trans-acting factors involved is essential for the optimization of current approaches and/or for the identification of new drug targets. Two different models, which are not necessarily incompatible, were previously proposed to explain the different splicing patterns of SMN1 and SMN2: loss of an SF2/ASF–dependent enhancer in SMN1 (Cartegni and Krainer 2002) or gain of an hnRNP A1–dependent silencer in SMN2 (Kashima and Manley 2003). We have now systematically assessed the roles of hnRNP A1 and SF2/ASF in SMN1/2 exon 7 splicing and conclude that the primary determinant of the extent of exon 7 inclusion is the SF2/ASF–dependent ESE. Interestingly, hnRNP A1 can antagonize SF2/ASF and, indeed, promote exon 7 skipping. However, this activity of hnRNP A1 is not specific to SMN2 but rather reflects the general inhibitory effect of hnRNP A1 on many alternatively spliced exons (reviewed in Cartegni et al. 2002), illustrating the antagonism between these two splicing effectors in a physiological context.
We have confirmed the loss of a putative ESE motif in SMN2 predicted by ESEfinder (Cartegni et al. 2003), using a very different ESE-prediction program, RESCUE-ESE (Fairbrother et al. 2004). However, the presence of a bona fide hnRNP A1 ESS motif in SMN2 remains uncertain. Two methods to identify ESS elements (Wang et al. 2004; Zhang and Chasin 2004) do not predict the creation of a silencer by the c6t transition (data not shown). Kashima and Manley (2003) compared the sequence of SMN2 with known or putative hnRNP A1–dependent ESSs from four genes. They noted that the sequence of SMN2 exon 7 at positions 6–11 is identical to CD44 (Entrez Gene accession number 960) exon v5 at positions 4–9 and that both have some resemblance to the hnRNP A1 SELEX motif (Burd and Dreyfuss 1994). In CD44, several linker-scan mutants throughout the entire exon v5, including one that overlaps the UAGACA sequence, result in higher levels of exon v5 inclusion (König et al. 1998). Later, it was shown that the activity of a dispersed silencer within exon v5 is controlled by hnRNP A1, but this ESS appears to be a composite of multiple elements located in subdomains throughout the exon, and no specific high-affinity binding site could be identified (Matter et al. 2000). Therefore, we cannot infer that positions 4–9 in CD44 exon v5 and, by analogy, positions 6–11 in SMN2 define an hnRNP A1–binding site, especially considering that this particular sequence fails to inhibit splicing when analyzed in a heterologous context (Kashima and Manley 2003). Similar to the situation in CD44 exon v5, we found that hnRNP A1 plays a role in SMN2 exon 7 skipping but does so in a general manner, perhaps dependent on other exon features or on the surrounding intronic regions rather than on the proposed hnRNP A1 motif.
In contrast to the general inhibitory effect of hnRNP A1 on CD44 and SMN2 splicing, bona fide examples of hnRNP A1–specific ESS elements have been identified (Cartegni et al. 2002). One such element, ESS3, is found in HIV-1 tat exon 3 (Amendt et al. 1995; Staffa and Cochrane 1995; Si et al. 1998; Tange et al. 2001; Zhu et al. 2001; Damgaard et al. 2002; Marchand et al. 2002). ESS3 comprises the sequence UAGUGA, which matches the high-affinity hnRNP A1–binding hexamer at five positions. This motif is required for direct interaction with hnRNP A1 (Zhu et al. 2001) and for hnRNP A1–mediated inhibition of splicing in vivo (Staffa and Cochrane 1995) and in vitro (Si et al. 1998; Zhu et al. 2001; Damgaard et al. 2002). One of the SMN2 exon 7 mutants tested by Kashima and Manley (2003), SMN2-TG, introduces a sequence identical to the tat ESS3 motif. Despite the fact that this mutation creates a nearly perfect match to an hnRNP A1–binding site that is a proven hnRNP A1–dependent ESS in its original context, those authors reported that this mutation rescues exon 7 inclusion. When we analyzed an equivalent mutant, SMN2.9t10g (fig. 1), we found that this mutation resulted in exon 7 skipping, a finding consistent with the introduction of a genuine hnRNP A1–mediated silencer element in this case and also with the concomitant disruption of the SF2/ASF motif.
The most compelling previous evidence supporting a direct role for hnRNP A1 in SMN2 exon 7 skipping was the activation of exon 7 inclusion upon transient knockdown of hnRNP A1/A2 proteins by siRNAs (Kashima and Manley 2003). We confirmed this result, but when we included controls for specificity, using three different mutants (SMN1.PyD, SMN.t3, and SMN.t4) that result in exon 7 skipping without introducing an hnRNP A1 motif, we found that knocking down hnRNP A1/A2 led to a comparable increase in exon 7 inclusion in all cases (fig. 3). We conclude that repression of exon 7 inclusion by hnRNP A1/A2 is a generalized effect, independent of the C→T transition. Thus, most of the results in the Kashima and Manley (2003) report can be explained by the general property of hnRNP A1 to promote skipping of alternative exons (Mayeda et al. 1993), rather than by an SMN2-specific effect.
On the basis of our present results and of previous work, we propose a new model for SMN1 and SMN2 exon 7 splicing, which is depicted in figure 6. According to this model, hnRNP A1 acts in a general manner to block splicing of exon 7 in the absence of the dominant SF2/ASF–dependent ESE (fig. 6B). Consistent with the known RNA-binding and repressor properties of hnRNP A1 (Zhu et al. 2001; reviewed by Cartegni et al. [2002]), its binding to SMN1 and SMN2 RNAs may nucleate from as-yet-uncharacterized high-affinity sites at some distance from the ESE. The presence of the SF2/ASF–dependent ESE in SMN1 supersedes this inhibition (fig. 6A). The same is true when exon 7 definition in the SMN2 context is strengthened by various means, such as the reconstitution of a new ESE at a similar position (fig. 6C), the overexpression of proteins that bind to other enhancer elements in exon 7 (fig. 6D), the targeting of synthetic molecules that mimic SR protein functions—for example, ESSENCE (Cartegni and Krainer 2003) and TOES (Skordis et al. 2003) (fig. 6F)—or the reduction in the levels of inhibitory factors by siRNA treatment (fig. 6E).
Figure 6.
Models for the effects of the SMN2 C→T transition on exon 7 splicing. Multiple positive and negative determinants influence SMN1 and SMN2 splicing. In SMN1 (A), efficient splicing is driven by the interactions of SF2/ASF with the ESE in the +6c region and of other factors with a separate enhancer downstream. There may also be an interplay between the various elements and additional, perhaps redundant, factors (“SR?”) might be involved. In this strong exon definition context, a possible hnRNP A1/A2 role and/or the action of a putative repressor (“R?”), are not sufficient to inhibit splicing. The loss of the SF2/ASF–dependent ESE in SMN2 (B) allows a secondary, non–SMN2-specific inhibitory action of hnRNP A1/A2, possibly in collaboration with “R?”. Positive interactions downstream cannot counteract this inhibitory activity. Exon 7 inclusion in SMN2 can be rescued to SMN1-like levels by other means (white arrows), such as reconstitution of the SF2/ASF–dependent ESE (or other ESEs) (C), overexpression of some splicing activators (e.g., Tra2β1) (D), down-regulation of hnRNP A1/A2 by RNAi (E), or the presence of compounds that functionally replace SR proteins or ESEs (e.g., ESSENCE or TOES) (F). Fluctuations in different cells and tissues in the relative levels of SF2/ASF, hnRNP A1/A2, and/or the additional splicing factors involved would therefore determine the precise pattern of SMN2 exon 7 splicing. Green boxes represent known ESEs. Red and orange boxes represent putative ESS and ISS elements, respectively.
Other splicing factors, such as Tra2β1, SRp30c, and hnRNP G (UniProt accession numbers P62995-1, Q13242, and P38159, respectively), have also been shown to influence SMN1 and SMN2 exon 7 inclusion via elements in the central portion of exon 7 (Hofmann et al. 2000; Hofmann and Wirth 2002; Young et al. 2002), and additional regulatory elements, both in exon 7 (Singh et al. 2004a, 2004b) and in the flanking introns (Miyajima et al. 2002; Miyaso et al. 2003), appear to be important for exon 7 recognition (fig. 6). Because SMN1 and SMN2 exon 7 sequences are identical, with the exception of the C→T transition at position +6, regulatory sequences that do not encompass this position are unlikely to be directly involved in the SMN2 exon 7 splicing defect. However, they clearly play an important role in exon 7 splicing efficiency and may influence the overall effect of the C→T transition.
It is possible that another repressor (“R?,” fig. 6) different from hnRNP A1/A2 recognizes SMN2 sequences with some degree of specificity. In support of this notion, a recent study showed that, in the absence of the ESE, several mutations in the first 5 nt (GGTTT) of SMN2 exon 7 partially rescue splicing (Singh et al. 2004a), and this motif is also present in 11 of 141 recently described ESS decamer elements (Wang et al. 2004). However, this putative ESS at the beginning of the exon is not sufficient to overcome the effect of the SF2/ASF ESE (fig. 1 and table 1). Consistent with this finding, our reanalysis of the extensive mutational and iterative selection data of Singh and colleagues (2004a, 2004b) shows a good correlation between exon 7 inclusion and the presence of an SF2/ASF ESE motif (fig. A3). In particular, in the functional SELEX data obtained by randomization of the first 6 nt of exon 7 (Singh et al. 2004a), SF2/ASF–dependent ESE motifs are present in 33 of the 42 selected clones (fig. A3). Likewise, partial randomization of the entire exon (Singh et al. 2004b) resulted in high-score SF2/ASF motifs at the original position (6–12) in 25 of the 59 selected exons and in 40 of the 59 exons when partial overlaps with the original position were considered (data not shown). Because only ∼4% of all possible 16,384 heptamers represent high-score SF2/ASF motifs (Wang et al. 2005), it is clear that these motifs are greatly enriched among sequences selected for their ability to promote exon 7 inclusion, thus strongly reinforcing the notion that the SF2/ASF–dependent ESE is the primary splicing determinant in this region and that the activity from any putative inhibitory sites is secondary to the loss of the ESE.
Previous negative results from in vivo SF2/ASF–depletion experiments suggested that SF2/ASF is not strictly required for exon 7 inclusion in SMN1. The depletion of SF2/ASF from cells, either genetically in the chicken cell line DT-40 (Kashima and Manley 2003) or by use of siRNA-mediated knockdown in HeLa cells (L. Cartegni, M. L. Hastings, J. A. Calarco, E. de Stanchino, and A. R. Krainer, unpublished data), has little effect on SMN1 splicing. Although a decrease in SMN1 exon 7 inclusion in the absence of SF2/ASF is a logical expectation, there are several possible alternative explanations for the lack of such an effect. First, SF2/ASF, which is a general splicing factor, may be important for both exon 8 and exon 7 splicing. There are six high-score SF2/ASF motifs in exon 8 (data not shown) and, if any of them represent genuine ESEs, the depletion of SF2/ASF may equalize the competition between the 3′ splice sites preceding these two exons, thereby resulting in no effect (or even in a positive effect) on overall exon 7 inclusion. Indeed, blocking the 3′ splice site of exon 8 with antisense oligonucleotides increases the extent of exon 7 inclusion (Lim and Hertel 2001). Second, because the depletion of SF2/ASF compromises DT-40 cell viability (Wang et al. 1996), this approach may not provide a direct indication of normal SF2/ASF activity, since secondary effects associated with cellular stress and apoptosis are likely in play. Third, SR proteins are well known to have partially redundant functions (reviewed by Cartegni et al. [2002]). As previously suggested (Cartegni and Krainer 2002), additional SR protein(s) may recognize the ESE in the absence of SF2/ASF and substitute for it in ESE-dependent exon 7 splicing, so that a double depletion might be required to observe an effect. By analogy, in the case of hnRNP A1, the simultaneous depletion of the related protein hnRNP A2 is required to observe a strong effect on SMN2 exon 7 inclusion (fig. 3a) (Kashima and Manley 2003). Fourth, compensatory mechanisms might also be triggered or enhanced through auto- and cross-regulatory loops involving different splicing factors and their respective alternative splicing isoforms; many examples of such regulatory loops have been described elsewhere (reviewed by Bourgeois et al. [2004]). Thus, a reduction in the levels of SF2/ASF could directly or indirectly lead to changes in the levels of other splicing factors, which would, in turn, maintain efficient SMN1 exon 7 inclusion.
Any of these scenarios, or a combination of them, could account for the absence of an apparent effect of SF2/ASF depletion on SMN2 splicing. On the other hand, there is substantial evidence supporting a direct role for SF2/ASF in SMN1 exon 7 splicing. Mutational analysis shows a strong correlation between exon 7 inclusion and the presence of an ESE-dependent SF2/ASF motif (fig. 1), which also enhances splicing in a heterologous context (Cartegni and Krainer 2002). SF2/ASF preferentially interacts with SMN1 compared with SMN2 transcripts (fig. 2) and makes direct contact with the ESE site (Cartegni and Krainer 2002). SF2/ASF overexpression can promote SMN1 exon 7 inclusion in a weakened context that preserves the ESE (fig. 3D), and SF2/ASF, but not another SR protein, SC35, specifically activates SMN1 exon 7 splicing in an in vitro S100-complementation assay (Cartegni and Krainer 2002). As is the case for many genes (reviewed by Cartegni et al. [2002] and Black [2003]), SF2/ASF–dependent splicing is counteracted by hnRNPA1/A2 in an antagonistic relationship that is likely physiologically relevant for the modulation of SMN splicing as well.
Our results from overexpression of hnRNP A1 (fig. 4) indicate that changes in the cellular levels of hnRNP A1 can modulate the extent of exon 7 inclusion in both SMN1 and SMN2. Likewise, SF2/ASF overexpression experiments (fig. 3D) showed that exon 7 inclusion is sensitive to the cellular levels of SF2/ASF. These results suggest that exon 7 inclusion is determined in part by the cellular levels of these two proteins, which have opposite effects on splicing. The balance between SF2/ASF and hnRNP A1 activity in exon 7 inclusion appears to hinge upon the recognition of the ESE in exon 7 by SF2/ASF, as inactivation of this ESE in SMN2 results in the substantial loss of exon 7 inclusion. This hypothesis predicts that, in the absence of hnRNP A1, the ESE should become dispensable for the splicing of exon 7, which is, indeed, the case (figs. 3 and 5). Because we find no evidence that hnRNP A1 is acting via a specific binding site created in SMN2, we presume that hnRNP A1 inhibition of exon 7 splicing is mediated by as-yet-undetermined splicing silencer sequences. Putative high-affinity binding sites in and around exon 7, calculated with a newly derived hnRNP A1 matrix, are shown in appendix A (figs. A1 and A5).
The reason why SMN1 gene disruption causes the selective degeneration of spinal cord motor neurons without affecting other tissues is not known. A threshold level of SMN protein is essential for cell viability (reviewed by Frugier et al. [2002]) and, clearly, the low level of full-length transcripts produced from SMN2 is sufficient for the survival of most cell types in patients with SMA and mouse models. It is possible that motor neurons have a greater need for the SMN protein or that SMN performs additional, neuron-specific functions (reviewed by Sendtner [2001] and Gubitz et al. [2004]). Alternatively, SMN2 may produce less functional SMN protein in spinal motor neurons compared with other cells, which would only be detrimental in affected SMA individuals (Coovert et al. 1997). One intriguing possibility is that α-motor neurons express high levels of hnRNP A/B proteins and/or low levels of SF2/ASF or other relevant splicing factors, resulting in increased skipping of SMN2 exon 7. Although there is no direct evidence that this is, in fact, the case for motor neurons and SMA, the relative levels of hnRNP A1/A2, SF2/ASF, and other SR proteins do vary with cell type and developmental stage (Hanamura et al. 1998; reviewed by Dreyfuss et al. [2002]), and our results demonstrate that SF2/ASF and hnRNP A1 levels can determine the extent of exon 7 splicing.
Any treatment that improves the ratio of SMN full-length to SMNΔ7 from SMN2 might have therapeutic value, but targeting splicing factors could be problematic. For example, hnRNP A1 could be viewed as a possible therapeutic target, given its inhibitory effect on SMN2 exon 7 splicing (Kashima and Manley 2003). However, in light of its properties as a global splicing inhibitor and its involvement in many other cellular processes (reviewed by Dreyfuss et al. [2002]), targeting hnRNP A1 could lead to undesirable effects. Likewise, most SR proteins probably regulate the alternative splicing of numerous genes, and it may be difficult to identify small molecules that modulate their effects with a high degree of specificity and/or without associated toxicity. Nevertheless, drugs with even limited specificity could provide a favorable risk/benefit ratio, especially considering the severity of SMA and some of the other disorders associated with defective splicing. The recent identification of small-molecule inhibitors specific for individual SR proteins provided the first indication that this family of proteins can be targeted pharmacologically (Soret et al. 2005). Gene-specific strategies that rely on antisense base pairing to increase SMN2 exon 7 inclusion provide an intrinsically higher degree of specificity, although they face their own unique obstacles.
Mutations in cis-elements that result in splicing alterations are commonly observed (reviewed by Cáceres and Kornblihtt [2002], Faustino and Cooper [2003], and Pagani and Baralle [2004]). However, how such mutations exert their effects is not always obvious. Because cis-acting mutations that affect splicing may result in gain of function (e.g., creation of an ESE or ESS element), loss of function (e.g., destruction of an ESE or ESS element), or even a combination of both, in addition to potential alterations in RNA secondary structure (reviewed by Buratti and Baralle [2004]), a wide variety of approaches should be employed, whenever possible, to elucidate the underlying mechanisms.
Our findings demonstrate the complexity of the relationship between positive and negative splicing elements and the factors that recognize them, and they underscore the importance of exon identity determinants for both constitutive and alternative splicing. The results also illustrate the need for a thorough understanding of splicing mechanisms, to facilitate accurate phenotypic risk assessment, and for the development of effective therapeutic treatments for human genetic diseases.
Acknowledgments
We thank Ravi Sachidanandam for helpful advice on statistical tests. This work was supported by National Institutes of Health grant NS041621 (to A.R.K.). L.C. is supported by the W. Hearst Foundation.
Appendix A
hnRNP A1 Weight Matrix
A nonredundant set of 18 sequences from the original hnRNP A1 SELEX “winners” that Burd and Dreyfuss (1994) used to identify a consensus high-affinity binding site was realigned using Gibbs sampler, as described elsewhere (Liu et al. 1998). The resulting alignment (identical to the manual alignment used by Burd and Dreyfuss [1994]) was used to derive a position weight matrix for the hnRNP A1 motif, as was done elsewhere for the SF2/ASF functional SELEX winner sequences (Liu et al. 1998).
Figure A1 shows the matrices and the corresponding pictogram representations, with or without background correction (Pictogram). The background composition of the initial RNA library used for SELEX was not reported, so we derived a matrix by assuming a neutral initial composition (panel a, left matrix: A1 25%_bckgr, A: 25%; T: 25%; C: 25%; and G: 25%) or another matrix corrected for the compositional bias of the set of 18 nonredundant winner sequences, as described elsewhere (Liu et al. 1998) (panel a, right matrix: A1 Win_bckgr, A: 31.6%; T: 24.5%; C: 8.7%; and G: 35.2%). The two versions of the matrix are very similar.
Both versions of the position weight matrix were used to calculate hnRNP A1 motif scores for the 30 SMN mutants described in this article (table 1 and fig. 1). The values are shown in figure A2g and A2h, together with the number of “matches” to the canonical A1 consensus motif TAGGGA/T (fig. A2f) and with the ESEfinder 2.0 scores for SF2/ASF (fig. A2d, normalized to the initial pool composition). For unbiased comparison with figure A2g, SF2/ASF scores were also calculated using compositional background correction based on the SF2/ASF winner pool (SF2/ASF Win_bckgr, A: 21.4%; T: 16.4%; C: 23.7%; G: 38.5%) (Liu et al. 1998). The two background correction methods yield very similar results for SF2/ASF, and we therefore assume, by analogy, that figure A2g is the best available approximation in the case of hnRNP A1.
The data from figure A2c versus A2d–A2h) are presented as scatterplots in figure A4a–A4e. Figure A4a and A4c correspond to the scatterplots in figure 1 of the main article. Although the absolute numerical values differ, the distribution of hnRNP A1 motif scores versus SMN exon 7 inclusion is remarkably similar when either matrix is used (see fig. A2g and A2h and fig. A4d and A4e), and it does not show any significant correlation, similar to what we observed when using the match/mismatch approach described in the main article (fig. A2f and fig. A4c). In contrast, the observed positive correlation between SF2/ASF ESEfinder scores and SMN exon 7 inclusion is statistically significant, regardless of the background correction procedure used (fig. A2d and A2e and fig. A4a and A4b).
hnRNP A1 Motif Distribution
The position weight matrix above (adjusted for the composition of the hnRNP A1 SELEX winner pool) was used to find putative hnRNP A1–binding sites. Figure A5 shows all the sites with positive scores within a region comprising the last 2,000 nt of SMN2. At the present time, there is not enough information to define a threshold score that separates high- and low-affinity sites. However, as demonstrated by this work, four matches to the original consensus motif (corresponding to the SMN2 exon 7 score of 2.62) is not sufficient to mediate silencing, at least in the context of SMN minigenes (fig. 1). The lowest possible score of a 5/6 match to the original consensus motif is, according to our matrix, 3.43. This value is, therefore, used as a reference (red horizontal line in fig. A5), although its relevance as a threshold is not proven. The highest possible score is 6.609, corresponding to the hexamer TAGGGA.
There are many putative hnRNP A1 low-affinity sites around and within SMN1 and SMN2 exon 7 and a few that might function as high-affinity sites. Interestingly, two such putative high-affinity sites overlap the 3′ and 5′ splice sites, raising the possibility that the inhibitory action of hnRNP A1 on the splicing of both SMN1 and SMN2 pre-mRNAs is the combined result of nucleation from adjacent sites (as depicted in fig. 6) and direct competition with spliceosomal components for binding to splice-site sequences. The resemblance between the hnRNP A1 motif and parts of the consensus splice-site motifs was noted elsewhere (Burd and Dreyfuss 1994).
Figure A1.
Weight matrices and pictograms representing the relative frequency of nucleotides in the hnRNP A1 high-affinity motif derived by Burd and Dreyfuss (1994). a, Weight matrix values derived from the nonredundant set of hnRNP A1 SELEX winner motifs, calculated as by Liu et al. (1998). Positive values are in bold. b, Pictogram representations of the two versions of the hnRNP A1 motif. The height of each letter represents the relative frequency of the corresponding nucleotide at that position; colored and gray letters indicate higher-than-background and lower-than-background frequencies, respectively. T is used instead of U, for convenience.
Figure A2.
SF2/ASF and hnRNP A1 matrix scores. SF2/ASF scores were calculated using ESE finder 2.0 (a) or after adjustment for the compositional bias of the functional SELEX SF2/ASF winner pool (b). hnRNP A1 scores were calculated either as matches to the Burd and Dreyfuss consensus (c) or by use of the position weight matrices shown in figure A1a, which were adjusted for the compositional bias of the hnRNP A1 SELEX winner pool (d) or were assumed to have a neutral composition (e). In each case, the highest scoring value for each sequence is shown. SMN1 constructs have a C, and SMN2 constructs a blue T, at position +6. Mutations are indicated in red lowercase, and insertions are indicated in green lowercase. Statistical analysis was performed using Prism 4.0 (GraphPad software) with the settings given in the table. a and c, Scores corresponding to the scores given in table 1 in the main article.
Figure A3.
Analysis of the SMN exon 7 mutations described by Singh et al. (2004a). a, 66 independent mutants described by Singh et al. (2004a). Mutants 25–66 are SELEX winner clones, of which 33/42 maintain a high-score SF2/ASF motif (30 at the original position). These mutations impinge only on the first position of the original SF2/ASF heptamer, where either C or G maintains a high-score motif, and, therefore, only 21/42 clones would be expected to retain high scores at the original position by chance; the calculated binomial probability for ⩾30 clones is P<.004. b, Nucleotide sequence around the ESE/ESS motifs. The ESEfinder 2.0 SF2/ASF top-scoring motif for each sequence is shaded. The best fit to the hnRNP A1–binding consensus (TAGGGA/T) is underlined. SMN1 constructs have a C, and SMN2 constructs a blue T, at position +6. Mutations are indicated in red lowercase. Insertions are indicated in italics. c, Top SF2/ASF score. High-score motifs are shown in bold (threshold = 1.96). d, Number of nucleotide matches to the hnRNP A1 hexamer consensus, shown in bold when >3. e, Average inclusion percentages. w = SELEX winner sequence not directly tested (all 14 tested winner sequences show >94% inclusion [Singh et al. 2004a]).
Figure A4.
Correlation between exon 7 inclusion and SF2/ASF or hnRNP A1 motifs. Scores derived from figure A3 are plotted against the percentage of SMN exon 7 inclusion for the 30 mutants (fig. 1), by use of the different scoring methodologies: a, SF2/ASF ESEfinder 2.0 scores; b, SF2/ASF matrix corrected for the winner pool background; c, matches to the hnRNP A1 consensus TAGGGA/T; d, hnRNP A1 matrix corrected for the winner pool background; e, hnRNP A1 matrix with the assumption of neutral background. The data points for SMN1 and SMN2 in each plot are indicated.
Figure A5.
Putative hnRNP A1 low- and high-affinity sites. The orange boxes represent hnRNP A1 positive scores within the last 2,000 nt of SMN2; the height of each box indicates the motif score (calculated using the composition-adjusted matrix in fig. A1), and the width shows the position of the hexamer motif along the sequence. The yellow box indicates the score of the low-affinity site in SMN1. The green boxes indicate the position of SMN2 exons, and the red line indicates the minimum value (3.43) of a 5/6 match to the TAGGGA/T consensus sequence.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Entrez Gene, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene/ (for SMN1, SMN2, and CD44)
- ESEfinder 2.0, http://rulai.cshl.edu/tools/ESE/ (for identifying putative SR protein–dependent ESEs and calculating motif scores)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SMA types I, II, and III)
- Pictogram, http://genes.mit.edu/pictogram.html/ (for pictogram representations of position weight matrices)
- RESCUE-ESE, http://genes.mit.edu/burgelab/rescue-ese/ (for identifying candidate ESEs)
- UniProt, http://pir.uniprot.org/index.shtml/ (for SMN, SMNΔ7, hnRNP A1, hnRNP A2, SF2/ASF, Tra2β1, SRp30c, and hnRNP G)
References
- Amendt BA, Si ZH, Stoltzfus CM (1995) Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol Cell Biol 15:4606–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72:291–336 10.1146/annurev.biochem.72.121801.161720 [DOI] [PubMed] [Google Scholar]
- Bourgeois CF, Lejeune F, Stévenin J (2004) Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog Nucleic Acid Res Mol Biol 78:37–88 [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE (2004) Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol 24:10505–10514 10.1128/MCB.24.24.10505-10514.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G (1994) RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J 13:1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Kornblihtt AR (2002) Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet 18:186–193 10.1016/S0168-9525(01)02626-9 [DOI] [PubMed] [Google Scholar]
- Cáceres JF, Stamm S, Helfman DM, Krainer AR (1994) Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265:1706–1709 [DOI] [PubMed] [Google Scholar]
- Caputi M, Mayeda A, Krainer AR, Zahler AM (1999) hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J 18:4060–4067 10.1093/emboj/18.14.4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3:285–298 10.1038/nrg775 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 30:377–384 10.1038/ng854 [DOI] [PubMed] [Google Scholar]
- ——— (2003) Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol 10:120–125 10.1038/nsb887 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 31:3568–3571 10.1093/nar/gkg616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH (1997) The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet 6:1205–1214 10.1093/hmg/6.8.1205 [DOI] [PubMed] [Google Scholar]
- Damgaard CK, Tange TO, Kjems J (2002) hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA 8:1401–1415 10.1017/S1355838202023075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM (1999) Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev 13:1102–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N (2002) Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3:195–205 10.1038/nrm760 [DOI] [PubMed] [Google Scholar]
- Eperon IC, Makarova OV, Mayeda A, Munroe SH, Cáceres JF, Hayward DG, Krainer AR (2000) Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol Cell Biol 20:8303–8318 10.1128/MCB.20.22.8303-8318.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Yeo GW, Yeh R, Goldstein P, Mawson M, Sharp PA, Burge CB (2004) RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res 32:W187–W190 10.1093/nar/gnh176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA (2003) Pre-mRNA splicing and human disease. Genes Dev 17:419–437 10.1101/gad.1048803 [DOI] [PubMed] [Google Scholar]
- Frugier T, Nicole S, Cifuentes-Diaz C, Melki J (2002) The molecular bases of spinal muscular atrophy. Curr Opin Genet Dev 12:294–298 10.1016/S0959-437X(02)00301-5 [DOI] [PubMed] [Google Scholar]
- Gennarelli M, Lucarelli M, Capon F, Pizzuti A, Merlini L, Angelini C, Novelli G, Dallapiccola B (1995) Survival motor neuron gene transcript analysis in muscles from spinal muscular atrophy patients. Biochem Biophys Res Commun 213:342–348 10.1006/bbrc.1995.2135 [DOI] [PubMed] [Google Scholar]
- Gubitz AK, Feng W, Dreyfuss G (2004) The SMN complex. Exp Cell Res 296:51–56 10.1016/j.yexcr.2004.03.022 [DOI] [PubMed] [Google Scholar]
- Habets WJ, Hoet MH, De Jong BA, Van der Kemp A, Van Venrooij WJ (1989) Mapping of B cell epitopes on small nuclear ribonucleoproteins that react with human autoantibodies as well as with experimentally-induced mouse monoclonal antibodies. J Immunol 143:2560–2566 [PubMed] [Google Scholar]
- Hanamura A, Cáceres JF, Mayeda A, Franza BR Jr, Krainer AR (1998) Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4:430–444 [PMC free article] [PubMed] [Google Scholar]
- Hastings ML, Krainer AR (2001) Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol 13:302–309 10.1016/S0955-0674(00)00212-X [DOI] [PubMed] [Google Scholar]
- Hockensmith JW, Kubasek WL, Vorachek WR, Evertsz EM, von Hippel PH (1991) Laser cross-linking of protein-nucleic acid complexes. Methods Enzymol 208:211–236 [DOI] [PubMed] [Google Scholar]
- Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B (2000) Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2). Proc Natl Acad Sci USA 97:9618–9623 10.1073/pnas.160181697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann Y, Wirth B (2002) hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum Mol Genet 11:2037–2049 10.1093/hmg/11.17.2037 [DOI] [PubMed] [Google Scholar]
- Jarecki J, Chen X, Bernardino A, Coovert DD, Whitney M, Burghes A, Stack J, Pollok BA (2005) Diverse small-molecule modulators of SMN expression found by high-throughput compound screening: early leads towards a therapeutic for spinal muscular atrophy. Hum Mol Genet 14:2003–2018 10.1093/hmg/ddi205 [DOI] [PubMed] [Google Scholar]
- Kashima T, Manley JL (2003) A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 34:460–463 10.1038/ng1207 [DOI] [PubMed] [Google Scholar]
- König H, Ponta H, Herrlich P (1998) Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J 17:2904–2913 10.1093/emboj/17.10.2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, Le Paslier D, Frézal J, Cohen D, Weissenbach J, Munnich A, Melki J (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155–165 10.1016/0092-8674(95)90460-3 [DOI] [PubMed] [Google Scholar]
- Lim SR, Hertel KJ (2001) Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J Biol Chem 276:45476–45483 10.1074/jbc.M107632200 [DOI] [PubMed] [Google Scholar]
- Liu HX, Cartegni L, Zhang MQ, Krainer AR (2001) A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet 27:55–58 [DOI] [PubMed] [Google Scholar]
- Liu HX, Zhang M, Krainer AR (1998) Identification of functional splicing enhancer motifs recognized by individual SR proteins. Genes Dev 12:1998–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson CL, Androphy EJ (2000) An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum Mol Genet 9:259–265 10.1093/hmg/9.2.259 [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B (1999) A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA 96:6307–6311 10.1073/pnas.96.11.6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand V, Mereau A, Jacquenet S, Thomas D, Mougin A, Gattoni R, Stévenin J, Branlant C (2002) A Janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J Mol Biol 323:629–652 10.1016/S0022-2836(02)00967-1 [DOI] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH (2005) The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272:2118–2131 10.1111/j.1742-4658.2005.04653.x [DOI] [PubMed] [Google Scholar]
- Matter N, Marx M, Weg-Remers S, Ponta H, Herrlich P, König H (2000) Heterogeneous ribonucleoprotein A1 is part of an exon-specific splice-silencing complex controlled by oncogenic signaling pathways. J Biol Chem 275:35353–35360 10.1074/jbc.M004692200 [DOI] [PubMed] [Google Scholar]
- Mayeda A, Helfman DM, Krainer AR (1993) Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol 13:2993–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68:365–375 10.1016/0092-8674(92)90477-T [DOI] [PubMed] [Google Scholar]
- Meister G, Eggert C, Fischer U (2002) SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol 12:472–478 10.1016/S0962-8924(02)02371-1 [DOI] [PubMed] [Google Scholar]
- Miyajima H, Miyaso H, Okumura M, Kurisu J, Imaizumi K (2002) Identification of a cis-acting element for the regulation of SMN exon 7 splicing. J Biol Chem 277:23271–23277 10.1074/jbc.M200851200 [DOI] [PubMed] [Google Scholar]
- Miyaso H, Okumura M, Kondo S, Higashide S, Miyajima H, Imaizumi K (2003) An intronic splicing enhancer element in survival motor neuron (SMN) pre-mRNA. J Biol Chem 278:15825–15831 10.1074/jbc.M209271200 [DOI] [PubMed] [Google Scholar]
- Monani UR, Coovert DD, Burghes AH (2000) Animal models of spinal muscular atrophy. Hum Mol Genet 9:2451–2457 10.1093/hmg/9.16.2451 [DOI] [PubMed] [Google Scholar]
- Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD (1999) A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet 8:1177–1183 10.1093/hmg/8.7.1177 [DOI] [PubMed] [Google Scholar]
- Pagani F, Baralle FE (2004) Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet 5:389–396 10.1038/nrg1327 [DOI] [PubMed] [Google Scholar]
- Parsons DW, McAndrew PE, Monani UR, Mendell JR, Burghes AHM, Prior TW (1996) An 11 base pair duplication in exon 6 of the SMN gene produces a type I spinal muscular atrophy (SMA) phenotype: further evidence for SMN as the primary SMA-determining gene. Hum Mol Genet 5:1727–1732 10.1093/hmg/5.11.1727 [DOI] [PubMed] [Google Scholar]
- Sendtner M (2001) Molecular mechanisms in spinal muscular atrophy: models and perspectives. Curr Opin Neurol 14:629–634 10.1097/00019052-200110000-00012 [DOI] [PubMed] [Google Scholar]
- Si ZH, Rauch D, Stoltzfus CM (1998) The exon splicing silencer in human immunodeficiency virus type 1 tat exon 3 is bipartite and acts early in spliceosome assembly. Mol Cell Biol 18:5404–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Androphy EJ, Singh RN (2004a) An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem Biophys Res Commun 315:381–388 10.1016/j.bbrc.2004.01.067 [DOI] [PubMed] [Google Scholar]
- ——— (2004b) In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA 10:1291–1305 10.1261/rna.7580704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F (2003) Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc Natl Acad Sci USA 100:4114–4119 10.1073/pnas.0633863100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soret J, Bakkour N, Maire S, Durand S, Zekri L, Gabut M, Fic W, Divita G, Rivalle C, Dauzonne D, Nguyen CH, Jeanteur P, Tazi J (2005) Selective modification of alternative splicing by indole derivatives that target serine-arginine-rich protein splicing factors. Proc Natl Acad Sci USA 102:8764–8769 10.1073/pnas.0409829102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staffa A, Cochrane A (1995) Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol Cell Biol 15:4597–4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange TO, Damgaard CK, Guth S, Valcárcel J, Kjems J (2001) The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J 20:5748–5758 10.1093/emboj/20.20.5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Smith PJ, Krainer AR, Zhang MQ (2005) Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res 33:5053–5062 10.1093/nar/gki810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Takagaki Y, Manley JL (1996) Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev 10:2588–2599 [DOI] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB (2004) Systematic identification and analysis of exonic splicing silencers. Cell 119:831–845 10.1016/j.cell.2004.11.010 [DOI] [PubMed] [Google Scholar]
- Young PJ, DiDonato CJ, Hu D, Kothary R, Androphy EJ, Lorson CL (2002) SRp30c-dependent stimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2 beta 1. Hum Mol Genet 11:577–587 10.1093/hmg/11.5.577 [DOI] [PubMed] [Google Scholar]
- Zhang XH, Chasin LA (2004) Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev 18: 1241–1250 10.1101/gad.1195304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Mayeda A, Krainer AR (2001) Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell 8:1351–1361 10.1016/S1097-2765(01)00409-9 [DOI] [PubMed] [Google Scholar]