Abstract
The human N-acetyltransferase genes NAT1 and NAT2 encode two phase-II enzymes that metabolize various drugs and carcinogens. Functional variability at these genes has been associated with adverse drug reactions and cancer susceptibility. Mutations in NAT2 leading to the so-called slow-acetylation phenotype reach high frequencies worldwide, which questions the significance of altered acetylation in human adaptation. To investigate the role of population history and natural selection in shaping NATs variation, we characterized genetic diversity through the resequencing and genotyping of NAT1, NAT2, and the pseudogene NATP in a collection of 13 different populations with distinct ethnic backgrounds and demographic pasts. This combined study design allowed us to define a detailed map of linkage disequilibrium of the NATs region as well as to perform a number of sequence-based neutrality tests and the long-range haplotype (LRH) test. Our data revealed distinctive patterns of variability for the two genes: the reduced diversity observed at NAT1 is consistent with the action of purifying selection, whereas NAT2 functional variation contributes to high levels of diversity. In addition, the LRH test identified a particular NAT2 haplotype (NAT2*5B) under recent positive selection in western/central Eurasians. This haplotype harbors the mutation 341T→C and encodes the “slowest-acetylator” NAT2 enzyme, suggesting a general selective advantage for the slow-acetylator phenotype. Interestingly, the NAT2*5B haplotype, which seems to have conferred a selective advantage during the past ∼6,500 years, exhibits today the strongest association with susceptibility to bladder cancer and adverse drug reactions. On the whole, the patterns observed for NAT2 well illustrate how geographically and temporally fluctuating xenobiotic environments may have influenced not only our genome variability but also our present-day susceptibility to disease.
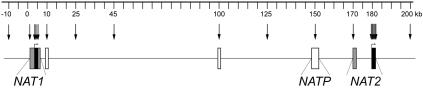
The two human N-acetyltransferase genes, NAT1 (MIM 108345) and NAT2 (MIM 243400), represent one of the first and clearest examples of the importance of genetic variation among individuals and across populations in drug response (Weber 1987). The two homologous genes are situated within a 200-kb region in 8p22, together with the NATP pseudogene (fig. 1). Both genes encode phase-II enzymes named “arylamine N-acetyltransferases” (NATs), which catalyze the transfer of an acetyl group to different arylhydrazines and arylamine drugs (Blum et al. 1990). Both genes carry functional polymorphisms whose effects on enzymatic activity have been well studied (Hein et al. 2000). Whereas the variants associated with reduced activity attain only low frequencies in NAT1, they constitute common polymorphisms in NAT2 (Upton et al. 2001). Two main classes of NAT2 phenotypes are therefore observed: the “fast-acetylation” phenotype, which refers to the wild-type acetylation activity, and the “slow-acetylation” phenotype, which results in reduced protein activity. In addition, NAT1 and NAT2 metabolize numerous common carcinogens, and variation in these genes can result in varying susceptibility to cancer (for a review, see the work of Hein [2002]). For example, the slow-acetylator NAT2 phenotype has been associated with side effects to the commonly used antitubercular isoniazid (Huang et al. 2002) and with higher risk for bladder cancer (Cartwright et al. 1982; Garcia-Closas et al. 2005). Nevertheless, most NAT2 mutations leading to the slow phenotype are found at high frequencies worldwide, calling into question the role of altered acetylation in human adaptation. Moreover, the function of NATs in mediating the interactions between humans and their xenobiotic environment, which varies depending on diet and lifestyle, makes them excellent targets for the action of natural selection. Indeed, several studies have identified the signature of different selective pressures in genes involved in the metabolism of exogenous substances, including the members of the CYP3A family (Thompson et al. 2004), CYP1A2 (Wooding et al. 2002), LCT (Bersaglieri et al. 2004), TAS2R16 (Soranzo et al. 2005), PTC (Wooding et al. 2004), HFE (Toomajian and Kreitman 2002; Toomajian et al. 2003), MDR1 (Tang et al. 2004), and MRP1 (Wang et al. 2005).
Figure 1.
Schematic representation of the NATs region spanning >200 kb. Sequenced loci are represented by boxes (black boxes = coding regions; gray boxes = flanking regions; white boxes = intergenic regions), and arrows indicate the positions of genotyped SNPs.
The main objective of the present study was to investigate the evolutionary history of the NATs region by unraveling the relative influences of natural selection and human demography in determining its present-day variability. With this goal in mind, we first resequenced NAT1, NAT2, and the pseudogene NATP in a multiethnic panel of 80 individuals (referred to as the “resequencing panel”). To further investigate the global linkage disequilibrium (LD) patterns in the NATs region, we selected 21 SNPs—including 5 NAT1 and 7 NAT2 SNPs retrieved from the initial sequence-based data set as well as 9 intergenic SNPs—to cover the entire 200-kb region (fig. 1). These markers were all genotyped in an extended collection of 563 individuals (referred to as the “genotyping panel”) originating from 13 different ethnologically well-defined human populations. Coalescent methods, sequence-based neutrality tests, and the long-range haplotype (LRH) test (Sabeti et al. 2002) were performed to provide insight into the role of these genes in human adaptation to geographically and historically fluctuating xenobiotic environments.
Subjects and Methods
DNA Samples
The resequencing panel consisted of 80 individuals (160 chromosomes) from eight populations representing major geographic regions; sub-Saharan African chromosomes were represented by Bakola Pygmies from Cameroon (20) and by Bantu speakers from Gabon (20); western Eurasian samples were represented by Ashkenazi Jews (20), Sardinians (12), French (20), and Saami from Finland (20); and eastern Eurasian samples were represented by Indians from Gujarat (20) and by Thai (28). One chimpanzee (Pan troglodytes) was also sequenced to define the ancestral state of each mutation. The genotyping panel consisted of 563 individuals (1,126 chromosomes) from 13 populations. Sub-Saharan African chromosomes were represented by Bakola Pygmies (80) and Baka Pygmies (60) from Cameroon, Ateke Bantu speakers from Gabon (100), and Somali (48); North African and western and central Eurasian samples were represented by Morrocans (88), Ashkenazi Jews (80), Sardinians (98), Swedes (100), Saami from Finland (96), and Turkmen from Uzbekistan (100); and eastern Eurasian samples were represented by Gujarati from India (100), Chinese from the Hunan and Zhejang regions (88), and Thai (88). All individuals were healthy donors from whom informed consent was obtained.
PCR and Sequence Determination
Six different regions were PCR amplified, for a total of ∼8.5 kb per chromosome (fig. 1): the entire coding exon of the NAT1 gene (870 bp) and 1,735 bp of noncoding flanking parts (1,122 bp in 5′ end and 613 bp in 3′ end); the entire coding exon of the NAT2 gene (870 bp) and 1,950 bp of noncoding flanking parts, including 1,603 bp surrounding its first noncoding exon; the pseudogene NATP (2,145 bp); and two intergenic noncoding regions at 10 kb and 100 kb (1,068 bp) from NAT1 5′ end. Details about PCR and sequencing conditions are available on request. As a measure of quality control for the data, individuals presenting singletons or ambiguous polymorphisms were reamplified and resequenced. Sequences were analyzed using the GENALYS software (Takahashi et al. 2003).
Selection and Genotyping of Polymorphisms
The newly discovered sequence-based variation was used to determine the minimal number of SNPs able to distinguish the haplotypic diversity (haplotype-tagging SNPs [htSNPs]) of NAT1 and NAT2 loci in a given population. Five SNPs and one (TAA)n microsatellite were typed in NAT1 by either genotyping or sequencing, and seven SNPs were genotyped in the NAT2 coding region. In addition, we genotyped nine intergenic SNPs selected because they were polymorphic in all human populations (fig. 1). These SNPs were chosen either from dbSNP (when dbSNPs met the previous criterion) or from the intergenic regions sequenced here. Genotyping was performed by either fluorescence polarization (VICTOR-2TM technology) or TaqMan (ABI Prism-7000 Sequence Detection System) assays.
Sequence-Based Data Analysis
Allele frequencies were determined by gene counting, and deviations from Hardy-Weinberg equilibrium were tested by Arlequin v.2.001 (Schneider et al. 2000). Haplotype reconstruction was performed using the Bayesian method implemented in PHASE v.2.1.1 (Stephens and Donnelly 2003), and htSNPs were defined using BEST v.1.0 (Sebastiani et al. 2003), after the exclusion of singletons because they could not be positioned with certainty on a given haplotypic context. With the use of phased data, the neutral parameter θML and the time since the most recent common ancestor (TMRCA) were estimated by maximum likelihood with GENETREE (Griffiths and Tavaré 1994), under a standard coalescent model. Since this model assumes no recombination, for this particular analysis we had to exclude a few SNPs or rare recombinant haplotypes (in NAT1, the first four 5′ SNPs; in NATP, three singleton haplotypes; in NAT2, two singleton haplotypes). Time, scaled in 2Ne units, was converted into years by use of a 25-year generation time and an Ne value obtained as θML divided by 4μ. The mutation rate per gene per generation (μ) was deduced from Dxy, the average number of nucleotide substitutions per site between human and chimpanzee (Nei 1987, equation 10.20), calculated by DnaSP v.4.0 (Rozas et al. 2003), with consideration that the two species diverged 200,000 generations ago. Simulations were performed to estimate the probability of a TMRCA greater than a given value, under a Wright-Fisher model. Fifty thousand simulations were performed using a version of the MS program modified to obtain TMRCA values (R. Hudson, personal communication).
Using DnaSP, we calculated the nucleotide diversity (π) and Watterson’s estimator of θ (θW) (Watterson 1975), and we performed a number of statistical tests: Tajima’s D (TD) (Tajima 1989), Fu and Li’s F* (Fu and Li 1993), Fay and Wu’s H (Fay and Wu 2000), KA/KS (Kimura 1968), the Hudson-Kreitman-Aguadé (HKA) test (Hudson et al. 1987), and the McDonald-Kreitman (MK) test (McDonald and Kreitman 1991). A neutrality test based on the expected heterozygosity was also performed with the Bottleneck program (Cornuet and Luikart 1996) on the NAT1 3′ UTR microsatellite, by the use of coalescent simulations (10,000 runs) and with the assumption of different mutational models (stepwise mutation model and two-phased mutation model with 0%–40% of multistep changes).
Genotyping-Based Data Analysis
Pairwise LD between the 21 genotyped SNPs was estimated after the exclusion, in each population, of SNPs with a minor-allele frequency (MAF) <0.10. Using DnaSP, we calculated the statistics D′ (Lewontin 1964) and r2 (Hill and Robertson 1968) and tested their statistical significance, using a Fisher’s exact test followed by Bonferroni corrections. To perform the LRH test, we selected two core regions (in NAT1, SNPs 445, 1088, 1095, and 1191; in NAT2, SNPs 341, 481, 590, 803, and 857) identified as haplotype blocks, following the criteria of Gabriel et al. (2002), and we assessed, for each core haplotype, its relative extended haplotype homozygozity (REHH) 200 kb apart. To test the significance of potentially selected core haplotypes, we first compared our sub-Saharan African and non-African data sets with coalescent simulations of 1-Mb regions, assuming a neutral model of evolution with recombination (Hudson 2002). Model parameters (including demography and recombination rate) were consistent with current estimates for African and non-African populations (Schaffner et al. 2005). Similarly, our sub-Saharan African and non-African data sets were compared with the empirical distribution of “core haplotype frequencies versus REHH” obtained from the screening of the entire chromosome 8 in Yoruban and European-descent populations, respectively (HapMap database).
To infer the population growth rate, r, and the age, g, of NAT2 nonsynonymous mutations, we used a joint maximum-likelihood estimation of these parameters, as described in Austerlitz et al. (2003). We compared these results with coalescent-based estimations of the two parameters: the growth rate estimation of Slatkin and Bertorelle (2001) and the Reeve and Rannala (2002) age estimation using the DMLE+ v.2.2 software. One million iterations were performed for each estimation. The recombination parameter required for these analyses was estimated by comparing deCODE and Marshfield genetic and physical distances in the NATs region (UCSC Genome Bioinformatics). The coefficient of selection, s, of the NAT2 mutation 341T→C was estimated using the deterministic equation 3.29 of Wright (1969), which relates the frequency of an allele in generation t+1 to its frequency in generation t. We stated the degree of dominance, h, to 0.0 (recessivity) and 0.5 (codominance). We assumed the frequency, p0, of the C allele before selection to vary between 0.05 and 0.15 (corresponding to the allele frequency in Pygmies and eastern Eurasians). Making these assumptions, we calculated the s values that would yield a frequency of 0.50 (the present-day frequency of the 341C allele in western Eurasians) from its initial p0 frequency in g generations.
Results
NATs Nucleotide Sequence Variation
The initial sequencing screening of the resequencing panel yielded a total of 111 mutations, including 68 transitions, 34 transversions, 8 insertions/deletions, and 1 triallelic microsatellite (table 1) (GenBank accession numbers DQ305496–DQ305975). In NAT1, we observed 2 nonsynonymous and 4 synonymous SNPs in its coding region and 26 SNPs and the triallelic (TAA)n microsatellite in its flanking regions. In the NAT2 coding region, we found two synonymous and eight nonsynonymous mutations, three of which were newly identified (L24I, T193M, and Y208H). These three variants were singletons and were restricted to sub-Saharan samples. In addition, 14 SNPs and 3 indels in NAT2 flanking regions were observed. In NATP, we identified 32 SNPs and 5 indels. For all the SNPs, only 1.54% of the tests departed significantly from Hardy-Weinberg equilibrium. However, these few tests would become nonsignificant after a correction for multiple testing.
Table 1.
Polymorphisms Identified through the Resequencing Survey of the NATs Region
|
Derived-Allele Frequency in Population(%) |
|||||||||||
| Polymorphism | dbSNP ReferenceNumber | Allele(s)(Ancestral/Derived) | SNP Type | Bakola(2N=20) | Bantu(2N=20) | Ashkenazi(2N=20) | Sardinian(2N=12) | French(2N=20) | Saami(2N=20) | Gujarati(2N=20) | Thai(2N=28) |
| NAT1−1112 | rs8190842 | G/A | Intron | 5.0 | 10.0 | .0 | .0 | 5.0 | .0 | 5.0 | 7.1 |
| NAT1−1048 | rs8190843 | G/C | Intron | .0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT1−943 | rs8190844 | C/T | Intron | .0 | .0 | .0 | .0 | 5.0 | .0 | .0 | .0 |
| NAT1−929 | rs8190845 | G/A | Intron | 45.0 | 50.0 | 5.0 | 33.3 | 15.0 | 20.0 | 5.0 | 25.0 |
| NAT1−868 | rs8190846 | G/A | Intron | .0 | .0 | .0 | .0 | 5.0 | .0 | 5.0 | 7.1 |
| NAT1−844 | rs8190847 | G/A | Intron | .0 | .0 | .0 | .0 | 5.0 | 0.0 | 5.0 | 7.1 |
| NAT1−826 | rs8190848 | C/T | Intron | 100.0 | 100.0 | 100.0 | 100.0 | 95.0 | 100.0 | 95.0 | 92.9 |
| NAT1−720 | NA | T/A | Intron | 100.0 | 100.0 | 100.0 | 100.0 | 95.0 | 100.0 | 95.0 | 92.9 |
| NAT1−706 | rs8190851 | G/A | Intron | 100.0 | 100.0 | 100.0 | 100.0 | 95.0 | 100.0 | 95.0 | 92.9 |
| NAT1−688 | rs8190852 | G/C | Intron | 100.0 | 100.0 | 100.0 | 100.0 | 95.0 | 100.0 | 95.0 | 92.9 |
| NAT1−685 | rs8190853 | C/T | Intron | 100.0 | 100.0 | 100.0 | 100.0 | 95.0 | 100.0 | 95.0 | 92.9 |
| NAT1−621 | NA | A/G | Intron | .0 | .0 | .0 | .0 | .0 | .0 | .0 | 3.6 |
| NAT1−565 | rs8190854 | A/G | Intron | 10.0 | .0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT1−433 | rs8190856 | T/C | Intron | .0 | .0 | .0 | .0 | 5.0 | .0 | 5.0 | 7.1 |
| NAT1−344 | rs4986988 | T/C | Intron | 100.0 | 100.0 | 100.0 | 100.0 | 95.0 | 100.0 | 95.0 | 92.9 |
| NAT1−278 | rs17126356 | T/A | Intron | 5.0 | 10.0 | .0 | 8.3 | .0 | .0 | .0 | .0 |
| NAT1−40 | rs4986989 | T/A | Intron | 100.0 | 100.0 | 100.0 | 100.0 | 95.0 | 100.0 | 95.0 | 92.9 |
| NAT1−36 | rs8190857 | A/T | Intron | .0 | 10.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT1 21 | rs4986992 | T/G | Silent mutation | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT1 342 | NA | T/C | Silent mutation | .0 | .0 | .0 | .0 | .0 | .0 | .0 | 3.6 |
| NAT1 445 | rs4987076 | A/G | Missense mutation V149I | 100.0 | 100.0 | 100.0 | 100.0 | 95.0 | 100.0 | 95.0 | 92.9 |
| NAT1 459 | rs4986990 | G/A | Silent mutation | .0 | .0 | .0 | .0 | 5.0 | .0 | 5.0 | 7.1 |
| NAT1 640 | rs4986783 | G/T | Missense mutation S214A | 100.0 | 100.0 | 100.0 | 100.0 | 95.0 | 100.0 | 95.0 | 92.9 |
| NAT1 777 | rs4986991 | T/C | Silent mutation | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT1 (TAA)9 | NA | ins(TAA) | 3′ UTR | .0 | 0.0 | .0 | .0 | .0 | .0 | .0 | 17.9 |
| NAT1 (TAA)5 | NA | del(TAATAATAA) | 3′ UTR | .0 | .0 | .0 | .0 | 5.0 | .0 | 5.0 | 7.1 |
| NAT1 1088 | rs1057126 | T/A | 3′ UTR | 35.0 | 65.0 | 20.0 | 33.3 | 20.0 | 25.0 | 15.0 | 46.4 |
| NAT1 1095 | rs15561 | A/C | 3′ UTR | 60.0 | 35.0 | 80.0 | 58.3 | 70.0 | 65.0 | 75.0 | 46.4 |
| NAT1 1191 | rs4986993 | T/G | 3′ UTR | 60.0 | 35.0 | 80.0 | 58.3 | 70.0 | 65.0 | 75.0 | 46.4 |
| NAT1 1236 | rs4987077 | A/G | Intergenic/unknown | 35.0 | .0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT1 1277 | NA | A/G | Intergenic/unknown | .0 | .0 | .0 | .0 | 5.0 | .0 | 5.0 | 7.1 |
| NAT1 1345 | rs8190862 | G/C | Intergenic/unknown | 100.0 | 100.0 | 100.0 | 100.0 | 95.0 | 100.0 | 95.0 | 92.9 |
| NAT1 1377 | rs8190863 | C/T | Intergenic/unknown | .0 | .0 | .0 | .0 | 5.0 | .0 | 5.0 | 7.1 |
| NAT1 1454 | rs8190864 | C/T | Intergenic/unknown | .0 | .0 | .0 | 8.3 | .0 | .0 | .0 | .0 |
| 10KB 455 | NA | G/A | Intergenic/unknown | 5.0 | .0 | .0 | .0 | .0 | NA | .0 | .0 |
| 10KB 546 | NA | G/T | Intergenic/unknown | .0 | 5.0 | .0 | .0 | .0 | NA | .0 | .0 |
| 10KB 633 | rs4921583 | C/A | Intergenic/unknown | 15.0 | 45.0 | 30.0 | 33.3 | 50.0 | NA | 25.0 | 30.0 |
| 10KB 648 | rs4921585 | C/T | Intergenic/unknown | 15.0 | 45.0 | 30.0 | 41.7 | 50.0 | NA | 25.0 | 30.0 |
| 10KB 666 | NA | C/A | Intergenic/unknown | .0 | .0 | .0 | .0 | 5.0 | NA | .0 | .0 |
| 10KB 884 | NA | G/A | Intergenic/unknown | .0 | .0 | .0 | .0 | .0 | NA | .0 | 5.0 |
| 10KB 904 | rs1389110 | T/C | Intergenic/unknown | 38.9 | 60.0 | 30.0 | 50.0 | 50.0 | NA | 25.0 | 45.0 |
| 100KB 314 | NA | G/T | Intergenic/unknown | .0 | 11.1 | .0 | .0 | .0 | NA | .0 | .0 |
| 100KB 409 | NA | T/C | Intergenic/unknown | 5.0 | 16.7 | .0 | .0 | .0 | NA | .0 | .0 |
| 100KB 411 | NA | T/G | Intergenic/unknown | .0 | .0 | .0 | .0 | 5.6 | NA | 5.0 | .0 |
| 100KB 455 | NA | G/C | Intergenic/unknown | 10.0 | .0 | .0 | .0 | .0 | NA | .0 | .0 |
| 100KB 507 | rs12541267 | A/G | Intergenic/unknown | 45.0 | 20.0 | 35.0 | 33.3 | 38.9 | NA | 40.0 | 45.0 |
| 100KB 530 | rs13259523 | A/G | Intergenic/unknown | .0 | .0 | .0 | .0 | 5.6 | NA | 5.0 | .0 |
| 100KB 595 | NA | G/T | Intergenic/unknown | 5.0 | .0 | .0 | .0 | .0 | NA | .0 | .0 |
| NATP −10 | NA | del(GAAA…TAGT) | Intergenic/unknown | .0 | .0 | .0 | .0 | .0 | 5.0 | .0 | .0 |
| NATP 49 | NA | A/G | Intergenic/unknown | 10.0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 52 | NA | C/T | Intergenic/unknown | 5.0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 362 | NA | C/T | Intergenic/unknown | 100.0 | 90.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| NATP 414 | NA | del(T) | Intergenic/unknown | .0 | 10.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 417 | rs12334336 | T/C | Intergenic/unknown | 5.0 | 10.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 520 | NA | A/G | Intergenic/unknown | .0 | 10.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 631 | NA | G/C | Intergenic/unknown | 100.0 | 90.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| NATP 685 | NA | T/C | Intergenic/unknown | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 698 | rs10088180 | A/G | Intergenic/unknown | 75.0 | 70.0 | 75.0 | 66.7 | 85.0 | 45.0 | 60.0 | 60.7 |
| NATP 733 | NA | T/G | Intergenic/unknown | .0 | .0 | .0 | .0 | .0 | .0 | .0 | 3.6 |
| NATP 745 | NA | del(G) | Intergenic/unknown | 25.0 | 35.0 | 35.0 | 16.7 | 50.0 | 10.0 | 25.0 | 42.9 |
| NATP 754 | NA | G/A | Intergenic/unknown | 90.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| NATP 828 | NA | T/C | Intergenic/unknown | 15.0 | 15.0 | .0 | .0 | .0 | 5.0 | 0.0 | .0 |
| NATP 859 | NA | ins(T) | Intergenic/unknown | 85.0 | 85.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| NATP 876 | NA | T/C | Intergenic/unknown | .0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 911 | NA | G/A | Intergenic/unknown | .0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 1007 | NA | G/A | Intergenic/unknown | .0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 1153 | NA | C/T | Intergenic/unknown | .0 | .0 | 10.0 | .0 | .0 | .0 | .0 | .0 |
| NATP 1198 | NA | T/C | Intergenic/unknown | 10.0 | 15.0 | 20.0 | 16.7 | 10.0 | 15.0 | 10.0 | 28.6 |
| NATP 1251 | NA | G/C | Intergenic/unknown | 5.0 | 10.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 1257 | NA | ins(A) | Intergenic/unknown | .0 | .0 | .0 | .0 | .0 | .0 | .0 | 3.6 |
| NATP 1433 | NA | T/C | Intergenic/unknown | 10.0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 1581 | NA | G/C | Intergenic/unknown | 10.0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 1602 | rs2172426 | T/C | Intergenic/unknown | 60.0 | 35.0 | 45.0 | 25.0 | 55.0 | 50.0 | 55.0 | 53.6 |
| NATP 1616 | NA | C/A | Intergenic/unknown | .0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 1686 | NA | T/C | Intergenic/unknown | 15.0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 1770 | rs12548816 | T/C | Intergenic/unknown | .0 | .0 | 10.0 | 8.3 | 5.0 | 30.0 | 10.0 | 10.7 |
| NATP 1794 | NA | G/A | Intergenic/unknown | .0 | 10.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 1827 | NA | T/G | Intergenic/unknown | 15.0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 1829 | NA | T/G | Intergenic/unknown | 15.0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 1851 | NA | G/A | Intergenic/unknown | 15.0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 1881 | rs13254216 | T/C | Intergenic/unknown | .0 | .0 | 10.0 | 8.3 | 5.0 | .0 | 5.0 | .0 |
| NATP 1903 | NA | A/C | Intergenic/unknown | 15.0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 2085 | rs17126565 | C/G | Intergenic/unknown | .0 | 10.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NATP 2115G | rs17126568 | T/G | Intergenic/unknown | 85.0 | 95.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| NATP 2115C | NA | T/C | Intergenic/unknown | 10.0 | .0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT2.1−1146 | rs4646243 | T/C | Intergenic/unknown | 10.0 | 15.0 | 5.0 | 8.3 | 10.0 | 20.0 | 5.0 | 28.6 |
| NAT2.1−1049 | NA | del(AG) | Intergenic/unknown | 50.0 | 60.0 | 45.0 | 33.3 | 65.0 | 30.0 | 40.0 | 75.0 |
| NAT2.1−1037 | rs4646244 | T/A | Intergenic/unknown | 15.0 | 25.0 | 40.0 | 25.0 | 55.0 | 15.0 | 35.0 | 46.4 |
| NAT2.1−949 | NA | del(TA) | Intergenic/unknown | 15.0 | .0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT2.1−842 | rs4646267 | A/G | Intergenic/unknown | 10.0 | 10.0 | 5.0 | 8.3 | 10.0 | 20.0 | 5.0 | 28.6 |
| NAT2.1−815 | NA | A/G | Intergenic/unknown | .0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT2.1−547 | rs4345600 | A/G | Intergenic/unknown | .0 | .0 | 10.0 | 8.3 | 5.0 | 30.0 | 30.0 | 10.7 |
| NAT2.1−542 | rs4345601 | A/T | Intergenic/unknown | .0 | .0 | 10.0 | 8.3 | 5.0 | 30.0 | 30.0 | 10.7 |
| NAT2.1−521 | rs5889794 | del(AATT) | Intergenic/unknown | 55.0 | 60.0 | 45.0 | 33.3 | 65.0 | 30.0 | 40.0 | 75.0 |
| NAT2.1−487 | rs4271002 | G/C | Intergenic/unknown | .0 | .0 | 10.0 | 8.3 | 5.0 | 30.0 | 30.0 | 10.7 |
| NAT2.1−327 | NA | A/G | Intergenic/unknown | .0 | .0 | .0 | .0 | .0 | .0 | .0 | 7.1 |
| NAT2.1−94 | rs4646246 | A/G | Intergenic/unknown | 25.0 | 20.0 | 5.0 | 8.3 | 10.0 | 20.0 | 25.0 | 28.6 |
| NAT2.1 46 | NA | A/G | Silent mutation | .0 | .0 | 5.0 | .0 | .0 | .0 | .0 | .0 |
| NAT2.1 112 | NA | C/T | Intron | .0 | .0 | .0 | .0 | .0 | .0 | .0 | 3.6 |
| NAT2 70 | NA | T/A | Missense mutation L24I | .0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT2 191 | rs1801279 | G/A | Missense mutation R64Q | .0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT2 282 | rs1041983 | C/T | Silent mutation | 30.0 | 30.0 | 50.0 | 25.0 | 40.0 | 30.0 | 40.0 | 57.1 |
| NAT2 341 | rs1801280 | T/C | Missense mutation I114T | 10.0 | 55.0 | 40.0 | 58.3 | 30.0 | 35.0 | 30.0 | 14.3 |
| NAT2 481 | rs1799929 | C/T | Silent mutation | 10.0 | 50.0 | 40.0 | 58.3 | 30.0 | 35.0 | 30.0 | 14.3 |
| NAT2 578 | NA | C/T | Missense mutation T193M | .0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT2 590 | rs1799930 | G/A | Missense mutation R197Q | 15.0 | 10.0 | 50.0 | 25.0 | 45.0 | 15.0 | 35.0 | 42.9 |
| NAT2 622 | NA | T/C | Missense mutation Y208H | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT2 803 | rs1208 | A/G | Missense mutation K268R | 60.0 | 65.0 | 40.0 | 58.3 | 30.0 | 35.0 | 30.0 | 14.3 |
| NAT2 857 | rs1799931 | G/A | Missense mutation G286E | .0 | .0 | .0 | .0 | .0 | 15.0 | 5.0 | 10.7 |
| NAT2 1021 | rs2552 | T/C | 3′ UTR | .0 | .0 | .0 | .0 | 5.0 | 5.0 | .0 | 3.6 |
| NAT 1085 | NA | G/A | 3′ UTR | 30.0 | .0 | .0 | .0 | .0 | .0 | .0 | .0 |
| NAT2 1101 | NA | C/G | 3′ UTR | .0 | 5.0 | .0 | .0 | .0 | .0 | .0 | .0 |
The nucleotide diversity, π, in the NAT1 and NAT2 coding and flanking regions as well as in the pseudogene NATP is reported in table 2. Interestingly, the two duplicated NAT genes, which share 87.3% of nucleotide identity in their coding region, showed completely different diversity levels, with the NAT2 coding region (π=0.275%) being 13 times more diverse than the NAT1 coding region (π=0.021%). To evaluate whether these differences were due to the local variation in substitution rates, we estimated the mutation rates per generation per nucleotide of NAT1 and NAT2 coding regions as well as that of NATP, which equaled 3.73 × 10−8, 3.96 × 10−8, and 5.94 × 10−8, respectively.
Table 2.
Diversity Indices of NAT1, NAT2, and NATP Sequences[Note]
|
NAT1 CodingRegion(870 bp) |
NAT1 Codingand FlankingRegions(2,605 bp) |
NAT2 CodingRegion(870 bp) |
NAT2 Codingand FlankingRegions(2,820 bp) |
NATP Pseudogene(2,145 bp) |
|||||||||||
| Population | θW(%) | π(%) | HD | θW(%) | π(%) | HD | θW(%) | π(%) | HD | θW(%) | π(%) | HD | θW(%) | π(%) | HD |
| Bakola | .065 | .023 | .100 | .108 | .118 | .821 | .194 | .195 | .768 | .110 | .113 | .863 | .263 | .223 | .863 |
| Bantu | .000 | .000 | .000 | .087 | .101 | .811 | .259 | .283 | .789 | .140 | .136 | .837 | .368 | .247 | .884 |
| Ashkenazi | .000 | .000 | .000 | .043 | .043 | .353 | .162 | .295 | .611 | .130 | .143 | .742 | .092 | .107 | .821 |
| Sardinian | .000 | .000 | .000 | .076 | .091 | .667 | .190 | .277 | .621 | .141 | .135 | .636 | .093 | .085 | .758 |
| French | .097 | .034 | .100 | .238 | .126 | .516 | .162 | .270 | .721 | .130 | .136 | .789 | .079 | .079 | .653 |
| Saami | .000 | .000 | .000 | .043 | .065 | .563 | .194 | .278 | .747 | .140 | .179 | .853 | .092 | .100 | .789 |
| Gujarati | .097 | .034 | .100 | .227 | .110 | .432 | .194 | .277 | .732 | .130 | .171 | .837 | .079 | .089 | .837 |
| Thai | .012 | .056 | .204 | .227 | .170 | .733 | .177 | .227 | .728 | .146 | .164 | .812 | .084 | .106 | .870 |
| Total | .012 | .021 | .073 | .217 | .112 | .662 | .203 | .275 | .767 | .151 | .157 | .841 | .297 | .134 | .855 |
Note.— HD = haplotype diversity.
Haplotype Diversity and TMRCA Estimates of NAT Loci
Haplotypes of the NAT genes were reconstructed first by use of the sequence data obtained from the resequencing panel (tables 3 and 4). The most unusual observation was the haplotype diversity of the NAT1 locus, made of two highly divergent haplotype clusters separated by 17 mutations. One cluster contained most of NAT1 haplotype diversity (97.5%), whereas the other contained a unique haplotype, called “NAT1*11A,” that was observed in just three non-African individuals (one heterozygous French, one heterozygous Gujarati, and one homozygous Thai). Consequently, diversity estimates of the NAT1 locus were inflated in these three populations, as attested by the much higher θW values in the French, Gujarati, and Thai samples, as compared with all other populations (table 2).
Table 3.
Allelic Composition and Frequency of NAT1 Haplotypes in Our Resequencing Panel[Note]
|
Polymorphism |
Haplotype Frequency in Population |
|||||||||||||||||||||||||||||||||||||||||
| Haplotype | −1112 | −1048 | −943 | −929 | −868 | −844 | −826 | −720 | −706 | −688 | −685 | −621 | −565 | −433 | −344 | −278 | −40 | −36 | 21 | 342 | 445 | 459 | 640 | 777 | (TAA)n | 1088 | 1095 | 1191 | 1236 | 1277 | 1345 | 1377 | 1454 | Bakola | Bantu | Ashkenazi | French | Sardinian | Saami | Gujarati | Thai | Total |
| Ancestral (chimpanzee) | G | G | C | G | G | G | C | T | G | G | C | A | A | T | T | T | T | A | T | T | A | G | G | T | 4 | T | A | T | A | A | G | C | C | |||||||||
| NAT1*10A | . | . | . | . | . | . | T | A | A | C | T | . | . | . | C | . | A | . | . | . | G | . | T | . | 8 | A | . | . | . | . | C | . | . | 3 | 3 | 2 | 1 | 3 | 3 | 7 | 22 | |
| NAT1*10B | . | . | T | . | . | . | T | A | A | C | T | . | . | . | C | . | A | . | . | . | G | . | T | . | 8 | A | . | . | . | . | C | . | . | 1 | 1 | |||||||
| NAT1*10C | . | . | . | . | . | . | T | A | A | C | T | . | . | . | C | . | A | . | . | C | G | . | T | . | 8 | A | . | . | . | . | C | . | . | 1 | 1 | |||||||
| NAT1*10D | . | . | . | A | . | . | T | A | A | C | T | . | . | . | C | . | A | . | . | . | G | . | T | . | 8 | A | . | . | . | . | C | . | . | 4 | 5 | 1 | 1 | 2 | 2 | 1 | 16 | |
| NAT1*10E | A | . | . | A | . | . | T | A | A | C | T | . | . | . | C | A | A | . | . | . | G | . | T | . | 8 | A | . | . | . | . | C | . | . | 1 | 2 | 3 | ||||||
| NAT1*10F | . | C | . | A | . | . | T | A | A | C | T | . | . | . | C | . | A | . | . | . | G | . | T | . | 8 | A | . | . | . | . | C | . | . | 1 | 1 | |||||||
| NAT1*10G | . | . | . | A | . | . | T | A | A | C | T | . | G | . | C | . | A | . | . | . | G | . | T | . | 8 | A | . | . | . | . | C | . | . | 2 | 2 | |||||||
| NAT1*10H | . | . | . | A | . | . | T | A | A | C | T | . | . | . | C | A | A | . | . | . | G | . | T | . | 8 | A | . | . | . | . | C | . | . | 1 | 1 | |||||||
| NAT1*10I | . | . | . | A | . | . | T | A | A | C | T | . | . | . | C | . | A | T | . | . | G | . | T | . | 8 | A | . | . | . | . | C | . | . | 2 | 2 | |||||||
| NAT1*26D | . | . | . | . | . | . | T | A | A | C | T | . | . | . | C | . | A | . | . | . | G | . | T | . | 9 | A | . | . | . | . | C | . | . | 1 | 1 | |||||||
| NAT1*26E | . | . | . | A | . | . | T | A | A | C | T | . | . | . | C | . | A | . | . | . | G | . | T | . | 9 | A | . | . | . | . | C | . | . | 3 | 3 | |||||||
| NAT1*4A | . | . | . | . | . | . | T | A | A | C | T | . | . | . | C | . | A | . | . | . | G | . | T | . | 8 | . | C | G | . | . | C | . | . | 4 | 7 | 16 | 14 | 6 | 13 | 15 | 12 | 87 |
| NAT1*4C | . | . | . | . | . | . | T | A | A | C | T | . | . | . | C | . | A | . | . | . | G | . | T | . | 8 | . | C | G | G | . | C | . | . | 7 | 7 | |||||||
| NAT1*4D | . | . | . | . | . | . | T | A | A | C | T | . | . | . | C | . | A | . | . | . | G | . | T | . | 8 | . | C | G | . | . | C | . | T | 1 | 1 | |||||||
| NAT1*26C | . | . | . | A | . | . | T | A | A | C | T | G | . | . | C | . | A | . | . | . | G | . | T | . | 9 | . | C | G | . | . | C | . | . | 1 | 1 | |||||||
| NAT1*27 | . | . | . | A | . | . | T | A | A | C | T | . | . | . | C | . | A | . | G | . | G | . | T | C | 8 | . | C | G | . | . | C | . | . | 1 | 1 | |||||||
| NAT1*3A | . | . | . | . | . | . | T | A | A | C | T | . | . | . | C | . | A | . | . | . | G | . | T | . | 8 | . | . | . | . | . | C | . | . | 1 | 1 | |||||||
| NAT1*3B | . | . | . | A | . | . | T | A | A | C | T | . | . | . | C | . | A | . | . | . | G | . | T | . | 8 | . | . | . | . | . | C | . | . | 1 | 1 | 1 | 2 | 5 | ||||
| NAT1*11A | A | . | . | A | A | A | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | A | . | . | 5 | . | . | . | . | G | . | T | . | 1 | 1 | 2 | 4 | |||||
| Total | 20 | 20 | 20 | 20 | 12 | 20 | 20 | 28 | 160 | |||||||||||||||||||||||||||||||||
Note.— The ancestral state of each genotyped polymorphism was deduced from the chimpanzee and is represented here as a dot. NAT1 haplotypes were named in accordance with NAT Nomenclature.
Table 4.
Allelic Composition and Frequency of NAT2 Haplotypes in Our Resequencing Panel[Note]
|
Polymorphism |
Haplotype Frequency in Population |
|||||||||||||||||||||
| Haplotype | 70 | 191 | 282 | 341 | 481 | 578 | 590 | 622 | 803 | 857 | 1021 | 1085 | 1101 | Bakola | Bantu | Ashkenazi | French | Sardinian | Saami | Gujarati | Thai | Total |
| Ancestral (chimpanzee) | T | G | C | T | C | C | G | T | A | G | T | G | C | |||||||||
| NAT2*4 | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | 3 | 2 | 5 | 2 | 7 | 6 | 8 | 35 |
| NAT2*13 | . | . | T | . | . | . | . | . | . | . | . | . | . | 3 | 1 | 1 | 5 | |||||
| NAT2*12B | . | . | T | . | . | . | . | . | G | . | . | . | . | 1 | 1 | |||||||
| NAT2*12E | . | . | T | . | . | T | . | . | G | . | . | . | . | 1 | 1 | |||||||
| NAT2*12F | . | . | . | . | . | . | . | C | G | . | . | . | . | 1 | 1 | |||||||
| NAT2*12G | . | . | . | . | . | . | . | . | G | . | . | A | . | 6 | 6 | |||||||
| NAT2*12A | . | . | . | . | . | . | . | . | G | . | . | . | . | 3 | 3 | |||||||
| NAT2*5B | . | . | . | C | T | . | . | . | G | . | . | . | . | 2 | 9 | 8 | 5 | 7 | 6 | 6 | 3 | 46 |
| NAT2*5K | . | . | . | C | T | . | . | . | G | . | C | . | . | 1 | 1 | 1 | 3 | |||||
| NAT2*5L | A | . | . | C | T | . | . | . | G | . | . | . | G | 1 | 1 | |||||||
| NAT2*5C | . | . | . | C | . | . | . | . | G | . | . | . | . | 1 | 1 | |||||||
| NAT2*14B | . | A | T | . | . | . | . | . | . | . | . | . | . | 1 | 1 | |||||||
| NAT2*6B | . | . | . | . | . | . | A | . | . | . | . | . | . | 1 | 1 | |||||||
| NAT2*6A | . | . | T | . | . | . | A | . | . | . | . | . | . | 3 | 2 | 10 | 8 | 3 | 3 | 7 | 12 | 48 |
| NAT2*7B | . | . | T | . | . | . | . | . | . | A | . | . | . | 3 | 1 | 3 | 7 | |||||
| Total | 20 | 20 | 20 | 20 | 12 | 20 | 20 | 28 | 160 | |||||||||||||
Note.— The ancestral state of each genotyped polymorphism was deduced from the chimpanzee and is represented here as a dot. NAT1 haplotypes were named in accordance with NAT Nomenclature.
As for the genotyping panel, NAT1 and NAT2 haplotype frequencies are reported in tables 5 and 6. Two NAT1 haplotypes, NAT1*10 and NAT1*4, account for 85%–100% of NAT1 diversity. In addition, genotyping results confirmed the sequence data, in that the divergent and low-frequency NAT1*11A haplotype is restricted to Eurasian populations. As for NAT2, the ancestral haplotype NAT2*4 and the remaining haplotypes associated with the fast-acetylator phenotype (NAT2*12 and NAT2*13) were most frequent in Bakola Pygmies and eastern Eurasians. Among the derived haplotypes associated with the slow-acetylator phenotype, NAT2*14 (191G→A) was African specific, NAT2*7 (857G→A) was observed mainly in eastern Eurasians, NAT2*5 (341T→C) was common in western and central Eurasians as well as in sub-Saharan Africans (Pygmies excepted), and NAT2*6 (590G→A) was found ubiquitously at intermediate frequencies.
Table 5.
Allelic Composition and Frequency of NAT1 Haplotypes in Our Genotyping Panel[Note]
|
Polymorphism |
Haplotype Frequency in Population |
|||||||||||||||||||
| Haplotype | −929 | 445 | (TAA)n | 1088 | 1095 | 1191 | Baka(2N=60) | Bakola(2N=80) | Ateke(2N=100) | Somali(2N=48) | Moroccan(2N=88) | Ashkenazi(2N=80) | Sardinian(2N=98) | Swedish(2N=100) | Saami(2N=96) | Turkmen(2N=100) | Gujarati(2N=100) | Thai(2N=88) | Chinese(2N=88) | Total(2N=1,126) |
| Ancestral (chimpanzee) | G | A | 4 | T | A | T | ||||||||||||||
| NAT1*10A | . | G | 8 | A | . | . | .050 | .075 | .190 | .146 | .125 | .138 | .112 | .080 | .146 | .250 | .190 | .409 | .489 | .189 |
| NAT1*10D | A | G | 8 | A | . | . | .617 | .438 | .370 | .271 | .170 | .038 | .102 | .100 | .063 | .110 | .110 | .045 | .171 | |
| NAT1*10R | A | G | 8 | A | . | G | .017 | 9E-04 | ||||||||||||
| NAT1*26D | . | G | 9 | A | . | . | .011 | .023 | .003 | |||||||||||
| NAT1*26E | A | G | 9 | A | . | . | .013 | .010 | .042 | .010 | .068 | .034 | .012 | |||||||
| NAT1*4A | . | G | 8 | . | C | G | .300 | .413 | .360 | .479 | .625 | .800 | .776 | .740 | .708 | .570 | .550 | .432 | .420 | .563 |
| NAT1*4B | A | G | 8 | . | C | G | .017 | .05 | .040 | .063 | .034 | .021 | .015 | |||||||
| NAT1*26B | . | G | 9 | . | C | G | .060 | .005 | ||||||||||||
| NAT1*26C | A | G | 9 | . | C | G | .011 | 9E-04 | ||||||||||||
| NAT1*18 | . | G | 7 | . | C | G | .010 | 9E-04 | ||||||||||||
| NAT1*3A | . | G | 8 | . | . | . | .010 | .030 | .023 | .005 | ||||||||||
| NAT1*3B | A | G | 8 | . | . | . | .013 | .030 | .011 | .013 | .010 | .060 | .063 | .010 | .010 | .019 | ||||
| NAT1*11A | A | . | 5 | . | . | . | .034 | .013 | .020 | .040 | .040 | .023 | .011 | .015 | ||||||
Note.— The ancestral state of each genotyped polymorphism was deduced from the chimpanzee and is represented here as a dot. NAT1 haplotypes were named in accordance with NAT Nomenclature.
Table 6.
Allelic Composition and Frequency of NAT2 Haplotypes in Our Genotyping Panel[Note]
|
Polymorphism |
Haplotype Frequency in Population |
||||||||||||||||||||
| Haplotype | 191 | 282 | 341 | 481 | 590 | 803 | 857 | Baka(2N=60) | Bakola(2N=80) | Ateke(2N=100) | Somali(2N=48) | Moroccan(2N=88) | Ashkenazi(2N=80) | Sardinian(2N=98) | Swedish(2N=100) | Saami(2N=96) | Turkmen(2N=100) | Gujarati(2N=100) | Thai(2N=88) | Chinese(2N=88) | Total(2N=1,126) |
| Ancestral (chimpanzee) | G | C | T | C | G | A | G | ||||||||||||||
| Acetylation status | S | F | S | F | S | F | S | ||||||||||||||
| NAT2*4 | . | . | . | . | . | . | . | .100 | .163 | .090 | .063 | .148 | .113 | .204 | .110 | .240 | .310 | .130 | .295 | .523 | .198 |
| NAT2*13 | . | T | . | . | . | . | . | .067 | .100 | .060 | .011 | .010 | .011 | .011 | .020 | ||||||
| NAT2*12B | . | T | . | . | . | G | . | .083 | .075 | .020 | .012 | ||||||||||
| NAT2*12A | . | . | . | . | . | G | . | .283 | .425 | .100 | .146 | .045 | .010 | .030 | .067 | ||||||
| NAT2*5A | . | . | C | T | . | . | . | .010 | .010 | .060 | .042 | .010 | .012 | ||||||||
| NAT2*5B | . | . | C | T | . | G | . | .217 | .050 | .370 | .396 | .511 | .513 | .531 | .500 | .427 | .230 | .290 | .114 | .068 | .329 |
| NAT2*5C | . | . | C | . | . | G | . | .083 | .030 | .010 | .030 | .030 | .040 | .017 | |||||||
| NAT2*5M | . | T | C | . | . | . | . | .010 | 9E-04 | ||||||||||||
| NAT2*14A | A | . | . | . | . | . | . | .030 | .021 | .004 | |||||||||||
| NAT2*14B | A | T | . | . | . | . | . | .033 | .050 | .100 | .014 | ||||||||||
| NAT2*6A | . | T | . | . | A | . | . | .133 | .138 | .160 | .354 | .250 | .363 | .245 | .280 | .156 | .300 | .430 | .386 | .250 | .266 |
| NAT2*6C | . | T | . | . | A | G | . | .010 | 9E-04 | ||||||||||||
| NAT2*7B | . | T | . | . | . | . | A | .020 | .021 | .034 | .013 | .020 | .125 | .120 | .060 | .193 | .148 | .061 | |||
Note.— The ancestral state of each genotyped polymorphism was deduced from the chimpanzee and is represented here as a dot. The fast (F) or slow (S) status of NAT2-derived mutations is reported. NAT2 haplotypes were named in accordance with NAT Nomenclature.
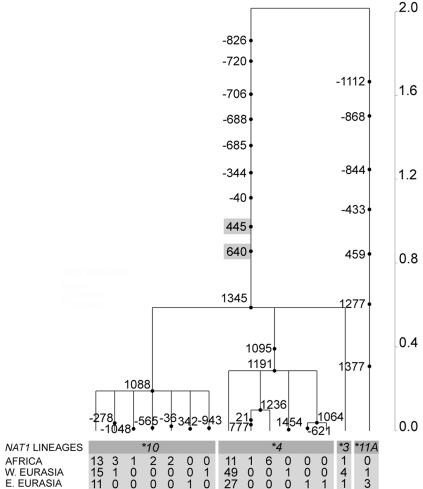
To investigate the tree topology and time depth of the three NAT loci, we next estimated the gene tree and the TMRCA of NAT1, NAT2, and NATP. The two divergent NAT1 lineages coalesced 2.01 ± 0.29 million years ago (MYA) (fig. 2), one of the highest estimated TMRCA values in the human genome (Excoffier 2002). By contrast, the TMRCA values of NAT2 (1.01 ± 0.27 MYA) and NATP (1.05 ± 0.24 MYA) were in agreement with neutral expectations, since most human neutral loci should coalesce ∼4Ne generations ago (i.e., ∼1 MYA) (Takahata 1993).
Figure 2.
NAT1 gene tree. Time is scaled in millions of years. Mutations are named for their physical positions along the NAT1 locus. Lineage absolute frequencies in Africa and western and eastern Eurasia are reported. Nonsynonymous mutations are highlighted in gray.
Population Variation in LD Patterns
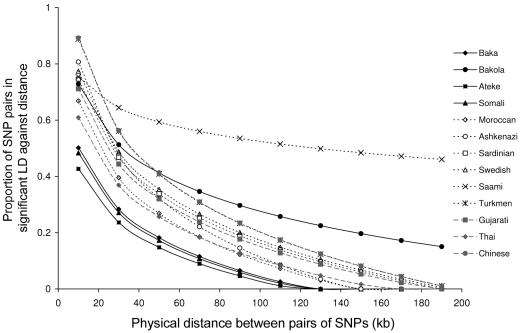
To determine global LD patterns in the 13 populations of our genotyping panel, we estimated D′ and r2 for the NATs region (data not shown). Both NAT1 and NAT2 genes showed significant and strong intragenic LD levels: the proportion of SNP pairs in significant LD equaled 87.5% and 89.6% in Africans and non-Africans, respectively, at the NAT1 locus and 73.7% and 84.0% at NAT2. The genomic structure of the entire 200-kb NATs region was made of two independent haplotype blocks, one corresponding to NAT1 and the other to NATP and NAT2. Further, we observed strong population variation in LD levels when plotting the proportion of SNP pairs in significant LD against physical distance for each population separately (fig. 3). Sub-Saharan Africans showed lower LD levels than non-Africans, with the clear exception of Bakola Pygmies. Both western and eastern Eurasians exhibited similar LD patterns, excluding the Saami, who were, by far, the population with the highest degree of allelic association.
Figure 3.
Proportion of SNP pairs in significant LD against physical distance in the NATs 200-kb region. In each population, genotyped SNPs were selected to have a MAF >10%. The Fisher’s exact test was used to assess LD significance, followed by Bonferroni corrections. SNP pairs were grouped into 20-kb bins.
Tests of the Standard Neutral Model
The absence of LD between the two NAT genes in all populations enabled us to study the evolutionary forces independently shaping NAT1 and NAT2 diversity. For the NAT1 coding region, tests were not feasible in four of eight populations because of a complete absence of exonic variation (table 2). In the remaining populations, most Tajima’s, Fu and Li’s, and Fay and Wu’s tests gave significant negative values (table 7). When these analyses were extended to NAT1 flanking regions, the same tests lost significance in Bakola Pygmies, whereas they turned out to be even more significant in French, Gujarati, and Thai because of an excess of singletons when mutations are not orientated for their ancestral state (TD and F*) or because of an excess of highly frequent derived variants when their ancestral state is considered (H). These results are mainly due to the presence of the low-frequency and highly divergent NAT1*11A haplotype in the three Eurasian populations (fig. 2). As for the NAT2 coding region, both Tajima’s D and Fu and Li’s F* in the Ashkenazi, Sardinians, and French and Fu and Li’s F* in the Saami were significantly positive (table 7). However, all tests were not significant anymore when both flanking and exonic NAT2 variation was considered. As for NATP, although it was found to be in LD with NAT2, all tests yielded nonsignificant values.
Table 7.
Sequenced-Based Neutrality Tests in NAT1, NAT2, and NATP
|
NAT1 Coding Regiona(870 bp) |
NAT1 Coding andFlanking Regions(2,605 bp) |
NAT2 Coding Region(870 bp) |
NAT2 Coding andFlanking Regions(2,820 bp) |
NATP Pseudogene(2,145 bp) |
|||||||||||
| Population | TD | F* | H | TD | F* | H | TD | F* | H | TD | F* | H | TD | F* | H |
| Bakola | −1.513b | −2.189b | .190 | .323 | −.298 | .579 | .007 | .456 | .653 | .090 | .852 | 1.726 | −.572 | .228 | −.947 |
| Bantu | NA | NA | NA | .563 | .828 | .632 | .303 | −.193 | .179 | −.091 | −.282 | 1.273 | −1.287 | −1.205 | −.011 |
| Ashkenazi | NA | NA | NA | −.044 | .131 | −1.674 | 2.516c | 1.797c | .505 | .374 | .080 | 1.557 | .545 | 1.255 | .284 |
| Sardinian | NA | NA | NA | .723 | .295 | .364 | 1.677b | 1.548b | −.091 | −.164 | −.523 | 1.090 | −.295 | .000 | .576 |
| French | −1.723b | −2.535b | −3.505b | −1.799b | −2.925b | −17.947c | 2.048b | 1.646b | 1.021 | .176 | .014 | 1.789 | .026 | −.119 | −1.011 |
| Saami | NA | NA | NA | 1.443 | 1.383 | −.305 | 1.365 | 1.480b | 1.358 | 1.017 | 1.281 | 3.136 | .290 | .135 | .926 |
| Gujarati | −1.723b | −2.535b | −3.505b | −1.974b | −3.143c | −18.600c | 1.355 | .895 | 1.242 | 1.135 | .653 | 2.673 | .393 | .582 | .326 |
| Thai | −1.384 | −.410 | −3.106b | −.904 | .560 | −15.016c | .818 | 1.281 | .767 | .441 | .828 | 2.386 | .809 | .206 | .344 |
NA = not applicable (no variation observed).
.01<P<.05.
P<.01.
To test significant differences in diversity levels among the three NAT loci, we performed the HKA test. The comparison between NAT1 and NATP was significant only among western and eastern Eurasians (P=.046 and .043, respectively), resulting from an excess of polymorphisms in NAT1, compared with fixed mutations. Again, these results are the consequence of the binary haplotype pattern observed at this locus (fig. 2). By contrast, the HKA tests comparing NAT1 versus NAT2 and NAT2 versus NATP yielded nonsignificant results. At the protein level, we compared the number of synonymous and nonsynonymous mutations in the two homologous genes. NAT1 exhibited a deficit in nonsynonymous mutations (KA/KS = 0.242). By contrast, NAT2 presented a KA/KS value closer to 1 (KA/KS = 0.802).
LRH Test for Recent Selection
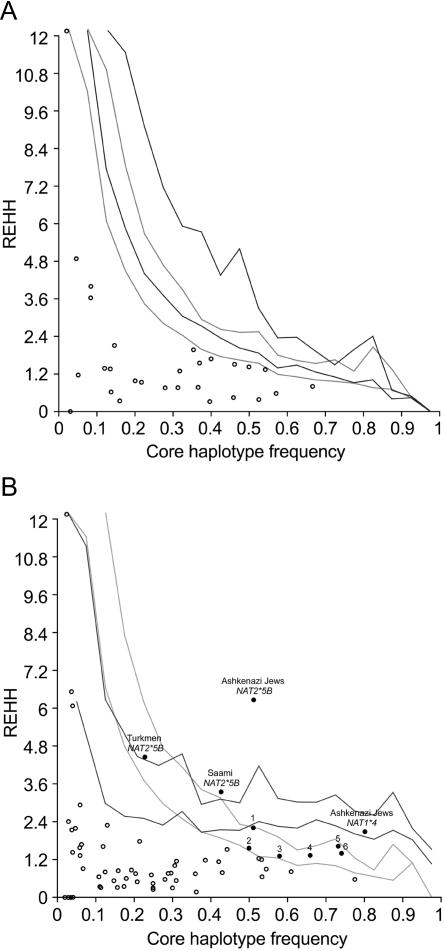
We next performed the LRH test, which is designed to identify mutations/haplotypes under recent positive selection by comparing the frequency of a given allele with the breakdown of LD around it (Sabeti et al. 2002). Our results for both NAT1 and NAT2 are reported separately for African and Eurasian populations (fig. 4A and 4B). P values were estimated for all core haplotypes in all populations against both simulations and the empirical distributions of the HapMap in Yoruban and European-descent populations. A single haplotype of NAT2 (NAT2*5B) appeared to deviate from neutrality in western and central Eurasian populations: Psim=.0001 (Pemp=.0006) in Ashkenazi Jews, .0062 (.0085) in Saami, .0363 (.0063) in Turkmen, .0124 (.0607) in Moroccans, and .0464 (.1836) in Swedish, with the same haplotype in Sardinians being close to significance (.0576 [.2178]). Interestingly, the NAT2*5B haplotype, which exhibited the highest frequencies (>50%) in western Eurasians (table 6), bears the nonsynonymous mutation 341T→C (I114T), which has been shown to lead to a slow-acetylation status (Zang et al. 2004). In addition, a single NAT1 haplotype (NAT1*4) appeared to deviate from the simulated distribution in the same populations that showed signals of positive selection for NAT2*5B: Psim=.0008 (Pemp=.0319) in Ashkenazi Jews, .0115 (.1346) in Saami, .0445 (.2498) in Turkmen, .0293 (.2694) in Moroccans, and .0090 (.1987) in Swedish. In contrast to NAT2*5B, the protein encoded by the NAT1*4 haplotype does not differ in enzyme activity with those encoded by the other NAT1 haplotypes observed in this study (e.g., NAT1*10, NAT1*3, and NAT1*11) (Hughes et al. 1998; de Leon et al. 2000).
Figure 4.
REHH plotted against core haplotype frequencies. Circles represent NAT1 and NAT2 core haplotypes. The 95th and 99th percentiles were calculated from both simulated data (gray lines) and an empirical distribution obtained from the screening of the entire chromosome 8 (black lines). Circles above the 95th percentile of simulated and/or empirical distributions are blackened. A, NAT1 and NAT2 sub-Saharan African core haplotypes are plotted against both simulated data and the empirical distribution of ∼40,000 Yoruban core haplotypes from the HapMap. B, NAT1 and NAT2 core haplotypes in western/central and eastern Eurasian populations are plotted against both the simulated data and the empirical distribution of ∼40,000 European-descent core haplotypes from the HapMap. Numbers affiliated with significant core haplotypes refer to: (1) Moroccan NAT2*5B, (2) Swedish NAT2*5B, (3) Turkmen NAT1*4, (4) Moroccan NAT1*4, (5) Saami NAT1*4, and (6) Swedish NAT1*4.
Growth Rate and Age of the NAT2 341T→C Mutation
The LD breakdown at the surrounding sites of a mutation is very informative for inferring allelic age estimates, through consideration of the recombination rate as a “genetic clock” (Labuda et al. 1997). The significant signal of selection detected for the NAT2 341T→C mutation, together with the functional consequences associated with this variant (i.e., reduced acetylation activity), prompted us to estimate the age of this mutation by use of both maximum-likelihood and coalescent-based methods, both of which gave similar results. To provide a comparison, we performed the same analyses for the 590G→A mutation, which is located 250 bp from 341T→C and is never observed on the same haplotype. Our results indicated that these two mutations started to increase in frequency at similar times and with comparable growth rates in all populations (table 8). The only exception to this pattern was the 341T→C mutation in western/central Eurasians. Our estimations showed that this mutation started to increase in frequency 6,315 years ago (95% CI 5,797–7,005 years) at a growth rate (0.062) twice as big as the values observed for the same mutation in eastern Eurasians (0.031) and for 590G→A in the entire Eurasian sample. Indeed, the growth rate of 341T→C in western/central Eurasians was significantly different from all the others, since their 95% CIs did not overlap (table 8).
Table 8.
Estimated Growth Rate and Age of the Mutations 341T→C and 590G→A (95% CI) of the NAT2 Gene[Note]
| Mutationand Parameter | Sub-Saharan Africans | Western/Central Eurasians | Eastern Eurasians |
| 341T→C: | |||
| p | .270 | .477 | .181 |
| r | .019 (.016–.024) | .062 (.055–.075) | .031 (.028–.037) |
| g | 15,652 (13,797–18,435) | 6,315 (5,797–7,005) | 12,627 (11,427–14,685) |
| 590G→A: | |||
| p | .185 | .262 | .362 |
| r | .023 (.020–.030) | .031 (.028–.037) | .034 (.031–.041) |
| g | 12,497 (10,932–14,958) | 12,762 (11,657–14,385) | 11,987 (10,940–13,643) |
Note.— p = relative frequency of the mutation; r = growth rate; g = age of the mutation in years.
Acetylation Phenotype Inference
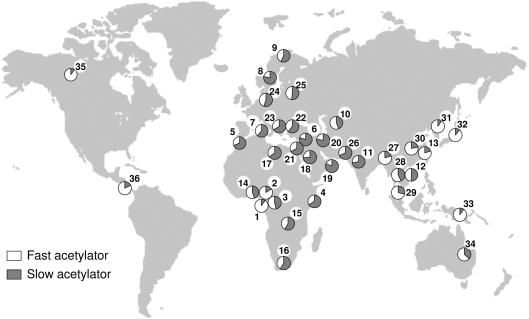
In view of the 95% NAT2 genotype-phenotype concordance (Cascorbi et al. 1995), we inferred the distribution of fast/slow–acetylation phenotypes across populations from our NAT2 genotyping panel and compared it with the phenotyping results of 23 healthy populations worldwide, reviewed for this occasion (fig. 5). To make both data sets comparable, we considered fast/slow heterozygotes to be fast acetylators because, even if they present a mean intermediate activity significantly different from that of fast homozygotes, they are mostly observed in the “fast acetylator activity peak” (Cascorbi et al. 1995). Phenotype frequencies showed strong variation among ethnic groups (fig. 5). The slow-acetylator phenotype is present at the lowest frequencies in eastern Asian and Native American populations as well as in the Pygmy groups studied here for the first time, whereas it exhibits the highest frequencies in Middle Eastern, European, central/south Eurasian, and African populations. The highest frequencies worldwide of the slow-acetylator phenotype are observed in the Ashkenazi population, in which it reaches 80%.
Figure 5.
Worldwide distribution of NAT2 acetylation phenotypes in healthy individuals. Each pie represents the population proportion of fast and slow acetylators. Populations numbered from 1 to 13 were analyzed in this study: (1) Bakola Pygmies, (2) Baka Pygmies, (3) Ateke Bantus, (4) Somali, (5) Morrocans, (6) Ashkenazi Jews, (7) Sardinians, (8) Swedes, (9) Saami, (10) Turkmen, (11) Gujarati, (12) Thai, and (13) Chinese. Numbers 14 to 36 refer to a reviewed population: (14) Yorubas (Jeyakumar and French 1981), (15) Zimbabweans (Nhachi 1988), (16) South Africans (Hodgkin et al. 1979), (17) Libyans (Karim et al. 1981), (18) Saudi Arabians (El-Yazigi et al. 1992), (19) Emiratis (Woolhouse et al. 1997), (20) Iranians (Sardas et al. 1993), (21) Jordanians (Irshaid et al. 1992), (22) Turkmen (Bozkurt et al. 1990), (23) Greeks (Asprodini et al. 1998), (24) Germans (Cascorbi et al. 1995), (25) Russians (Lil’in et al. 1984), (26) Pakistanis (Saleem et al. 1989), (27) Bangladeshi (Zaid et al. 2004), (28) Thai (Kukongviriyapan et al. 1984), (29) Malaysians (Ong et al. 1990), (30) Chinese (Zhao et al. 2000), (31) Koreans (Lee et al. 2002), (32) Japanese (Hashiguchi and Ebihara 1992), (33) Papua New Guineans (Hombhanje 1990), (34) Australian Aborigines (Ilett et al. 1993), (35) Eskimos (Eidus et al. 1974), and (36) Amerindians (Jorge-Nebert et al. 2002).
Discussion
The direct interaction of NAT1 and NAT2 gene products with the human chemical environment makes them potential targets of natural selection, provided that the exposure to xenobiotics has significantly influenced population fitness over time. Our population-based genetic study revealed distinctive patterns of variability for the two NAT genes, reflecting two very different evolutionary histories.
Reduced Variation and Deep-Rooting Genealogy in the NAT1 Gene
The NAT1 coding region is characterized by a reduced genetic diversity (π=0.021%), with four populations of eight showing no variation at all (table 2). In addition, the nearby 3′ UTR (TAA)n microsatellite of NAT1, which was also typed in the genotyping panel, presented low levels of heterozygosity (Hz=0.073), with the allele (TAA)8 accounting for 96.3% of the overall diversity (table 5). This deficit in heterozygosity was significant under both the stepwise mutation model and the two-phased mutation model (P<.05). Also, in strong contrast to NAT2, the functional mutations identified in NAT1 are present at very low frequencies (Upton et al. 2001). These observations, together with the KA/KS value of 0.242, are compatible with the action of purifying selection in shaping NAT1 diversity, in agreement with a previous study stating that the majority of human genes may be under weak negative selection (Bustamante et al. 2005). In this view, and considering that NAT1 is expressed in many tissues early in development and may play an additional role in the metabolism of folate (Sim et al. 2000), our genetic results suggest that its involvement in endogenous metabolic pathways might be more important than previously thought.
Purifying selection may not be the only evolutionary force that has influenced NAT1 diversity. Indeed, one of the most salient observations of this study is the highly divergent tree topology and high TMRCA (2.01 ± 0.29 MYA) of this locus (fig. 2). This binary pattern is translated into significant departures from neutrality in populations presenting the divergent haplotype NAT1*11A (see table 7 and results of the HKA test). The probability of finding such a high TMRCA under a Wright-Fisher model was found to be low (P=.029). Different hypotheses can be proposed to explain such long basal branches in the NAT1 gene tree. First, long-term balancing selection can result in divergent haplotype clusters, by maintaining two or more alleles over time, provided that they result in functional differences. Nevertheless, our data do not support this hypothesis, since the two nonsynonymous mutations separating the two clusters (fig. 2) have been shown to have no significant effects on the in vivo protein activity in human cells (Hein 2002) or on the stability and activity of the recombinant protein in yeast (Hughes et al. 1998). Any kind of selection due to a hitchhiking effect with neighbor genes is equally unlikely, because the two closest genes (ASAH1 located 5′ and NAT2 located 3′) behave as independent haplotype blocks (this study and the HapMap database). Furthermore, our sequence data from the NAT1 coding region are consistent with the action of purifying selection rather than balancing selection, with the first selective regime having a minor influence on tree topologies (Williamson and Orive 2002). Second, gene conversion could also lead to such divergent haplotype patterns by the replacement of a segment of NAT1 with a tract from its nearby paralogs (NAT2 and/or NATP). This alternative is unlikely, however, since the 17 SNPs separating the two divergent NAT1 lineages are not physically clustered (fig. 2) as one would expect after gene conversion between duplicated loci (Innan 2003). Thus, if gene conversion formed the basis of such a haplotype pattern, multiple conversion events must be invoked, with some tracts of lengths <5 bp. Yet, the conversion-tract lengths have been estimated to range from 55 bp to 290 bp, through sperm-typing analyses (Jeffreys and May 2004).
In this view, an alternative and most likely scenario to explain our data is a demographic event such as ancient population structure. A number of studies have recently reported gene genealogies that present not only unexpectedly old coalescent times (∼2 MYA) but also long basal branches (Harris and Hey 1999; Webster et al. 2003; Barreiro et al. 2005; Garrigan et al. 2005; Hayakawa et al. 2005). Our observations at NAT1, together with these studies, further support the view that some diversity in the genome of modern humans may have persisted from a structured ancestral population (Harding and McVean 2004). In addition, NAT1*11A appears to be absent in sub-Saharan Africa, since it was not detected in either our genotyping panel of 144 sub-Saharan Africans from distinct geographic locations or 600 African American individuals reported elsewhere (Upton et al. 2001). Therefore, the observation that the NAT1 gene tree is rooted in Eurasia questions the geographic location of such a structured ancestral population (Takahata et al. 2001). The origins of NAT1*11A could thus be placed either in sub-Saharan Africa, from where it must have subsequently disappeared, or in Eurasia. Should the latter be the case, the NAT1 gene tree is at odds with the commonly accepted replacement hypothesis (Lewin 1987) and is more parsimoniously explained by the occurrence of partial hybridization between modern humans expanding from Africa and preexisting hominids in Eurasia, as recently sustained by the RRM2P4 locus (Garrigan et al. 2005). However, such inferences require further support from the analyses of multiple independent loci in increased numbers of samples and human populations.
The NAT2 Gene: An Advantage to Be a Slow Acetylator?
The significantly positive values observed for most sequence-based neutrality tests of NAT2 in western Eurasians (table 7) suggest the action of natural selection. Likewise, population size reductions—such as that probably experienced by non-African populations during the out-of-Africa exit (Marth et al. 2003) or, more recently, by Ashkenazi Jews (Behar et al. 2004)—could have also inflated TD and F* values (Przeworski et al. 2000). However, these demographic events should have equally influenced neutrality statistics for other non-African populations (i.e., eastern Eurasians), which is not the case. These observations argue thus in favor of the action of natural selection. Conversely, interspecific tests (i.e., KA/KS and MK tests) do not rule out the neutral evolution of NAT2 diversity, which does not show any clear excess or lack of nonsynonymous mutations. In view of these apparently contradictory results, it is plausible that the frequencies, rather than the number of these mutations, have been influenced by selection, suggesting more subtle and recent fluctuations in the selective pressures operating on NAT2.
Such changes can be detected by the LRH test, which aims to identify haplotypes under recent positive selection. Moreover, this approach should be robust to the confounding effects of demography, since it corrects the extended haplotype homozygosity (EHH) of a given core haplotype by the EHH of all other haplotypes at the same core region (Sabeti et al. 2002). Overall, the LRH tests were more significant when NAT1 and NAT2 core haplotypes were compared with the simulated than with the empirical distribution. Manifestly, the empirical distribution also includes genes under selection that bias REHH toward higher values, and it allows the detection of selected haplotypes in a conservative way. Independently of which background distribution was used (e.g., simulated or empirical), the same NAT2 haplotype, NAT2*5B, was detected to depart from neutrality in western and central Eurasians (fig. 4B). These populations also showed significant P values for a single NAT1 haplotype, NAT1*4. It is worth mentioning that ∼80% of NAT2*5B haplotypes are associated with NAT1*4 in western/central Eurasians (vs. 43% and 60% in sub-Saharan Africans and eastern Eurasians, respectively). Thus, the signals detected in both NAT1 and NAT2 may be the result of a single event of selection targeting a long-range haplotype composed of both NAT1*4 and NAT2*5B. Several lines of evidence, however, strongly support that the NAT1 haplotype does not harbor the functional cause of such a selective event: P values are globally less significant for NAT1*4 than for NAT2*5B, and, most importantly, all the NAT1 haplotypes observed in this study encode proteins with identical enzymatic activity (Hughes et al. 1998; de Leon et al. 2000). In sharp contrast, NAT2*5B bears the 341T→C mutation that is well known to encode an altered slow-acetylator protein (Zang et al. 2004).
Further support for the action of selection on the slow-encoding 341T→C NAT2 mutation comes from its growth-rate estimate, which showed that this mutation has increased in frequency twice as quickly as expected (r=0.062) (table 8) only among western and central Eurasians. Previous estimations (Slatkin and Bertorelle 2001), together with our own, indicate that Eurasian populations have grown at a rate of ∼0.030. Because population growth and selection have additive effects on the growth rate of a mutation (Slatkin and Rannala 1997), these observations suggest that the 341C allele has been driven by selection with a selective coefficient of ∼0.062-0.030=0.032. Using a more accurate approach (Wright 1969), we estimated the 95% CI of this selective coefficient as 0.0124–0.0913. These figures reinforce the idea of a selective advantage of the 341C allele, even if weaker than other examples of recent but strong positive selection, such as lactase persistence (s ∼0.09–0.19) (Bersaglieri et al. 2004) or the G6PD A− alleles (s>0.1) (Saunders et al. 2005).
Altogether, both the LRH test and the growth rate of 341T→C argue in favor of positive selection acting on this slow-encoding mutation, which would imply a general selective advantage for the slow-acetylator phenotype. However, three other NAT2 variants (191G→A, 590G→A, and 857G→A) also encode slow proteins but do not show any significant departure from neutrality. Actually, the 341T→C mutation involves the greatest reduction in NAT2 enzymatic activity (Hein et al. 2000). In this context, it is very plausible that all NAT2 slow-acetylating variants have been subject to weak selective pressures, the signal of selection being detectable only in the mutation 341T→C causing the “slowest-acetylation” phenotype. Thus, the predicted increase in frequency of all slow mutations, in response to directional selection, would explain the observed excess of intermediate-frequency alleles in western Eurasian populations, as depicted by the significant positive values of TD (table 7).
The footprints of natural selection identified in western/central Eurasians raise the question of which event(s) may have provoked fluctuations in the spectrum of xenobiotics inactivated/activated by NAT2 (e.g., NAT2 activates heterocyclic carcinogens found in well-cooked meat [Hein et al. 2000; Hein 2002]) in these populations. In this context, given the geographic distribution of the slow-acetylator phenotype and the estimated expansion time of the slowest-encoding 341T→C mutation (5,797–7,005 years ago in western/central Eurasians), it is tempting to hypothesize that the emergence of agriculture in western Eurasia could be at the basis of such environmental changes. Indeed, there is accumulating evidence that this major transition resulted in a profound modification of human diets and lifestyles (Cordain et al. 2005) and, consequently, in the exposure of humans to chemical environments (Ferguson 2002). Moreover, the highest frequencies of slow acetylators are observed in the Middle East (fig. 5), one of the first regions where agriculture originated ∼10,000 years ago, and these frequencies decrease toward western Europe, North Africa, and India, three regions where agriculture was subsequently diffused from the Fertile Crescent (Harris 1996). However, the hypothesis that the transition to agriculture influenced both the human exposure to xenobiotic environments and, consequently, the selective pressures at NAT2 remains tentative and requires a better characterization of the naturally occurring substrates of the NAT2 enzyme.
Conclusion
The diversity patterns observed at the NATs region clearly illustrate our current vision of the human genome as a “mosaic of discrete segments,” each with its own individual evolutionary history (Pääbo 2003). Whereas NAT1 could belong to a small proportion of nuclear loci that kept traces of an ancient population structure, the NAT2 gene gives some insights into the evolutionary processes that could make some present-day detrimental mutations frequent. The theory of the “thrifty genotype” proposes that common diseases, such as obesity and diabetes, are the result of a past advantage to efficiently metabolize rare food sources that are no longer restricted (Neel 1962). By analogy, the slow NAT2 acetylator haplotype (NAT2*5B), which is found at high frequencies worldwide and which we propose conferred some selective advantage, at least in western/central Eurasian populations, exhibits today the strongest association with susceptibility to bladder cancer and adverse drug reactions (Hein et al. 2000; Hein 2002). This “evolutionary conflict” could be widespread among genes involved in carcinogen metabolism, since two major events in human history—the Neolithic transition from foraging to agriculture and the more recent Industrial Revolution—may have dramatically changed our exposure to the damaging effects of environmental carcinogens. Consequently, dissecting the evolutionary processes that have shaped patterns of diversity of the genes involved in drug metabolism may represent a major analytical tool not only to identify those having played a crucial role in the past adaptation of Homo sapiens but also to better understand our present-day susceptibility to disease.
Acknowledgments
We warmly acknowledge M. Slatkin for supplying source codes for growth-rate estimations, R. R. Hudson for the modifications of the MS program, E. Sim for invaluable advice in the beginning of this project, and two anonymous reviewers for constructive criticisms on the early version of the manuscript. We also thank A. Novelletto, A. Froment, J. M. Hombert, H. Rouba, and S. Santachiara-Benerecetti, for kindly providing us with DNA samples. This work was supported by CNRS and Institut Pasteur research funding. E.P. was supported by the French Ministry of Research Ph.D. program.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Arlequin v.2.001, http://lgb.unige.ch/arlequin/
- BEST v.1.0, http://genomethods.org/best/
- Bottleneck software, http://www.montpellier.inra.fr/CBGP/softwares/bottleneck/bottleneck.html
- dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/
- DMLE+ v.2.2, http://www.dmle.org/
- DnaSP v.4.0, http://www.ub.es/dnasp/
- GENALYS software, http://software.cng.fr/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for NAT1, NAT2, and NATP [accession numbers DQ305496–DQ305975])
- GENETREE software, http://www.stats.ox.ac.uk/~griff/software.html
- International HapMap Project, http://www.hapmap.org/
- MS program, http://home.uchicago.edu/~rhudson1/
- NAT Nomenclature, http://www.louisville.edu/medschool/pharmacology/NAT.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NAT1 and NAT2)
- PHASE v.2.1.1, http://www.stat.washington.edu/stephens/phase.html
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/
References
- Asprodini EK, Zifa E, Papageorgiou I, Benakis A (1998) Determination of N-acetylation phenotyping in a Greek population using caffeine as a metabolic probe. Eur J Drug Metab Pharmacokinet 23:501–506 [DOI] [PubMed] [Google Scholar]
- Austerlitz F, Kalaydjieva L, Heyer E (2003) Detecting population growth, selection and inherited fertility from haplotypic data in humans. Genetics 165:1579–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro LB, Patin E, Neyrolles O, Cann HM, Gicquel B, Quintana-Murci L (2005) The heritage of pathogen pressures and ancient demography in the human innate-immunity CD209/CD209L region. Am J Hum Genet 77:869–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Hammer MF, Garrigan D, Villems R, Bonne-Tamir B, Richards M, Gurwitz D, Rosengarten D, Kaplan M, Pergola SD, Quintana-Murci L, Skorecki K (2004) MtDNA evidence for a genetic bottleneck in the early history of the Ashkenazi Jewish population. Eur J Hum Genet 12:355–364 10.1038/sj.ejhg.5201156 [DOI] [PubMed] [Google Scholar]
- Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, Rhodes M, Reich DE, Hirschhorn JN (2004) Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet 74:1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M, Grant DM, McBride W, Heim M, Meyer UA (1990) Human arylamine N-acetyltransferase genes: isolation, chromosomal localization, and functional expression. DNA Cell Biol 9:193–203 [DOI] [PubMed] [Google Scholar]
- Bozkurt A, Basci NE, Kalan S, Tuncer M, Kayaalp SO (1990) N-acetylation phenotyping with sulphadimidine in a Turkish population. Eur J Clin Pharmacol 38:53–56 10.1007/BF00314803 [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, Civello D, Adams MD, Cargill M, Clark AG (2005) Natural selection on protein-coding genes in the human genome. Nature 437:1153–1157 10.1038/nature04240 [DOI] [PubMed] [Google Scholar]
- Cartwright RA, Glashan RW, Rogers HJ, Ahmad RA, Barham-Hall D, Higgins E, Kahn M (1982) A role of N-acetyltransferase phenotypes in bladder carcinogenesis: a pharmacogenetic epidemiological approach to bladder cancer. Lancet 2:842–846 10.1016/S0140-6736(82)90810-8 [DOI] [PubMed] [Google Scholar]
- Cascorbi I, Drakoulis N, Brockmoller J, Maurer A, Sperling K, Roots I (1995) Arylamine N-acetyltransferase (NAT2) mutations and their allelic linkage in unrelated Caucasian individuals: correlation with phenotypic activity. Am J Hum Genet 57:581–592 [PMC free article] [PubMed] [Google Scholar]
- Cordain L, Eaton SB, Sebastian A, Mann N, Lindeberg S, Watkins BA, O’Keefe JH, Brand-Miller J (2005) Origins and evolution of the Western diet: health implications for the 21st century. Am J Clin Nutr 81:341–354 [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon JH, Vatsis KP, Weber WW (2000) Characterization of naturally occurring and recombinant human N-acetyltransferase variants encoded by NAT1. Mol Pharmacol 58:288–299 [DOI] [PubMed] [Google Scholar]
- Eidus L, Hodgkin MM, Schaefer O, Jessamine AG (1974) Distribution of isoniazid inactivators determined in Eskimos and Canadian college students by a urine test. Rev Can Biol 33:117–123 [PubMed] [Google Scholar]
- El-Yazigi A, Johansen K, Raines DA, Dossing M (1992) N-acetylation polymorphism and diabetes mellitus among Saudi Arabians. J Clin Pharmacol 32:905–910 [DOI] [PubMed] [Google Scholar]
- Excoffier L (2002) Human demographic history: refining the recent African origin model. Curr Opin Genet Dev 12:675–682 10.1016/S0959-437X(02)00350-7 [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu CI (2000) Hitchhiking under positive Darwinian selection. Genetics 155:1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson LR (2002) Natural and human-made mutagens and carcinogens in the human diet. Toxicology 181–182:79–82 [DOI] [PubMed] [Google Scholar]
- Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- Garcia-Closas M, Malats N, Silverman D, Dosemeci M, Kogevinas M, Hein DW, Tardon A, Serra C, Carrato A, Garcia-Closas R, Lloreta J, Castano-Vinyals G, Yeager M, Welch R, Chanock S, Chatterjee N, Wacholder S, Samanic C, Tora M, Fernandez F, Real FX, Rothman N (2005) NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet 366:649–659 10.1016/S0140-6736(05)67137-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D, Mobasher Z, Severson T, Wilder JA, Hammer MF (2005) Evidence for archaic Asian ancestry on the human X chromosome. Mol Biol Evol 22:189–192 10.1093/molbev/msi013 [DOI] [PubMed] [Google Scholar]
- Griffiths RC, Tavaré S (1994) Sampling theory for neutral alleles in a varying environment. Philos Trans R Soc Lond B Biol Sci 344:403–410 [DOI] [PubMed] [Google Scholar]
- Harding RM, McVean G (2004) A structured ancestral population for the evolution of modern humans. Curr Opin Genet Dev 14:667–674 10.1016/j.gde.2004.08.010 [DOI] [PubMed] [Google Scholar]
- Harris DR (ed) (1996) The origins and spread of agriculture and pastoralism in Eurasia. Smithsonian Institution Press, Washington, DC [Google Scholar]
- Harris EE, Hey J (1999) X chromosome evidence for ancient human histories. Proc Natl Acad Sci USA 96:3320–3324 10.1073/pnas.96.6.3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi M, Ebihara A (1992) Acetylation polymorphism of caffeine in a Japanese population. Clin Pharmacol Ther 52:274–276 [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Aki I, Varki A, Satta Y, Takahata N (2005) Fixation of the human-specific CMP-N-acetylneuraminic acid hydroxylase pseudogene and implications of haplotype diversity for human evolution. Genetics (http://www.genetics.org/cgi/rapidpdf/genetics.105.046995v1) (electronically published November 4, 2005; accessed January 12, 2006) [DOI] [PMC free article] [PubMed]
- Hein DW (2002) Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res 506–507:65–77 [DOI] [PubMed] [Google Scholar]
- Hein DW, Doll MA, Fretland AJ, Leff MA, Webb SJ, Xiao GH, Devanaboyina US, Nangju NA, Feng Y (2000) Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol Biomarkers Prev 9:29–42 [PubMed] [Google Scholar]
- Hill WG, Robertson A (1968) The effects of inbreeding at loci with heterozygote advantage. Genetics 60:615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin MM, Eidus L, Bailey WC (1979) Isoniazid phenotyping of black as well as white patients. Can J Physiol Pharmacol 57:760–763 [DOI] [PubMed] [Google Scholar]
- Hombhanje F (1990) An assessment of acetylator polymorphism and its relevance in Papua New Guinea. P N G Med J 33:107–110 [PubMed] [Google Scholar]
- Huang YS, Chern HD, Su WJ, Wu JC, Lai SL, Yang SY, Chang FY, Lee SD (2002) Polymorphism of the N-acetyltransferase 2 gene as a susceptibility risk factor for antituberculosis drug-induced hepatitis. Hepatology 35:883–889 10.1053/jhep.2002.32102 [DOI] [PubMed] [Google Scholar]
- Hudson RR (2002) Generating samples under a Wright-Fisher neutral model. Bioinformatics 18:337–338 10.1093/bioinformatics/18.2.337 [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M (1987) A test of neutral molecular evolution based on nucleotide data. Genetics 116:153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes NC, Janezic SA, McQueen KL, Jewett MA, Castranio T, Bell DA, Grant DM (1998) Identification and characterization of variant alleles of human acetyltransferase NAT1 with defective function using p-aminosalicylate as an in-vivo and in-vitro probe. Pharmacogenetics 8:55–66 [DOI] [PubMed] [Google Scholar]
- Ilett KF, Chiswell GM, Spargo RM, Platt E, Minchin RF (1993) Acetylation phenotype and genotype in Aboriginal leprosy patients from the north-west region of western Australia. Pharmacogenetics 3:264–269 [DOI] [PubMed] [Google Scholar]
- Innan H (2003) A two-locus gene conversion model with selection and its application to the human RHCE and RHD genes. Proc Natl Acad Sci USA 100:8793–8798 10.1073/pnas.1031592100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irshaid Y, al-Hadidi H, Abuirjeie M, Latif A, Sartawi O, Rawashdeh N (1992) Acetylator phenotypes of Jordanian diabetics. Eur J Clin Pharmacol 43:621–623 10.1007/BF02284960 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, May CA (2004) Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet 36:151–156 10.1038/ng1287 [DOI] [PubMed] [Google Scholar]
- Jeyakumar LH, French MR (1981) Polymorphic acetylation of sulfamethazine in a Nigerian (Yoruba) population. Xenobiotica 11:319–321 [DOI] [PubMed] [Google Scholar]
- Jorge-Nebert LF, Eichelbaum M, Griese EU, Inaba T, Arias TD (2002) Analysis of six SNPs of NAT2 in Ngawbe and Embera Amerindians of Panama and determination of the Embera acetylation phenotype using caffeine. Pharmacogenetics 12:39–48 10.1097/00008571-200201000-00006 [DOI] [PubMed] [Google Scholar]
- Karim AK, Elfellah MS, Evans DA (1981) Human acetylator polymorphism: estimate of allele frequency in Libya and details of global distribution. J Med Genet 18:325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M (1968) Evolutionary rate at the molecular level. Nature 217:624–626 [DOI] [PubMed] [Google Scholar]
- Kukongviriyapan V, Lulitanond V, Areejitranusorn C, Kongyingyose B, Laupattarakasem P (1984) N-acetyltransferase polymorphism in Thailand. Hum Hered 34:246–249 [DOI] [PubMed] [Google Scholar]
- Labuda D, Zietkiewicz E, Labuda M (1997) The genetic clock and the age of the founder effect in growing populations: a lesson from French Canadians and Ashkenazim. Am J Hum Genet 61:768–771 [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Lee KA, Ki CS, Kwon OJ, Kim HJ, Chung MP, Suh GY, Kim JW (2002) Complete sequencing of a genetic polymorphism in NAT2 in the Korean population. Clin Chem 48:775–777 [PubMed] [Google Scholar]
- Lewin R (1987) Africa: cradle of modern humans. Science 237:1292–1295 [DOI] [PubMed] [Google Scholar]
- Lewontin RC (1964) The interaction of selection and linkage. II. Optimum models. Genetics 50:757–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lil’in ET, Korsunskaia MP, Meksin VA, Drozdov ES, Nazarov VV (1984) Distribution of acetylator phenotypes in the normal Moscow city population and in chronic alcoholism. Genetika 20:1557–1559 [PubMed] [Google Scholar]
- Marth G, Schuler G, Yeh R, Davenport R, Agarwala R, Church D, Wheelan S, Baker J, Ward M, Kholodov M, Phan L, Czabarka E, Murvai J, Cutler D, Wooding S, Rogers A, Chakravarti A, Harpending HC, Kwok PY, Sherry ST (2003) Sequence variations in the public human genome data reflect a bottlenecked population history. Proc Natl Acad Sci USA 100:376–831 10.1073/pnas.222673099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654 10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- Neel JV (1962) Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet 14:353–362 [PMC free article] [PubMed] [Google Scholar]
- Nei M (ed) (1987) Molecular evolutionary genetics. Columbia University Press, New York [Google Scholar]
- Nhachi CF (1988) Polymorphic acetylation of sulphamethazine in a Zimbabwe population. J Med Genet 25:29–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong ML, Mant TG, Veerapen K, Fitzgerald D, Wang F, Manivasagar M, Bosco JJ (1990) The lack of relationship between acetylator phenotype and idiopathic systemic lupus erythematosus in a South-east Asian population: a study of Indians, Malays and Malaysian Chinese. Br J Rheumatol 29:462–464 [DOI] [PubMed] [Google Scholar]
- Pääbo S (2003) The mosaic that is our genome. Nature 421:409–412 10.1038/nature01400 [DOI] [PubMed] [Google Scholar]
- Przeworski M, Hudson RR, Di Rienzo A (2000) Adjusting the focus on human variation. Trends Genet 16:296–302 10.1016/S0168-9525(00)02030-8 [DOI] [PubMed] [Google Scholar]
- Reeve JP, Rannala B (2002) DMLE+: Bayesian linkage disequilibrium gene mapping. Bioinformatics 18:894–895 10.1093/bioinformatics/18.6.894 [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497 10.1093/bioinformatics/btg359 [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, Ackerman HC, Campbell SJ, Altshuler D, Cooper R, Kwiatkowski D, Ward R, Lander ES (2002) Detecting recent positive selection in the human genome from haplotype structure. Nature 419:832–837 10.1038/nature01140 [DOI] [PubMed] [Google Scholar]
- Saleem M, Malik SA, Ahmed M, Saleem N (1989) Isoniazid acetylation and polymorphism in humans. J Pak Med Assoc 39:285–286 [PubMed] [Google Scholar]
- Sardas S, Lahijany B, Cok I, Karakaya AE (1993) N-acetylation phenotyping with sulfamethazine in an Iranian population. Pharmacogenetics 3:131–134 [PubMed] [Google Scholar]
- Saunders MA, Slatkin M, Garner C, Hammer MF, Nachman MW (2005) The span of linkage disequilibrium caused by selection on G6PD in humans. Genetics 171:1219–1229 10.1534/genetics.105.048140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D (2005) Calibrating a coalescent simulation of human genome sequence variation. Genome Res 15:1576–1583 10.1101/gr.3709305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L (2000) Arlequin version 2.000: a software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva [Google Scholar]
- Sebastiani P, Lazarus R, Weiss ST, Kunkel LM, Kohane IS, Ramoni MF (2003) Minimal haplotype tagging. Proc Natl Acad Sci USA 100:9900–9905 10.1073/pnas.1633613100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim E, Payton M, Noble M, Minchin R (2000) An update on genetic, structural and functional studies of arylamine N-acetyltransferases in eukaryotes and prokaryotes. Hum Mol Genet 9:2435–2441 10.1093/hmg/9.16.2435 [DOI] [PubMed] [Google Scholar]
- Slatkin M, Bertorelle G (2001) The use of intraallelic variability for testing neutrality and estimating population growth rate. Genetics 158:865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Rannala B (1997) Estimating the age of alleles by use of intraallelic variability. Am J Hum Genet 60:447–458 [PMC free article] [PubMed] [Google Scholar]
- Soranzo N, Bufe B, Sabeti PC, Wilson JF, Weale ME, Marguerie R, Meyerhof W, Goldstein DB (2005) Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr Biol 15:1257–1265 10.1016/j.cub.2005.06.042 [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Matsuda F, Margetic N, Lathrop M (2003) Automated identification of single nucleotide polymorphisms from sequencing data. J Bioinform Comput Biol 1:253–265 10.1142/S021972000300006X [DOI] [PubMed] [Google Scholar]
- Takahata N (1993) Allelic genealogy and human evolution. Mol Biol Evol 10:2–22 [DOI] [PubMed] [Google Scholar]
- Takahata N, Lee SH, Satta Y (2001) Testing multiregionality of modern human origins. Mol Biol Evol 18:172–183 [DOI] [PubMed] [Google Scholar]
- Tang K, Wong LP, Lee EJ, Chong SS, Lee CG (2004) Genomic evidence for recent positive selection at the human MDR1 gene locus. Hum Mol Genet 13:783–797 10.1093/hmg/ddh099 [DOI] [PubMed] [Google Scholar]
- Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A (2004) CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet 75:1059–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomajian C, Ajioka RS, Jorde LB, Kushner JP, Kreitman M (2003) A method for detecting recent selection in the human genome from allele age estimates. Genetics 165:287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomajian C, Kreitman M (2002) Sequence variation and haplotype structure at the human HFE locus. Genetics 161:1609–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton A, Johnson N, Sandy J, Sim E (2001) Arylamine N-acetyltransferases: of mice, men and microorganisms. Trends Pharmacol Sci 22:140–146 10.1016/S0165-6147(00)01639-4 [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang B, Tang K, Lee EJ, Chong SS, Lee CG (2005) A functional polymorphism within the MRP1 gene locus identified through its genomic signature of positive selection. Hum Mol Genet 14:2075–2087 10.1093/hmg/ddi212 [DOI] [PubMed] [Google Scholar]
- Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276 10.1016/0040-5809(75)90020-9 [DOI] [PubMed] [Google Scholar]
- Weber WW (1987) The acetylator genes and drug response. Oxford University Press, New York [Google Scholar]
- Webster MT, Clegg JB, Harding RM (2003) Common 5′ β-globin RFLP haplotypes harbour a surprising level of ancestral sequence mosaicism. Hum Genet 113:123–139 [DOI] [PubMed] [Google Scholar]
- Williamson S, Orive ME (2002) The genealogy of a sequence subject to purifying selection at multiple sites. Mol Biol Evol 19:1376–1384 [DOI] [PubMed] [Google Scholar]
- Wooding S, Kim UK, Bamshad MJ, Larsen J, Jorde LB, Drayna D (2004) Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet 74:637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding SP, Watkins WS, Bamshad MJ, Dunn DM, Weiss RB, Jorde LB (2002) DNA sequence variation in a 3.7-kb noncoding sequence 5′ of the CYP1A2 gene: implications for human population history and natural selection. Am J Hum Genet 71:528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse NM, Qureshi MM, Bastaki SM, Patel M, Abdulrazzaq Y, Bayoumi RA (1997) Polymorphic N-acetyltransferase (NAT2) genotyping of Emiratis. Pharmacogenetics 7:73–82 [DOI] [PubMed] [Google Scholar]
- Wright S (ed) (1969) Evolution and the genetics of population. University of Chicago Press, Chicago, p 33 [Google Scholar]
- Zaid RB, Nargis M, Neelotpol S, Hannan JM, Islam S, Akhter R, Ali L, Azad-Khan AK (2004) Acetylation phenotype status in a Bangladeshi population and its comparison with that of other Asian population data. Biopharm Drug Dispos 25:237–241 10.1002/bdd.403 [DOI] [PubMed] [Google Scholar]
- Zang Y, Zhao S, Doll MA, States JC, Hein DW (2004) The T341C (Ile114Thr) polymorphism of N-acetyltransferase 2 yields slow acetylator phenotype by enhanced protein degradation. Pharmacogenetics 14:717–723 10.1097/00008571-200411000-00002 [DOI] [PubMed] [Google Scholar]
- Zhao B, Seow A, Lee EJ, Lee HP (2000) Correlation between acetylation phenotype and genotype in Chinese women. Eur J Clin Pharmacol 56:689–692 10.1007/s002280000203 [DOI] [PubMed] [Google Scholar]