Abstract
Eukaryotic cells reprogram their global patterns of gene expression in response to stress. Recent studies in Schizosaccharomyces pombe showed that the RNA-binding protein Csx1 plays a central role in controlling gene expression during oxidative stress. It does so by stabilizing atf1+ mRNA, which encodes a subunit of a bZIP transcription factor required for gene expression during oxidative stress. Here, we describe two related proteins, Cip1 and Cip2, that were identified by multidimensional protein identification technology (MudPIT) as proteins that coprecipitate with Csx1. Cip1 and Cip2 are cytoplasmic proteins that have RNA recognition motifs (RRMs). Neither protein is essential for viability, but a cip1Δ cip2Δ strain grows poorly and has altered cellular morphology. Genetic epistasis studies and whole genome expression profiling show that Cip1 and Cip2 exert posttranscriptional control of gene expression in a manner that is counteracted by Csx1. Notably, the sensitivity of csx1Δ cells to oxidative stress and their inability to induce expression of Atf1-dependent genes are partially rescued by cip1Δ and cip2Δ mutations. This study emphasizes the importance of a modulated mRNA stability in the eukaryotic stress response pathways and adds new information to the role of RNA-binding proteins in the oxidative stress response.
INTRODUCTION
Reactive oxygen species (ROS) are found in all aerobically growing cells. Changes in the intracellular redox state actively regulate several signaling pathways that control essential biological processes (Finkel, 2003; Torres, 2003). However, cell survival requires tight control of the cellular redox state. When ROS increase beyond homeostatic concentrations, they react with a multitude of molecules (such as lipids, proteins, and nucleic acids), resulting in impaired cellular functions and formation of toxic species. In fact, oxidative stress is thought to be an important factor in aging, in the progression of cancer, and in many common neurodegenerative diseases such as amyotrophic lateral sclerosis, Alzheimer's disease, Huntington's disease, and Parkinson's disease (Migliore and Coppede, 2002; Barnham et al., 2004). Antioxidant proteins such as catalase, superoxide dismutase, glutathione peroxidase, and glutathione reductase are part of the defense mechanisms that help the cells to deal with oxidative stress.
The evolutionary conserved mitogen-activated protein kinase (MAPK) pathways control the expression of many genes in response to oxidative stress (reviewed in Martindale and Holbrook, 2002). In Schizosaccharomyces pombe, the Spc1 (Sty1, Phh1) MAPK pathway is essential for the cellular response to different forms of stress, including oxidative stress, hyperosmotic stress, heat, UV light, and nutrient limitation (Millar et al., 1995; Shiozaki and Russell, 1995; Degols et al., 1996; Degols and Russell, 1997). Spc1 is activated through phosphorylation by the MAPK kinase (MAPKK) Wis1, which in turn is activated through phosphorylation by two MAPKK kinases (MAPKKK), Wis4 and Win1 (Samejima et al., 1997; Shieh et al., 1997, 1998; Shiozaki et al., 1997; Samejima et al., 1998; Quinn et al., 2002). In addition to Wis1, the tyrosine phosphatases Pyp1 and Pyp2 participate in the negative regulation of Spc1 (Millar et al., 1995; Shiozaki and Russell, 1995; Samejima et al., 1997; Shieh et al., 1997, 1998; Shiozaki et al., 1997, 1998). Spc1 regulates stress-dependent transcription through the Atf1–Pcr1 heterodimeric bZip transcription factor complex (Toda et al., 1991; Takeda et al., 1995; Kumada et al., 1996; Wilkinson et al., 1996; Toone et al., 1998; Yamada et al., 1999; Nguyen et al., 2000; Quinn et al., 2002). Pap1, a bZip transcription factor whose activity seems to be indirectly influenced by Spc1, also participates in transcriptional regulation during oxidative stress (Vivancos et al., 2004, 2005). Pap1 is a homologue of c-Jun and activates target genes in response to low levels of H2O2, whereas the transcriptional response to higher concentrations of H2O2 and other kinds of stress is mediated by Atf1–Pcr1 (Toone et al., 1998; Quinn et al., 2002).
Components of the Spc1 MAPK cascade are functionally and structurally homologous to members of the HOG MAPK pathway in Saccharomyces cerevisiae and to the mammalian and Drosophila c-Jun NH2-terminal kinase (JNK) and p38 stress-activated protein kinase cascades (reviewed in Toone and Jones, 1998). In contrast to the HOG pathway, which almost exclusively senses and responds to osmotic stress, the Spc1, JNK and p38 pathways are activated by a wide range of stress stimuli. However, depending on the stimulus, different patterns of gene expression result.
The recent discovery of the RNA binding protein Csx1, which regulates global gene expression during oxidative stress in S. pombe, has helped to elucidate how cells tailor specific gene expression responses to each kind of stress. The cytoplasmic protein Csx1 contains three RNA recognition motifs (RRMs) and binds to atf1+ mRNA, stabilizing atf1+ mRNA and allowing cells to maintain normal levels of Atf1 protein under conditions of oxidative stress (Rodriguez-Gabriel et al., 2003). The sensitivity of Csx1-deficient cells to H2O2 is partly explained by a deregulated expression of Atf1-dependent genes. However, Csx1 also possesses Atf1-independent functions in the response to oxidative stress, because csx1Δ mutants are more sensitive to H2O2 than atf1Δ cells. Both Csx1 and Spc1 are necessary to maintain normal levels of atf1+ mRNA. Csx1 and Spc1 coordinately regulate expression of many genes. However, microarray analyses of cells subjected to oxidative stress show that many genes whose expression is Spc1-dependent are Csx1-independent and vice versa, indicating that Csx1 and Spc1 have certain nonoverlapping functions (Rodriguez-Gabriel et al., 2003).
A more thorough understanding of the precise mechanisms that participate in the posttranscriptional control of gene expression in H2O2-treated S. pombe cells relies on the identification and biochemical characterization of the specific molecules involved in this pathway. Here, we describe two novel RRM proteins of S. pombe that interact with Csx1 and participate in the control of gene expression under conditions of oxidative stress.
MATERIALS AND METHODS
Yeast Strains, Media, and General Methods
Standard procedures and growth media for S. pombe genetics have been described previously (Moreno et al., 1991). Cip1 and Cip2 were deleted by replacement of the entire open reading frame of each gene with the KanMx6 module as described previously (Bahler et al., 1998). Epitope-tagged Cip1 and Cip2 were also generated as described previously (Bahler et al., 1998), placing a FLAG, green fluorescent protein (GFP), or cyan fluorescent protein (CFP) epitope at the C terminus of each protein and marking the allele with the kanamycin resistance gene. The epitope-tagged alleles seemed to be functional because they did not rescue csx1Δ.
All strains used in these studies were ura4-D18 leu1-32. Their genotypes are PR 109, wild-type; VM3770, cip1-CFP-KanMx6; VM3771, cip1::KanMx6; VM2772, cip2::KanMx6; VM3773, cip2-GFP-KanMx6; VM3774, cip1::KanMx6 cip2::KanMx6; VM3775, cip2::KanMx6 sx1::KanMx6; VM3776, cip1::KanMx6 csx1::KanMx6; VM3777, cip1::KanMx6 cip2::KanMx6 csx1::KanMx6; VM3778, cip1-FLAG-KanMx6; VM3779, cip2-FLAG-KanMx6; VM3780, kanMx6-nmt1-cip2+; VM3781 cip1::KanMx6 atf1::ura4; VM3782, cip2::KanMx6 atf1::ura4; VM3783, spc1::ura4 cip1-FLAG-kanMx6; VM3784, spc1::ura4 cip2-FLAG; VM3785, csx1::kanMx6 cip1-FLAG-kanMx6; VM3786, csx1::kanMx6 cip2-FLAG-kanMx6; MR3213 csx1::kanMX6; KS1497, atf1::ura4, and KS1605, spc1::ura4 (this strain is leu1+).
For plate survival assays, serial dilutions of yeast culture were plated in media containing 0.6 or 0.8 mM H2O2. For cell survival assays, cells were grown in the presence of H2O2 for different times, plated in rich media, and colonies were counted after 4 d at 30°C.
RNA and Microarray Methods
RNA for Northern blots and microarray analysis was obtained as described in http://www.sanger.ac.uk/PostGenomics/S_pombe/protocols/. Sample labeling, microarray hybridization and data acquisition were performed as described previously (Lyne et al., 2003).
Mass Spectrometry and Protein Methods
Csx1-TAP protein was purified from fission yeast cells treated with 1 mM H2O2 using a previously described method (Saitoh et al., 2002). The resulting peptide mixture was analyzed by multidimensional protein identification technology (MudPIT) (MacCoss et al., 2002). MudPIT combines multidimensional chromatography with mass spectrometry, obviating the need for visualization and excision of protein bands from gels for peptide identification (Graumann et al., 2004). This approach has been successfully used in our laboratory for the identification of partners of other proteins (Boddy et al., 2001, 2003). For immunoblotting, the FLAG epitope was detected using mouse monoclonal antibodies. Immunoprecipitated Cip2-FLAG was used as substrate for λ phosphatase treatment.
Microscopy
CFP and GFP fluorescence was visualized in mid-log phase live cells. Cells were photographed using a Nikon Eclipse E800 microscope (Nikon, Tokyo, Japan) equipped with a Photometrics Quantix charge-coupled device camera (Photometrics, Tucson, AZ).
RESULTS
Identification of Cip1 and Cip2
For a better understanding of how Csx1 mediates posttranscriptional regulation of gene expression in response to oxidative stress, we sought to identify interaction partners by using MudPIT (see Materials and Methods for details). To this end, the csx1+ endogenous locus was engineered to encode a protein with a C-terminal TAP tag (Rodriguez-Gabriel et al., 2003).
Cells were then treated for 15 min with 1 mM H2O2 and subsequently processed as described previously (Saitoh et al., 2002). Before the MudPIT analysis, we confirmed by immunoblotting that the Csx1-TAP protein had been efficiently precipitated (our unpublished data). The percentage of coverage of the primary sequence and the number of peptides obtained for each of the proteins recovered was used as an indication of the relative abundance of each protein in the sample. We have performed many TAP purifications of nuclear and cytosolic proteins, and this allowed us to generate a list of the highly abundant proteins that commonly contaminate TAP purifications (e.g., metabolic enzymes, actin, and ribosomal proteins). The list of proteins identified in the Csx1-TAP purification was compared with this list to exclude the proteins that were nonspecifically purified.
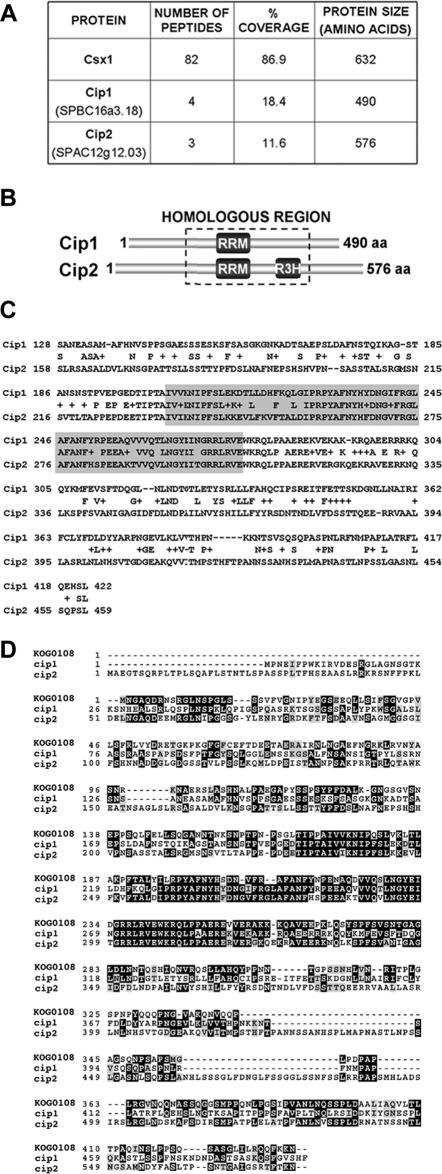
As expected, mass spectrometric analysis of the affinity-purified Csx1-TAP sample revealed extensive peptide coverage of Csx1 (86.9%; Figure 1A). We found two novel proteins that coprecipitated with Csx1-TAP, which we termed Cip1 and Cip2. Cip1 (“Csx1-interacting protein 1”) was identified by peptides covering 18.4% of its 490 amino acid primary sequence (Figure 1A). Peptides covering 11.6% of another novel protein, Cip2, were also obtained (Figure 1A). Cip2 is a 576-amino acid protein that showed significant homology to Cip1 (Figure 1, B and C). Both Cip1 and Cip2 were identified with greater than 98% confidence.
Figure 1.
Identification of Cip1 and Cip2. (A) Number of independent peptides and percent of primary sequence coverage obtained by mass spectrometry for each of the indicated proteins. The TAP purification was performed after treating the cells for 15 min with 1 mM H2O2. (B) RNA-binding domain distribution in Cip1 (SPBC16A3.18) and Cip2 (SPAC12G12.03). The region with the highest sequence identity between the two proteins is boxed. (C) Alignment of the homologous regions of Cip1 and Cip2. Shaded residues represent the RRM. The percentage of homology is 58%, with 41% of the amino acid residues being identical in both proteins. (D) Sequence comparison between Cip1 and Cip2 and the eukaryotic KOG0108 domain, corresponding to the mRNA cleavage and polyadenylation factor I complex, subunit 15. Each COG includes proteins that are inferred to be orthologues and represents an ancient conserved domain.
The sequence of both Cip1 and Cip2 contained an RNA recognition motif (RRM) (Figure 1, B and C). RRM domains are typically present in proteins involved in RNA processing, a relevant example being Csx1 (Rodriguez-Gabriel et al., 2003). Cip2 also harbored an R3H motif (Figure 1B). R3H motifs are thought to function in sequence-specific binding to single-stranded nucleic acids (Grishin, 1998). As observed in the sequence of Cip2, R3H motifs usually occur in association with other DNA- or RNA-binding domains (Letunic et al., 2002; Bateman et al., 2004).
Performing a BLAST search, most Cip1 and Cip2 homologues showed sequence homology only across the RRM domain. In addition, Rna15—a subunit of the cleavage and polyadenylation factor I complex in S. cerevisiae (Gross and Moore, 2001)—also shared common residues outside the RNA recognition motif with Cip1 and Cip2. Rna15, Cip1, and Cip2 seemed to be direct evolutionary counterparts because all of them were included in the same cluster of orthologous group (COG) (Figure 1D) (Marchler-Bauer et al., 2005). SPAC644.16, which is the closest homologue of Rna15 in fission yeast, was not present in the group of proteins coprecipitating with Csx1-TAP. The only S. pombe orthologue of the budding yeast cleavage and polyadenylation factor I complex identified in our TAP purification was the poly (A) binding protein Pabp, with six peptides covering 15% of its sequence.
Phenotypes of cip1Δ and cip2Δ Cells
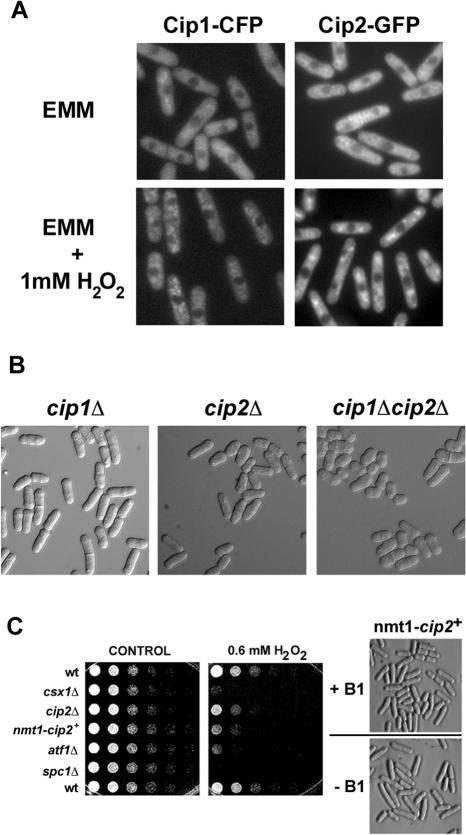
Csx1 is a cytoplasmic protein (Rodriguez-Gabriel et al., 2003). We predicted that to be able to interact with Csx1, Cip1 and Cip2 should also localize to the cytoplasm. To determine the localization of Cip1 and Cip2, we tagged each protein at its genomic locus with a C-terminal epitope, creating the fusion proteins Cip1-CFP and Cip2-GFP. Both fusion proteins were expressed from their endogenous promoters. Consistent with a possible role in the control of mRNA stabilization or degradation, both proteins were predominantly cytoplasmic at all stages of the cell cycle, and in the presence or absence of H2O2 (Figure 2A).
Figure 2.
Phenotype of cip1Δ and cip2Δ mutant strains. (A) Strains carrying Cip1 or Cip2 tagged at their genomic loci with CFP or GFP, respectively, were used to determine the localization of each protein. Cip1 and Cip2 show cytoplasmic localization in cells growing in minimal medium (EMM), without (top) or during treatment with 1 mM H2O2 for 15 min (bottom). (B) cip1Δ, cip2Δ and cip1Δcip2Δ strains were grown to mid-log phase at 30°C and analyzed under the microscope. (C) Left, spot assays of wild-type and mutant strains. Fourfold serial dilutions were plated on EMM plates (CONTROL) or EMM plates supplemented with 0.6 mM H2O2. No thiamine was added to the media to induce overexpression of Cip2. Pictures were taken after incubating the plates for 5 d at 30°C. Right, morphology of Cip2-overexpressing cells. Wild-type cells containing the endogenous copy of cip2+ under the control of the nmt1 promoter were grown in liquid EMM media in the absence (–B1) or presence (+B1) of thiamine. Pictures were taken after growing cells to mid-log phase for 21 h in the indicated media at 30°C.
To gain insight into the function of Cip1 and Cip2, we replaced the entire open reading frame of both genes with the kanMx6 cassette (Bahler et al., 1998). Precise replacement of the chromosomal copies of cip1+ and cip2+ by the kanamycin marker was confirmed by Southern blot and PCR analyses (our unpublished data). cip1Δ and cip2Δ cells were viable and grew at the same rate as wild-type cells. Microscopic analysis showed no significant difference between wild-type cells and cip1Δ (Figure 2B). In contrast, the morphology of cip2Δ cells was noticeably different from wild type, with many of the cip2Δ cells seeming to be swollen and shorter (Figure 2B). This phenotype was even more profound in cip1Δ cip2Δ double mutants. Conversely, overexpression of Cip2, by using the nmt1 promoter, caused an elongation of the cells (Figure 2C). Although deletion of cip2+ did not alter the cells' sensitivity to H2O2, overexpression of cip2+ led to a slightly increased H2O2 sensitivity. As expected, csx1Δ, atf1Δ and spc1Δ mutants were more susceptible to H2O2 (Figure 2C).
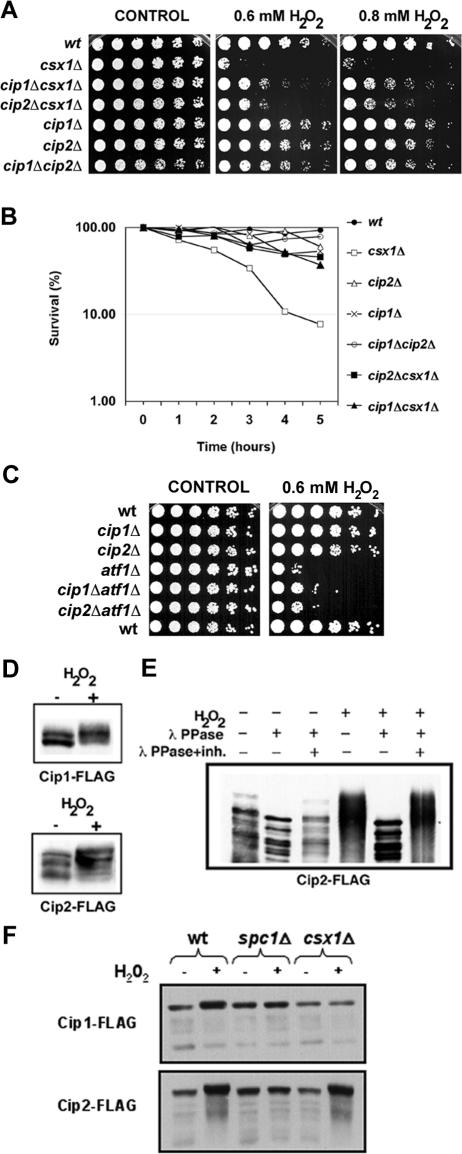
cip1Δ cip2Δ double mutant strains grew slower than wild-type cells (Figure 3A) and showed an enhanced version of the cip2Δ morphology phenotype (Figure 2B). The sensitivity of cip1Δ cip2Δ strains to oxidative stress, osmotic stress, UV light, γ irradiation, and hydroxyurea was comparable to the single mutants and to wild-type cells (Figure 3A; our unpublished data).
Figure 3.
Cip1 and Cip2 participate in the oxidative stress response. (A) Serial dilutions of wild-type and mutant strains were plated in rich YES (yeast extract, glucose, and supplements) medium (left) or YES media with 0.6 mM H2O2 (middle) or 0.8 mM H2O2 (right). Photographs were taken after incubating the plates for 4 d at 30°C. (B) Survival of wild-type and mutant strains after H2O2 treatment. Liquid cultures were incubated in the presence of 1 mM H2O2 for 0–5 h, and cells were then plated on YES plates. Colonies were counted after 4 d of incubation at 30°C. (C) Fourfold serial dilutions of wild-type and the indicated mutant strains were plated on YES plates (left) or YES plates supplemented with 0.6 mM H2O2 (right). Pictures were taken after incubating the plates for 4 d at 30°C. (D) Cip1 and Cip2 Western blots. Cip1-FLAG and Cip2-FLAG cultures were treated with 1 mM H2O2, and cells were collected after 60 min. The same amount (50 μg) of whole cell extract was loaded in each lane. Proteins were detected using anti-FLAG monoclonal antibodies. (E) Wild-type Cip2-FLAG cultures were treated with 1 mM H2O2 for 60 min, and whole cell extracts were prepared. After immunoprecipitation with anti-FLAG agarose, the pull-down was treated with λ phosphatase. Cip2-FLAG protein was detected by Western blotting with anti-FLAG monoclonal antibodies. (F) Cip1 and Cip2 Western blots. Wild-type, spc1Δ, and csx1Δ strains expressing Cip1-FLAG or Cip2-FLAG were treated with 1 mM H2O2, and cells were collected after 60 min. The same amount (50 μg) of whole cell extract was loaded in each lane. Proteins were detected using anti-FLAG monoclonal antibodies.
Cip1 and Cip2 Participate in the Response to Oxidative Stress
Cells deficient in Csx1 are sensitive to oxidative stress (Rodriguez-Gabriel et al., 2003; Figure 2C). We investigated whether the absence of Cip1 and/or Cip2 in a csx1Δ background changed sensitivity to oxidative stress. As expected, strains lacking Csx1 were incapable of growing in the presence of H2O2. However, cip1Δ csx1Δ and cip2Δ csx1Δ strains were less sensitive to oxidative stress treatments, both chronic and acute, compared with csx1Δ strains (Figure 3, A and B). This effect was specific for oxidative stress, because it was not possible to detect any other difference between the double mutants and the single mutant cxs1Δ in response to other forms of stress (our unpublished data). The rescue of the H2O2 sensitivity of csx1Δ mutants by elimination of Cip1 or Cip2 was stronger at higher H2O2 concentrations (Figure 3A). The differences in the subset of genes and transcription factors that are activated in response to low and high H2O2 levels could explain the variation of the sensitivity of cip1Δ csx1Δ and cip2Δ csx1Δ strains to different concentrations of H2O2.
Triple deletions cip1Δ cip2Δ csx1Δ were also generated. The simultaneous absence of Cip1 and Cip2 caused a rescue level of csx1Δ H2O2 sensitivity identical to the level obtained by the elimination of any of the proteins independently (our unpublished data). The partial rescue of the H2O2 sensitivity of csx1Δ strains points to a specific role of Cip1 and Cip2 in tolerance to oxidative stress in S. pombe and is consistent with the idea that Csx1, Cip1, and Cip2 have related functions in the response to oxidative stress.
The sensitivity of atf1Δ cells to H2O2 was not much improved by deletion of either Cip1 or Cip2 (Figure 3C), implying that Cip1 and Cip2 required Atf1 for their function. However, Cip1 and Cip2 must also have targets other than Atf1, because these cip1Δ atf1Δ and cip2Δ atf1Δ double mutants were slightly less sensitive to H2O2 compared with atf1Δ single mutant cells.
Whole genome expression profiling has shown that the abundance of cip1+ mRNA increases by about twofold in response to H2O2 treatment, whereas the amount of cip2+ mRNA is unaffected (Chen et al., 2003). We monitored protein abundance by immunoblotting with an anti-FLAG antibody. Immunoblot analyses detected multiple electrophoretic mobility species of Cip1 and Cip2, but no significant increase in protein levels after treatment with H2O2 (Figure 3, D and E). It is possible that increased expression of cip1+ mRNA during oxidative stress is required to compensate for reduced mRNA translation or accelerated turnover of Cip1 protein. Interestingly, oxidative stress caused Cip1 and Cip2 proteins to have reduced electrophoretic mobility (Figure 3D), indicating that Cip1 and Cip2 might become phosphorylated in the presence of H2O2. We therefore treated a Cip2 immunoprecipitate with λ phosphatase and analyzed the samples using SDS-PAGE conditions that accentuated changes in the electrophoretic mobility of proteins. This analysis showed that the Cip2 mobility shift induced by oxidative stress was caused by phosphorylation (Figure 3E). It has been previously shown that phosphorylation of Csx1 depends on the Spc1 MAPK (Rodriguez-Gabriel et al., 2003). In a similar way, deletion of spc1+ abolished the change in Cip1 and Cip2 electrophoretic mobility (Figure 3F). In contrast to Cip2, Cip1 phosphorylation was also dependent on Csx1 (Figure 3F), suggesting that Cip1 and Cip2 might be regulated differently during the oxidative stress response. Based on these results, Spc1 seems to be controlling directly or indirectly the phosphorylation status of these three RRM-containing proteins (Csx1, Cip1, and Cip2). Further studies will be required to determine the functional significance of this phosphorylation.
Role of Cip1 in the Global Transcriptional Response to Oxidative Stress
The fact that cip1Δ csx1Δ and cip2Δ csx1Δ mutants were more resistant to H2O2 treatment than the csx1Δ mutant suggested that Cip1 and Cip2 might participate in the control of gene expression during oxidative stress. Therefore, we decided to analyze whether Cip1 modulated the transcriptional response to oxidative stress. We focused our studies on Cip1 because the cip1Δ mutation suppressed the oxidative stress phenotype of csx1Δ cells without altering cell morphology (see above).
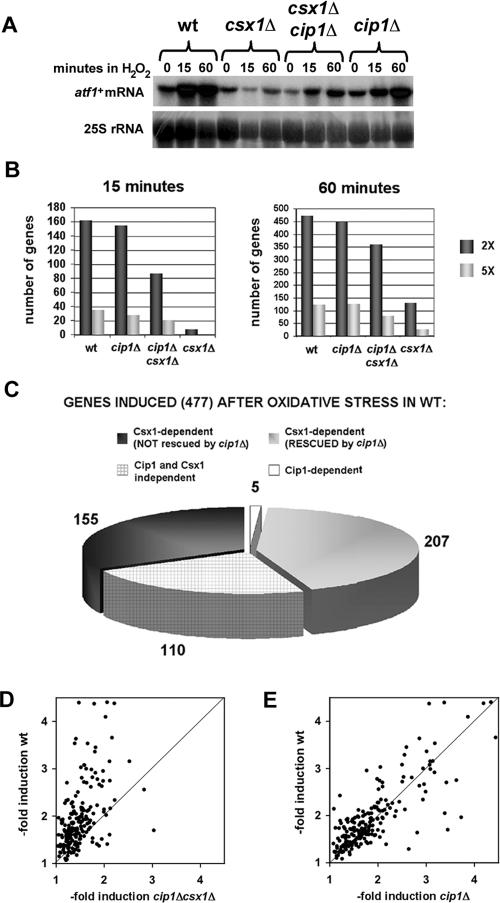
We isolated total RNA from wild-type, csx1Δ, cip1Δ, and csx1Δ cip1Δ strains without stress or 15 or 60 min after treatment with 1 mM H2O2. atf1+ mRNA accumulated after H2O2 treatment in wild-type and cip1Δ cells (Figure 4A), consistent with the observation that cip1Δ mutants were not sensitive to oxidative stress (Figure 3, A and B). The H2O2-sensitive csx1Δ cells, however, failed to accumulate atf1 mRNA after oxidative stress (Figure 4A; Rodriguez-Gabriel et al., 2003). Interestingly, csx1Δ cip1Δ double mutants were able to induce expression of atf1+ during the oxidative stress response, although the increase in atf1+ mRNA levels was not as strong as the one detected in wild-type or cip1Δ cells (Figure 4A). This latter finding correlates with the H2O2 sensitivity of csx1Δ cip1Δ cells, which was intermediate between the one of wild-type and csx1Δ cells (Figure 3, A and B).
Figure 4.
Influence of Cip1 on global gene expression after oxidative stress. (A) Northern blot of atf1+ mRNA before (time 0) and after treatment of wild-type, csx1Δ, cip1Δ, and cip1Δ csx1Δ cells with 1 mM H2O2 for 15 or 60 min. 25S rRNA is shown as loading control. (B) Comparison of the total number of genes with two- or fivefold induction in wild-type, cip1Δ, cip1Δ csx1Δ, or csx1Δ cells 15 and 60 min after treatment with 1 mM H2O2. (C) Role of Cip1 and Csx1 in controlling the expression of genes induced at least twofold at one or both time-points (15 and/or 60 min) in wild-type cells. (D and E) Scatter chart analyses of the induction levels of the genes shown in Figure 4B as Csx1-dependent (RESCUED by cip1Δ). Comparisons were made between wild-type cells and cip1Δ csx1Δ mutants (D) or wild-type and cip1Δ cells (E).
To analyze the global effect of Cip1 in the expression profile of the fission yeast genome, RNAs isolated from wild-type, csx1Δ, cip1Δ, and csx1Δ cip1Δ strains without treatment or 15 and 60 min after treatment with 1 mM H2O2 were labeled during reverse transcription. The resulting cDNA was hybridized onto DNA microarrays containing probes of all known and predicted fission yeast genes. After eliminating the genes that failed to give measurable data in all the samples, we were able to monitor the expression of ∼3500 genes. We first compared the total number of genes whose expression was induced two- or fivefold in wild-type, cip1Δ, cip1Δ csx1Δ, and csx1Δ cells, 15 or 60 min after treatment with 1 mM H2O2. This analysis showed that the global pattern of induction of gene expression in wild-type and cip1Δ strains was very similar (Figure 4B). The magnitude of the transcriptional response to H2O2 was considerably weaker in cip1Δ csx1Δ and more so in csx1Δ mutants (Figure 4B).
We studied the expression of the 477 genes that were induced twofold or more in at least one time point in the wild-type strain. In cip1Δ mutants, 406 of these genes were induced as well, indicating that the oxidative stress response in cip1Δ and wild-type cells followed similar patterns. Interestingly, 362 of the genes induced in wild-type cells were not induced in csx1Δ mutants compared with 155 genes in cip1Δ csx1Δ double deficient mutants (Figure 4C). From these results, we conclude that elimination of Cip1 in a csx1Δ background partially restores the defect in gene expression induced by H2O2. The fact that 90 (43.5%) of the 207 genes whose induction is partially restored in cip1Δ csx1Δ mutants are Atf1-independent indicates that part of the effect of Cip1 in the oxidative stress response does not require Atf1.
We further evaluated whether the level of induction of the 207 genes “rescued” in cip1Δ csx1Δ mutants was similar to the one observed in wild-type cells. In at least one of the time points, 85% of these genes were induced stronger in wild-type cells than in cip1Δ csx1Δ cells (Figure 4D). This defective induction together with the lack of induction in the remaining 155 genes (Figure 4C) could explain why cip1Δ csx1Δ mutants show higher H2O2 sensitivity than wild-type cells. Consistent with this idea we found that just 59% of the genes belonging to this group showed reduced induction upon H2O2 treatment in cip1Δ mutants compared with wild-type cells (Figure 4E).
Similar comparisons were performed for the 110 genes that were induced after oxidative stress in all the strains, i.e., wild type, cip1Δ, csx1Δ, and cip1Δ csx1Δ. Almost all of them (>97%) were expressed less in csx1Δ mutants compared with wild-type cells (Figure 5A). The differences were much smaller when the comparison was done between cip1Δ csx1Δ mutants or cip1Δ mutants and wild-type cells (Figure 5, B and C). These results indicate that the cip1Δ mutation reverses some of the expression defects observed in csx1Δ cells after H2O2 treatment. Collectively, we conclude that Cip1 and Csx1 have counteractive roles in controlling gene expression in response to oxidative stress.
Figure 5.
Importance of Cip1 in the control of the expression of oxidative-stress induced genes. Scatter chart analyses of the induction levels of the subset of genes (110) whose expression increased at least twofold (at 15 and/or 60 min) after oxidative stress wild-type, cip1Δ, cip1Δ csx1Δ, or csx1Δ cells. (A) Comparison of the induction levels between wild-type and csx1Δ strains. (B) Comparison of the induction levels between wild-type cells and cip1Δ csx1Δ double mutants. (C) Comparison of the levels of induction between wild-type and cip1Δ cells.
DISCUSSION
In this report, we have analyzed the roles of two novel proteins, Cip1 and Cip2, in the cellular response to oxidative stress in S. pombe. Cip1 and Cip2 were discovered by Mud-PIT analysis of TAP-tagged Csx1. The physiological significance of this interaction is indicated by the analysis of cip1+ and cip2+ deletions. Notably, these mutations ameliorate the oxidative stress phenotype of csx1Δ cells, a specific and striking genetic interaction that could not have been predicted beforehand. Furthermore, analysis of global patterns of gene expression has shown cip1Δ to partially correct the defect in oxidative-stress induced gene expression in csx1Δ cells. This effect potentially explains the suppression of csx1Δ by cip1Δ and cip2Δ mutations.
Involvement of Cip1 and Cip2 in the Response to Oxidative Stress
We have identified Cip1 and Cip2 as two proteins that coprecipitate with Csx1-TAP analyzed by MudPIT. The detection of these related proteins is highly specific for Csx1-TAP because they have not been identified in >30 other TAP purifications done in our laboratory; however, it should be noted that they do not coprecipitate when analyzed by conventional immunoprecipitation analysis. This finding suggests that Csx1, Cip1, and Cip2 do not form a stable complex, and their interaction might be bridged through their association with mRNA.
Although cip1Δ and cip2Δ single and double mutants are not abnormally sensitive to H2O2, both mutations are able to partially rescue the oxidative stress-sensitive phenotype of the csx1Δ strain. Genetic analysis revealed that Csx1 functions in the pathway that connects Spc1 with Atf1 (Rodriguez-Gabriel et al., 2003). We tested whether the sensitivity of spc1Δ mutants to different treatments, including H2O2 treatment, was rescued by independent or simultaneous deletion of cip1+ and/or cip2+; however, we could not observe any rescue (our unpublished data). In addition, cip1Δ and cip2Δ mutants were not sensitive to osmotic stress, UV light, γ irradiation, or hydroxyurea treatment, although they show a slight sensitivity to arsenic (our unpublished data), which has been related to the production of ROS (Harris and Shi, 2003). Thus, the function of these two putative RNA-binding proteins in survival to stress is only obvious in csx1Δ mutants treated with H2O2.
Interestingly, Cip1 and Cip2 seem to become heavily phosphorylated in response to oxidative stress but not in response to osmotic stress (our unpublished data). This phosphorylation is dependent on Spc1 MAPK (Figure 3F). The phosphorylation state of many RNA-binding proteins determines the fate of their associated transcripts and is controlled by MAPKs. For example, phosphorylation of the mRNA-destabilizing protein TPP increases the half-life of its target mRNA, whereas phosphorylation of the RNA stabilizing protein HuR helps to prevent degradation of interleukin-3 mRNA (reviewed in Bevilacqua et al., 2003). In fission yeast, phosphorylation of the RNA-binding proteins Rnc1 and Csx1 has been shown to be dependent on the Pmk1 and Spc1 MAPKs, respectively (Rodriguez-Gabriel et al., 2003; Sugiura et al., 2003). Similar regulation could also control Cip1 and Cip2 activities.
Consequences of the Absence of Cip1 and Csx1 in the Global Transcription Response to Oxidative Stress
In S. pombe, the levels of proteins required for survival under stress conditions are fine-tuned by posttranscriptional mechanisms. csx1Δ mutants were unable to induce expression of most of the genes necessary for tolerance to H2O2 (Figure 4B). cip1Δ cells, on the contrary, were very well able to elaborate a response almost identical to wild-type strains. Accordingly, cip1Δ mutants were as resistant as wild-type cells to H2O2, whereas csx1Δ mutants were highly sensitive to such treatment. Interestingly, the induction level of the genes activated in csx1Δ when treated with H2O2 was always lower than the level found for those same genes in wild-type, cip1Δ, and cip1Δ csx1Δ cells. Therefore, the sensitivity of csx1Δ mutants to oxidative stress could derive from the combination of two factors: absence of induction of crucial genes, such as the transcription factor atf1+, and reduced levels of induction of other genes, such as the phosphatase pyp2+.
Microarray analyses of the cip1Δ csx1Δ cells were consistent with Csx1 and Cip1 (and possibly also its homologue Cip2) participating in opposite processes in RNA metabolism after H2O2 stress. The absence of Cip1 restored the induction of more than half of the genes, which were not induced in csx1+ null mutants, atf1+ among them. In many cases, however, the level of induction was still lower compared with wild-type cells, which could explain why the sensitivity to oxidative stress was only partially rescued in cip1Δ csx1Δ double mutants compared with wild-type cells. The genes with the most impaired induction in csx1Δ mutants (e.g., pyp2+) recover to greatest extent the normal induction levels when cip1+ is eliminated.
Possible Roles of Cip1 and Cip2 in the Control of mRNA Stability
RRMs occur in several proteins involved in all the steps of RNA processing: from nuclear events, such as transcriptional regulation, splicing, and 3′ processing to nuclear export, localization in the cytoplasm, translation, and stability (reviewed in Dreyfuss et al., 2002). The cytoplasmic localization of Cip1 and Cip2 and their interaction with Csx1 suggests that they participate in the control of mRNA stability. As mentioned, Cip1 and Cip2 share homology throughout their sequences with Rna15, a protein that interacts with poly (A) sequences and one of the components of the multisubunit complex that forms the 3′ ends of mRNAs in S. cerevisiae (Gross and Moore, 2001). In contrast, the human poly (A) ribonuclease DAN (Korner and Wahle, 1997; Korner et al., 1998) and xPARN, a deadenylating nuclease from Xenopus (Copeland and Wormington, 2001), contain R3H domains in their sequences. Deadenylation is the first step of the major pathway of mRNA decay in yeast.
Recent studies indicate that in yeast and human cells, mRNA decay occurs in cytoplasmic processing bodies (Sheth and Parker, 2003; Cougot et al., 2004). After oxidative stress, Cip1 and Cip2 localized to phase-dense cytoplasmic sites, which could link these proteins to the mRNA decay pathways. It is tempting to propose that oxidative stress could activate destabilizing factors or lead to mRNA degradation pathways, which are promoted by Cip1 and Cip2 and counteracted by Csx1. Alternatively, oxidants could directly affect the mRNA stability and Csx1 binding would protect mRNAs from degradation. It is known that after treatments causing stress, mammalian mRNAs are dynamically sorted into so-called “stress granules” in which RNA-binding proteins such as TIA-1, TIAR, and HuR control mRNA translation and stability (Kedersha and Anderson, 2002; Stoecklin et al., 2004). Perhaps the function of Cip1 and Cip2 is related to the assembly of these structures or the regulation of the transition of mRNAs between the different cytoplasmic compartments after oxidative stress.
This work contributes to understanding the detailed mechanism of the oxidative stress response in eukaryotic cells. However, more studies are needed to address the question of how general this type of regulation is and which specific roles RNA-binding proteins play during the posttranscriptional control of gene expression after cell stress.
Acknowledgments
We thank members of the Russell laboratory and the University of California-San Diego Superfund Basic Research Program for input and encouragement. We also thank O. Boyman for critical reading of the manuscript. V. M acknowledges financial support from a postdoctoral fellowship granted by the Spanish Ministry of Education, Culture and Sports. The project was supported by Grant ES10337 from the National Institute of Environmental Health Sciences/National Institutes of Health (to P. R.) and by grants from Cancer Research UK (to J. B.), MERK-MGRI-241 (to W.H.M.), National Institutes of Health (EY1328801), and MERK-MGRI-241 (to J.R.Y.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of Environmental Health Sciences/National Institutes of Health.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–09–0847) on January 11, 2006.
References
- Bahler, J., Wu, J. Q., Longtine, M. S., Shah, N. G., McKenzie, A., 3rd, Steever, A. B., Wach, A., Philippsen, P., and Pringle, J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Barnham, K. J., Masters, C. L., and Bush, A. I. (2004). Neurodegenerative diseases and oxidative stress. Nat. Rev. Drug Discov. 3, 205–214. [DOI] [PubMed] [Google Scholar]
- Bateman, A., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32, D138–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua, A., Ceriani, M. C., Capaccioli, S., and Nicolin, A. (2003). Posttranscriptional regulation of gene expression by degradation of messenger RNAs. J. Cell. Physiol. 195, 356–372. [DOI] [PubMed] [Google Scholar]
- Boddy, M. N., Gaillard, P. H., McDonald, W. H., Shanahan, P., Yates, J. R., 3rd, and Russell, P. (2001). Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107, 537–548. [DOI] [PubMed] [Google Scholar]
- Boddy, M. N., Shanahan, P., McDonald, W. H., Lopez-Girona, A., Noguchi, E., Yates, I. J., and Russell, P. (2003). Replication checkpoint kinase Cds1 regulates recombinational repair protein Rad60. Mol. Cell. Biol. 23, 5939–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Toone, W. M., Mata, J., Lyne, R., Burns, G., Kivinen, K., Brazma, A., Jones, N., and Bahler, J. (2003). Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14, 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland, P. R., and Wormington, M. (2001). The mechanism and regulation of deadenylation: identification and characterization of Xenopus PARN. RNA 7, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot, N., Babajko, S., and Seraphin, B. (2004). Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degols, G., and Russell, P. (1997). Discrete roles of the Spc1 kinase and the Atf1 transcription factor in the UV response of Schizosaccharomyces pombe. Mol. Cell. Biol. 17, 3356–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degols, G., Shiozaki, K., and Russell, P. (1996). Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol. Cell. Biol. 16, 2870–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss, G., Kim, V. N., and Kataoka, N. (2002). Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 3, 195–205. [DOI] [PubMed] [Google Scholar]
- Finkel, T. (2003). Oxidant signals and oxidative stress. Curr. Opin. Cell Biol. 15, 247–254. [DOI] [PubMed] [Google Scholar]
- Graumann, J., Dunipace, L. A., Seol, J. H., McDonald, W. H., Yates, J. R., 3rd, Wold, B. J., and Deshaies, R. J. (2004). Applicability of tandem affinity purification MudPIT to pathway proteomics in yeast. Mol. Cell Proteomics 3, 226–237. [DOI] [PubMed] [Google Scholar]
- Grishin, N. V. (1998). The R3H motif: a domain that binds single-stranded nucleic acids. Trends Biochem. Sci. 23, 329–330. [DOI] [PubMed] [Google Scholar]
- Gross, S., and Moore, C. L. (2001). Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol. 21, 8045–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, G. K., and Shi, X. (2003). Signaling by carcinogenic metals and metal-induced reactive oxygen species. Mutat. Res. 533, 183–200. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., and Anderson, P. (2002). Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30, 963–969. [DOI] [PubMed] [Google Scholar]
- Korner, C. G., and Wahle, E. (1997). Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem. 272, 10448–10456. [DOI] [PubMed] [Google Scholar]
- Korner, C. G., Wormington, M., Muckenthaler, M., Schneider, S., Dehlin, E., and Wahle, E. (1998). The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 17, 5427–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumada, K., Yanagida, M., and Toda, T. (1996). Caffeine-resistance in fission yeast is caused by mutations in a single essential gene, crm1+. Mol. Gen. Genet 250, 59–68. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Goodstadt, L., Dickens, N. J., Doerks, T., Schultz, J., Mott, R., Ciccarelli, F., Copley, R. R., Ponting, C. P., and Bork, P. (2002). Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne, R., Burns, G., Mata, J., Penkett, C. J., Rustici, G., Chen, D., Langford, C., Vetrie, D., and Bahler, J. (2003). Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCoss, M. J., et al. (2002). Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. USA 99, 7900–7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer, A., et al. (2005). CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33, D192–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale, J. L., and Holbrook, N. J. (2002). Cellular response to oxidative stress: signaling for suicide and survival. J. Cell Physiol. 192, 1–15. [DOI] [PubMed] [Google Scholar]
- Migliore, L., and Coppede, F. (2002). Genetic and environmental factors in cancer and neurodegenerative diseases. Mutat. Res. 512, 135–153. [DOI] [PubMed] [Google Scholar]
- Millar, J. B., Buck, V., and Wilkinson, M. G. (1995). Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 9, 2117–2130. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Nguyen, A. N., Lee, A., Place, W., and Shiozaki, K. (2000). Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol. Biol. Cell 11, 1169–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn, J., Findlay, V. J., Dawson, K., Millar, J. B., Jones, N., Morgan, B. A., and Toone, W. M. (2002). Distinct regulatory proteins control the graded transcriptional response to increasing H(2)O(2) levels in fission yeast Schizosaccharomyces pombe. Mol. Biol. Cell 13, 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Gabriel, M. A., Burns, G., McDonald, W. H., Martin, V., Yates, J. R., 3rd, Bahler, J., and Russell, P. (2003). RNA-binding protein Csx1 mediates global control of gene expression in response to oxidative stress. EMBO J. 22, 6256–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, S., Chabes, A., McDonald, W. H., Thelander, L., Yates, J. R., and Russell, P. (2002). Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109, 563–573. [DOI] [PubMed] [Google Scholar]
- Samejima, I., Mackie, S., and Fantes, P. A. (1997). Multiple modes of activation of the stress-responsive MAP kinase pathway in fission yeast. EMBO J. 16, 6162–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima, I., Mackie, S., Warbrick, E., Weisman, R., and Fantes, P. A. (1998). The fission yeast mitotic regulator win1+ encodes an MAP kinase kinase kinase that phosphorylates and activates Wis1 MAP kinase kinase in response to high osmolarity. Mol. Biol. Cell 9, 2325–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U., and Parker, R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh, J. C., Wilkinson, M. G., Buck, V., Morgan, B. A., Makino, K., and Millar, J. B. (1997). The Mcs4 response regulator coordinately controls the stress-activated Wak1-Wis1-Sty1 MAP kinase pathway and fission yeast cell cycle. Genes Dev. 11, 1008–1022. [DOI] [PubMed] [Google Scholar]
- Shieh, J. C., Wilkinson, M. G., and Millar, J. B. (1998). The Win1 mitotic regulator is a component of the fission yeast stress-activated Sty1 MAPK pathway. Mol. Biol. Cell 9, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki, K., and Russell, P. (1995). Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature 378, 739–743. [DOI] [PubMed] [Google Scholar]
- Shiozaki, K., Shiozaki, M., and Russell, P. (1997). Mcs4 mitotic catastrophe suppressor regulates the fission yeast cell cycle through the Wik1-Wis1-Spc1 kinase cascade. Mol. Biol. Cell 8, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki, K., Shiozaki, M., and Russell, P. (1998). Heat stress activates fission yeast Spc1/StyI MAPK by a MEKK-independent mechanism. Mol. Biol. Cell 9, 1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin, G., Stubbs, T., Kedersha, N., Wax, S., Rigby, W. F., Blackwell, T. K., and Anderson, P. (2004). MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23, 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, R., Kita, A., Shimizu, Y., Shuntoh, H., Sio, S. O., and Kuno, T. (2003). Feedback regulation of MAPK signalling by an RNA-binding protein. Nature 424, 961–965. [DOI] [PubMed] [Google Scholar]
- Takeda, T., Toda, T., Kominami, K., Kohnosu, A., Yanagida, M., and Jones, N. (1995). Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 14, 6193–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda, T., Shimanuki, M., and Yanagida, M. (1991). Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 5, 60–73. [DOI] [PubMed] [Google Scholar]
- Toone, W. M., and Jones, N. (1998). Stress-activated signalling pathways in yeast. Genes Cells 3, 485–498. [DOI] [PubMed] [Google Scholar]
- Toone, W. M., Kuge, S., Samuels, M., Morgan, B. A., Toda, T., and Jones, N. (1998). Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 12, 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M. (2003). Mitogen-activated protein kinase pathways in redox signaling. Front Biosci. 8, d369–391. [DOI] [PubMed] [Google Scholar]
- Vivancos, A. P., Castillo, E. A., Biteau, B., Nicot, C., Ayte, J., Toledano, M. B., and Hidalgo, E. (2005). A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc. Natl. Acad. Sci. USA 102, 8875–8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivancos, A. P., Castillo, E. A., Jones, N., Ayte, J., and Hidalgo, E. (2004). Activation of the redox sensor Pap1 by hydrogen peroxide requires modulation of the intracellular oxidant concentration. Mol. Microbiol. 52, 1427–1435. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M. G., Samuels, M., Takeda, T., Toone, W. M., Shieh, J. C., Toda, T., Millar, J. B., and Jones, N. (1996). The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 10, 2289–2301. [DOI] [PubMed] [Google Scholar]
- Yamada, K., Nakagawa, C. W., and Mutoh, N. (1999). Schizosaccharomyces pombe homologue of glutathione peroxidase, which does not contain selenocysteine, is induced by several stresses and works as an antioxidant. Yeast 15, 1125–1132. [DOI] [PubMed] [Google Scholar]