Abstract
Insect parasitoids are a major component of global biodiversity and affect the population dynamics of their hosts. However, identification of insect parasitoids is often difficult, and they are suspected to contain many cryptic species. Here, we ask whether the cytochrome c oxidase I DNA barcode could function as a tool for species identification and discovery for the 20 morphospecies of Belvosia parasitoid flies (Diptera: Tachinidae) that have been reared from caterpillars (Lepidoptera) in Area de Conservación Guanacaste (ACG), northwestern Costa Rica. Barcoding not only discriminates among all 17 highly host-specific morphospecies of ACG Belvosia, but it also raises the species count to 32 by revealing that each of the three generalist species are actually arrays of highly host-specific cryptic species. We also identified likely hybridization among Belvosia by using a variable internal transcribed spacer region 1 nuclear rDNA sequence as a genetic covariate in addition to the strategy of overlaying barcode clusters with ecological information. If general, these results will increase estimates of global species richness and imply that tropical conservation and host–parasite interactions may be more complex than expected.
Keywords: Area de Conservación Guanacaste, Belvosia, cytochrome coxidase I, internal transcribed spacer region 1, species richness
Insect parasitoids are often a major cause of mortality for many insect species, but the biology of many tropical parasitoids is poorly known. This is in large part because of the very large number of morphologically similar species and the ensuing difficulty of identifying them. Understanding parasitoid host-specificity and species-richness (1) is particularly affected by these problems of identification. Although parasitoids are currently believed to constitute 8–25% of all insect species (1, 2), there may be many more species if host-specificity has been underestimated (3, 4).
A tachinid fly larva is an endoparasitoid of an insect. The fly larva hides or feeds inside the host larva while it feeds and grows and then rapidly eats the host in the late larval or pupal stage, eventually killing it. With ≈8,500 described species, the Tachinidae are among the most species-rich of Diptera families (2, 5–7). The New World and largely Neotropical tachinid genus Belvosia (7) contains as many as 78 morphologically defined species, of which ≈25 occur in Costa Rica. Of these, 20 have been reared from wild-caught caterpillars by the ongoing caterpillar and parasitoid inventory of Area de Conservación Guanacaste (ACG), a 1,100-km2 area of dry forest, rain forest, and cloud forest in northwestern Costa Rica (http://janzen.sas.upenn.edu). The bumblebee-sized, black and yellow fly lays single eggs on the foliage near a moth caterpillar, which swallows the egg(s) when eating the leaf. The fly larva(e) remains dormant until after the caterpillar has pupated and then, days to many months later, develops, consumes the host, and pupates and then ecloses shortly thereafter to repeat the cycle.
How host-specific are Belvosia in a complex tropical habitat? It is widely believed that tachinid parasitoids are relatively generalist (polyphagous) in their host selection (6–8), but the 27-year inventory of 400-plus species of tachinids reared from 3,500-plus species of ACG caterpillars indicates that the great majority of tropical species are, indeed, highly host-specific (9). The three species of ACG Belvosia that appear to be generalists are exceptional (http://janzen.sas.upenn.edu). Here, we ask whether the units identified by DNA barcoding (10–15) are consistent with morphological species identification for all the species and whether the three generalists really are polyphagous.
Results
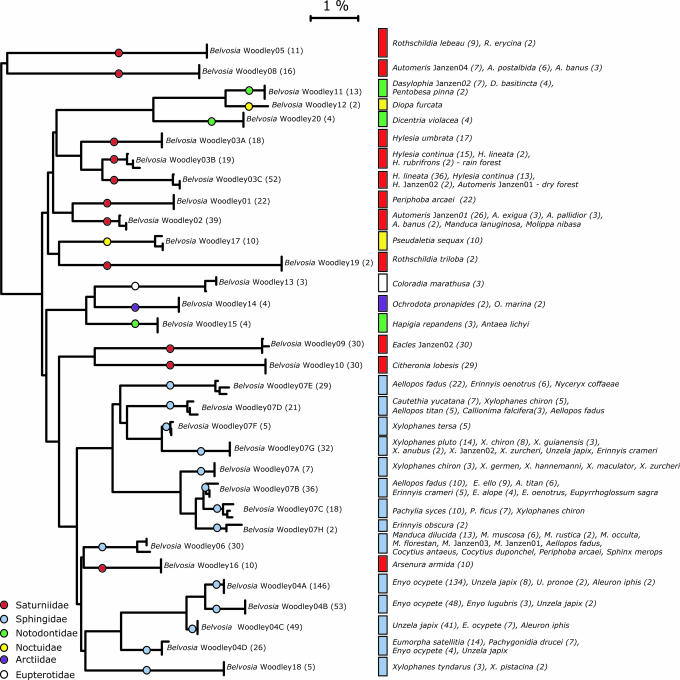
The 3,000-plus Belvosia flies reared from 1,800-plus wild-caught ACG caterpillars (of the total sample of 300,000-plus wild-caught caterpillars) were morphologically separated into 20 species by an experienced fly taxonomist (N.E.W.), working without knowledge of the species of caterpillar from which each fly was reared and before the application of barcoding. Because the majority of these Belvosia cannot yet be linked with certainty to previously described species, we treat them here with alphanumeric interim names (Belvosia Woodley01, Belvosia Woodley02, etc.), where the naming reflects the order in which they were encountered by the inventory. There were no questionable identifications, and 17 of the species matched a reasonable and very narrow array of host caterpillar species (see host records portrayed in Fig. 1; and see Appendices 1 and 2, which are published as supporting information on the PNAS web site), despite the availability in the same habitat of 2,000-plus other species of caterpillars large enough to be hosts for Belvosia (http://janzen.sas.upenn.edu). However, as expected of tachinids, three species were found to be more generalist in their host selection: B. Woodley03 uses all commonly reared species of ACG Hylesia (Saturniidae), B. Woodley04 uses six species in four genera of Sphingidae feeding on Dilleniaceae and Vitaceae, and B. Woodley07 uses 24 species of nine genera of Sphingidae feeding on many plant families.
Fig. 1.
NJ tree of genetic distance (K2P) for 93 representative Belvosia specimens, with host information mapped onto tree. The NJ tree contains three specimens per species (where sample size permitted). The total sample size for each Belvosia species is shown in parentheses on the tree (Left). The host species and number of rearings of Belvosia from that species are shown (Right). Cases where numbers on the left- and right-hand sides are not equivalent are because of wild-caught adult flies where the host is unknown. See Appendix 1 for an NJ tree containing all >500-bp barcoded individuals.
Next, we added the sequence data from DNA barcoding to the morphologically described units already recognized, to determine whether the 20 species could be identified by their DNA barcodes. This protocol for species recognition has proven effective for Lepidoptera, spiders, ants, Collembola and Ephemeroptera, fish, and birds (10, 12, 13, 15–18). When we barcoded one fly from each of 759 Belvosia rearings, we found that all 20 morphological species were readily distinguishable by their DNA barcodes (Fig. 1 and Appendices 1 and 2), with each species represented by a distinct, nonoverlapping cluster of sequences in the neighbor-joining (NJ) tree. The barcode clusters in the NJ tree (Appendix 1) usually had >0.5% sequence divergence among them. However, the three morphospecies of Belvosia thought to be more generalist were found to consist of three, four, and eight cryptic species, each using a set of hosts as narrowly circumscribed as the other 17, thereby raising the total number of species to 32 (Fig. 1 and Appendices 1 and 2). Intraspecific sequence divergences for the 13,921 pairwise NJ comparisons within these 32 taxa average 0.17% and range from 0% to 3.021%. The average interspecific divergence was 5.781% (SE = 2.01). The discovery of highly host-specific relationships within an apparent generalist mirrors what happened when an ACG skipper butterfly (Lepidoptera: Hesperiidae) with a generalist caterpillar was extensively barcoded (10).
DNA barcodes for individuals from the three generalist morphospecies separated into 15 groups in the NJ tree (Fig. 1 and Appendices 1 and 2). Each of these groups, except for one pair, uses different host caterpillars, most are highly sympatric, and the parapatric species have occasional incursions into each other’s population range.
The morphologically defined B. Woodley03 is comprised of three apparent species, one using only Hylesia umbrata, one using rain forest Hylesia spp., and the other using dry forest Hylesia spp. but with the latter two species overlapping at the margins of their parapatric microgeographic distributions (all hosts are very spiny and social small Saturniidae caterpillars). All three species have been reared from caterpillars found in the same hectare. Pairwise intraspecific sequence divergences for these three taxa ranged from 0% to 1.54% (average, 0.278) and pairwise interspecific divergences from 1.64% to 4.918% (average, 3.254).
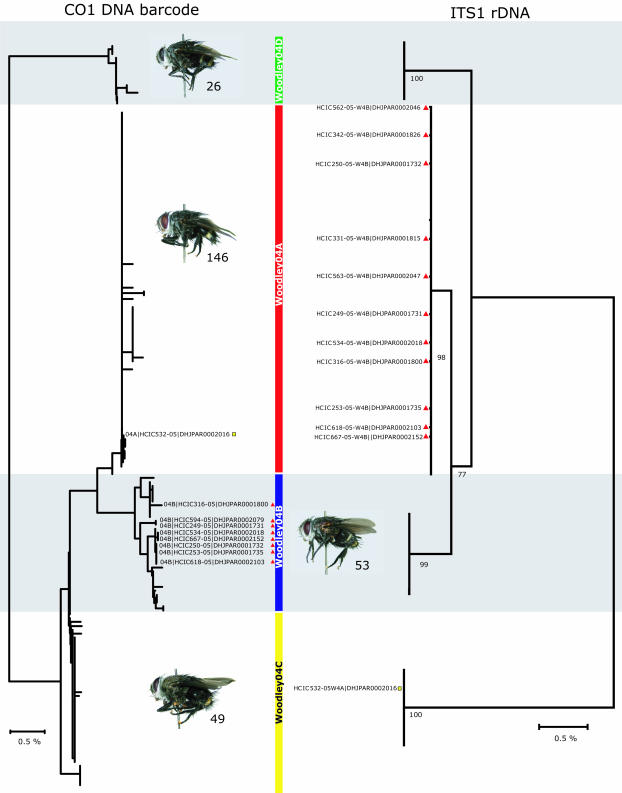
The morphologically defined B. Woodley04 contains four lineages, one using almost entirely Unzela japix caterpillars, one using almost entirely Eumorpha satellitia and Pachygonidia drucei caterpillars, and two using Enyo ocypete caterpillars. The latter two are differentiated from each other by eight signature nucleotide positions within the cytochrome coxidase I (CO1) barcode. Intraspecific barcode divergences for the four members of this group ranged from 0% to 1.47% (average, 0.137), whereas interspecific distances ranged between 0.496% and 4.5% (average, 1.831). Although B. Woodley04A and B. Woodley04B show the same degree of barcode separation from each other as from the others, they use the same species of host caterpillars (E. ocypete) feeding on the same food plants (vines in the Dilleniaceae) in both ACG dry forest and rain forest. For these two sympatric species exposed by barcoding, we also amplified the first internal transcribed spacer region (ITS1) of the ribosomal DNA. Individual variation among conspecifics in rDNA is often homogenized by concerted evolution, whereas interspecific variation continually accumulates (19). We found significant differences in this nuclear marker that strongly suggest that this pair is two species (Fig. 2). The ITS1 data also support the same separation of the entire B. Woodley04 complex into the same four groups as does the CO1 barcode. However, we discovered discordance between nuclear and mitochondrial sequences for ≈50% of the individuals assigned to B. Woodley04B as identified by the CO1 barcode. These individuals are allocated to B. Woodley04A if the NJ tree is based on the ITS1 sequence (Fig. 2). When the bidirectionally sequenced electropherograms for these individuals (04BCO1–04AITS1) were examined, the characteristic B. Woodley04B ITS1 nucleotide base pairs were often also visible at a lower frequency than those of B. Woodley04A. Our favored conclusion is that these 04BCO1–04AITS1 individuals are hybrid offspring of B. Woodley04B females and B. Woodley04A males, although the 04BCO1–04AITS1 individual could also result from the retention of a shared ancestral polymorphism.
Fig. 2.
Relationships within the Belvosia Woodley04 complex using nuclear ITS1 (Right) and mitochondrial CO1 (Left). Full specimen accessions are listed for those species that gave divergent species associations by using nuclear or mitochondrial markers. Yellow square, Woodley04ACO1–Woodley04CITS1; red triangles, Woodley04BCO1–Woodley04AITS1.
Before barcoding, morphospecies Belvosia Woodley07 was the sole species that displayed sufficient within-species morphological variation to generate doubt as to whether it was a single biological entity. When barcoded, it was found to contain eight species, each largely to entirely using a different group of Sphingidae and, sometimes, microhabitat (Fig. 1 and Appendices 1 and 2). These species had pairwise intraspecific divergences ranging from 0% to 3.53% (average, 0.43%), whereas interspecific divergences ranged from 0.34% to 7.632% (average, 3.887). One species pair (B. Woodley07B and B. Woodley07C) showed <1.5% interspecific divergence but next to no overlap in host use.
There were ≈20 cases, each confirmed by barcoding, where a Belvosia successfully parasitized a species of caterpillar that is normally used by another species of Belvosia or a species not normally parasitized by any species of Belvosia. Each occurrence was likely because of one of three causes: (i) a fly laid eggs intended for a host caterpillar but a nontarget caterpillar swallowed them while feeding on the same plant, (ii) the caterpillar was fed its usual foliage species in captivity, but this foliage was contaminated by eggs laid in the wild by a different Belvosia species before collection, and/or (iii) a fly was apparently sufficiently stimulated by a nontarget host caterpillar to lay eggs near it, even though the caterpillar was on a food plant species never fed on by any of the fly’s usual hosts.
The 32 species of Belvosia range from sympatric to parapatric, and, were they to hybridize, the mitochondrial DNA barcode would identify the offspring as the mother’s species, whereas the ovipositional behavior may include hosts of both parental species. In the one unambiguous case of hybridization described above, evidence from nuclear rDNA suggests that B. Woodley04A and B. Woodley04B are capable of interbreeding. This second genetic marker also helped identify a single possible hybridization between two morphologically distinct Belvosia species living in the same ecosystem: a single individual of B. Woodley11 barcoded as B. Woodley12. This was the only case in the entire study of a COI barcode identification differing from a morphology-based identification of a morphologically distinct Belvosia.
We also avoided mistaking successful non-target-host use as examples of hybridization by using a variable nuclear marker as a genetic covariate. For example, four specimens that barcoded as B. Woodley04C were reared from E. ocypete, the target host for B. Woodley04A and B. Woodley04B. Using barcodes and collection records alone, we could have interpreted this as evidence for hybridization between B. Woodley04C and B. Woodley04A or B. Woodley04B. However, ITS1 sequence data from these four specimens showed unambiguously [owing to unique indels and substitutions (see Appendix 3, which is published as supporting information on the PNAS web site)] that the individuals were B. Woodley04C that had successfully emerged from a nontarget host. Similarly, of the eight specimens barcoded as B. Woodley04A that emerged from the primary host of B. Woodley04C (U. japix), seven were further categorized as B. Woodley04A by using the nuclear ITS1 region, whereas the remaining individual was determined to be a likely hybrid between B. Woodley04A and B. Woodley04C (Fig. 2).
The high level of host-specificity disclosed by rearing records and barcoding is being generated by female Belvosia depositing their eggs on foliage where (and when) the host caterpillar is feeding (D.H.J., unpublished field observations). It is clear that ovipositing female Belvosia have a well developed ability to recognize their hosts among a very large array of other species of caterpillars, but this finding may also be coupled with the unstudied process of larval survival once consumed by the caterpillar. However, the nontarget records mentioned above and detailed in Appendix 2 show clearly that, at least on some occasions, a Belvosia can successfully emerge from a nontarget caterpillar. It is unknown whether the rarity of this event is because of careful oviposition by the fly or a high level of failure of the larvae to develop to eclosion in nontarget caterpillars.
This study demonstrates that DNA barcoding can facilitate species discovery and identification within a taxon of parasitoid insects that is rich in morphologically similar species, and companion studies have revealed that this approach is similarly effective in identifying their host Lepidoptera (10, 20). It also changed the categorization of three parasitoid species from relative generalist to host-specific as are their congeners. Current taxonomic systems for most parasitoids rely largely on adult morphology. Although not the focus of this study, our results also suggest that barcoding immature parasitoids could remove the necessity of delaying identification until eclosion. Although none of the ACG Belvosia can be assigned with assurance to a described species, the acquisition of DNA barcodes from type specimens may assist in making that assignment. If cryptic host-specificity, such as that encountered here, is commonplace, there may be a need for a significant increase in global species-richness estimates. Furthermore, species-based tropical conservation efforts and natural history studies will become yet more complex.
Methods
Collection and Preservation.
As described in the Methodology section of the caterpillar and parasitoid inventory of Costa Rica’s ACG dry forest, rain forest, and cloud forest (http://janzen.sas.upenn.edu), the project parataxonomists (21) rear individual wild-caught caterpillars in plastic bags at ambient temperatures until the insects die, produce a parasitoid, or produce an adult moth or butterfly. Newly eclosed flies are killed by freezing, pinned, oven-dried on-site, and individually databased and uniquely coded. Morphological identification (by N.E.W.) is done without knowledge of the host data, and the dry flies are deposited as permanent vouchers at ambient temperature in the Diptera Section of the National Museum of Natural History, Smithsonian Institution, Washington, DC. More than one fly may be produced by a single caterpillar, and, because these flies have a high probability of being siblings, only one fly from a rearing was used in the current barcode analysis (except in a single case, where three flies from one caterpillar of Rothschildia triloba were barcoded). A single dry leg was plucked from each fly with cleaned forceps, dropped into a dry, coded tube in a Matrix box (TrakMates microplate system; Matrix Technologies, Hudson, NH), and couriered to the Biodiversity Institute of Ontario at the University of Guelph, for barcoding. Each delegged fly is indicated with a yellow tag “Legs away for DNA” and recorded as such in its individual record at http://janzen.sas.upenn.edu. Each fly (and barcode sequence) bears the voucher alphanumeric code of the caterpillar from which it was reared (e.g., 98-SRNP-8653) as well as its own unique voucher alphanumeric code (e.g., DHJPAR0001833), either of which can be used to access its data at http://janzen.sas.upenn.edu. Three wild-caught adult ACG Belvosia were also included in the analysis.
Genetic Analysis.
Total genomic DNA was extracted from small pieces (≤1 mm long) of fly leg by using the NucleoSpin 96 Tissue kit (Macherey–Nagel, Duren, Germany) following manufacturer’s protocols. Extracts were resuspended in 30 μl of distilled H2O, and a 658-bp region near the 5′ terminus of the CO1 gene was amplified following standard protocol (13). Briefly, full-length CO1 sequences were amplified by using primers LepF1–5′-ATTCAACCAATCATAAAGATATTGG-3′ and LepR1 5′-TAAACTTCTGGATGTCCAAAAAATCA-3′ (10). In cases where a 658-bp product was not successfully generated, internal primer pairs (LepF1–mLepR-5′-CCTGTTCCAGCTCCATTTT-3′ and mLepF1-5′-GCTTTCCCACGAATAAATAATA-3′) (22) (LepR1) were used to generate shorter overlapping sequences that allowed the creation of a composite sequence (contig). PCRs were carried out in 96-well plates in 12.5-μl reaction volumes containing 2.5 mM MgCl2, 5 pmol of each primer, 20 μM dNTPs, 10 mM Tris·HCl (pH 8.3), 50 mM KCl, 10–20 ng (1–2 μl) of genomic DNA, and 1 unit of TaqDNA polymerase using a thermocycling profile of one cycle of 2 min at 94°C; five cycles of 40 sec at 94°C, 40 sec at 45°C, and 1 min at 72°C; followed by 35 cycles of 40 sec at 94°C, 40 sec at 51°C, and 1 min at 72°C, with a final step of 5 min at 72°C. Products were visualized on a 2% agarose E-Gel 96-well system (Invitrogen), and samples containing clean single bands were bidirectionally sequenced by using bigdye 3.1 on an ABI 3730 DNA Analyzer (Applied Biosystems). Contigs were assembled by using sequencher 4.0.5 (Gene Codes) and were subsequently aligned by eye in bioedit (23). Sequence divergences were calculated by using the K2P distance model (24), and an NJ tree of distances (25) was created to provide a graphic representation of the among-species divergences by using mega3.1 (26) and bold (www.barcodinglife.org). Unless otherwise stated, all genetic distances are corrected. When a member of a morphospecies showed deep genetic divergences within its sequence cluster in the NJ tree, more specimens were sequenced to provide a better understanding of the distribution of this variation and its relationship to morphology and natural history. Sample sizes of <10 flies per species indicate that not enough flies had been reared to achieve 10 nonsibling barcoded members of that species.
For the Belvosia Woodley04 species complex, (04A, 04B, 04C, and 04D) and for B. Woodley11 and B. Woodley12, we amplified the ITS1 region of the ribosomal DNA. The full ITS1 region was amplified by using primers CAS18Fs1–5′-TACACACCGCCCGTCGCTACTA-3′ and CAS5p8sB1d 5′-ATGTGCGTTCRAAATGTCGATGTTCA-3′ (27). PCRs were carried out in 12.5-μl reaction volumes containing 2.5 mM MgCl2, 25 pmol of each primer, 50 μM dNTPs, 10 mM Tris·HCl (pH 8.3), 50 mM KCl, 10–20 ng (1–2 μl) of genomic DNA, and 1 unit of TaqDNA polymerase using a thermocycling profile of one cycle of 2 min at 94°C and 40 cycles of 40 sec at 94°C, 40 sec at 67°C, and 2 min at 72°C, with a final step of 5 min at 72°C. Contigs were assembled by using sequencher 4.0.5 (Gene Codes) and were subsequently aligned by using clustalw (28) and then by eye by using bioedit (23).
COI sequences were recovered from 96.96% (736) of the 759 specimens analyzed. Full-length PCR products (>500 bp) were amplified from 85.4% (629). Of these, 69% (439) were created from overlapping shorter amplicons. Shorter, potentially nonoverlapping amplicons (<500 bp) were recovered from 14.6% (107) of the specimens, and many of these were from collections >15 years old.
To compare barcode clusters with host taxonomy, host records for all individuals with a sequence >500 bp were mapped onto an NJ tree of sequence divergences (Appendix 2). The proposed size for a full-length CO1 barcode is 648 bp (13, 14), but a decision regarding a standard barcode length is currently being assessed by the scientific review committee of the Consortium for the Barcode of Life (CBOL; www.barcoding.si.edu). Currently, a CO1 sequence >500 bp in the 5′ end of the CO1 gene is categorized in GenBank (29) as a DNA barcode, and we have found in this study that 500 bp gives qualitatively identical results to an analysis restricted to 648-bp sequences. When distinct barcode clusters within B. Woodley03, B. Woodley04, and B. Woodley07 were separated by COI sequence divergences >0.5% and/or derived from different host(s), they were treated as different species. All 20 morphologically different Belvosia were treated as different species. Levels of barcode variation between and within species were calculated after recognition of all 32 species and using all individuals whose sequence length was >500 bp. Sequences and other specimen information are available in the “ACG Belvosia” file in the Completed Projects section of the Barcode of Life website (www.barcodinglife.org). Additional collection information is deposited at http://janzen.sas.upenn.edu, and the sequences have been deposited in GenBank (accession nos. DQ3480895 –DQ348780 and DQ347662–DQ347669) (see Appendix 2 for detailed matching of voucher specimens with their GenBank accession nos.).
Supplementary Material
Acknowledgments
We thank Taika von Königslöw, Stephanie Kirk, Heather Cole, Jeremy deWaard, and Angela Holliss for laboratory assistance; Teri Crease for discussions regarding rDNA; Sujeevan Ratnasingham and Rob Dooh for database management; Tanya Dapkey and Gary Oullette for delegging and photographing specimens; Cathy Hulshof for image processing; the 21 ACG parataxonomists for collecting, rearing, and databasing caterpillars and parasitoids; and J. M. Burns, J. B. Whitfield, P. Price, M. G. Pogue, and A. L. Norrbom for constructive editing of the manuscript. This work was supported by grants from the Gordon and Betty Moore Foundation, the Natural Sciences and Engineering Research Council (Canada), and the Canada Research Chairs program (to P.D.N.H.), a Fonds Québécois de la Recherche sur la Nature et les Technologies B3 postdoctoral fellowship (to M.A.S.), National Science Foundation Grants DEB 0072730 and 0515699 (to D.H.J.), and grants from the Guanacaste Dry Forest Conservation Fund and Area de Conservación Guanacaste (to D.H.J.).
Abbreviations
- ACG
Area de Conservación Guanacaste
- CO1
cytochrome coxidase I
- ITS
internal transcribed spacer region
- NJ
neighbor-joining.
Footnotes
References
- 1.Godfray H. C. J., Shimada M. Popul. Ecol. 1999;41:3–10. [Google Scholar]
- 2.Godfray H. C. J. Parasitoids: Behavioral and Evolutionary Ecology. Princeton: Princeton Univ. Press; 1994. p. 16. [Google Scholar]
- 3.Bensch S., Perez-Tris J., Waldenstrom J., Hellgren O. Evolution. 2004;58:1617–1621. doi: 10.1111/j.0014-3820.2004.tb01742.x. [DOI] [PubMed] [Google Scholar]
- 4.Westenberger S. J., Sturm N. R., Yanega D., Podlipaev S. A., Zeledon R., Campbell D. A., Maslov D. A. Parasitology. 2004;129:537–547. doi: 10.1017/s003118200400592x. [DOI] [PubMed] [Google Scholar]
- 5.Gaston K. J. Conserv. Biol. 1991;5:283–296. [Google Scholar]
- 6.Stireman J. O., O’Hara J. E., Wood D. M. Annu. Rev. Entomol. 2006;51:525–555. doi: 10.1146/annurev.ento.51.110104.151133. [DOI] [PubMed] [Google Scholar]
- 7.Wood D. M. In: Manual of Nearctic Diptera. McAlpine J. F., editor. Vol. 2. Ottawa: Agriculture Canada; 1987. pp. 1193–1269. [Google Scholar]
- 8.Stireman J. O., III, Dyer L. A., Janzen D. H., Singer M. S., Lill J. T., Marquis R. J., Ricklefs R. E., Gentry G. L., Hallwachs W., Coley P. D., et al. Proc. Natl. Acad. Sci. USA. 2005;102:17384–17387. doi: 10.1073/pnas.0508839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janzen D. H. The Tachinid Times. 1995;8:2–5. [Google Scholar]
- 10.Hebert P. D. N., Penton E. H., Burns J. M., Janzen D. H., Hallwachs W. Proc. Natl. Acad. Sci. USA. 2004;101:14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Criscione C. D., Poulin R., Blouin M. S. Mol. Ecol. 2005;14:2247–2257. doi: 10.1111/j.1365-294X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- 12.Hebert P. D. N., Stoeckle M. Y., Zemlak T. S., Francis C. M. PLoS Biol. 2004;2:e312. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hebert P. D. N., Cywinska A., Ball S. L., deWaard J. R. Proc. R. Soc. London B; 2003. pp. 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hebert P. D. N., Ratnasingham S., deWaard J. R. Proc. R. Soc. London B; 2003. pp. S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith M. A., Fisher B. L., Hebert P. D. N. Philos. Trans. R. Soc. London B. 2005;360:1825–1834. doi: 10.1098/rstb.2005.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ball S. L., Hebert P. D. N., Burian S. K., Webb J. M. J. North Am. Benthol. Soc. 2005;24:508–524. [Google Scholar]
- 17.Hogg I. D., Hebert P. D. N. Can. J. Zool. 2004;82:749–754. [Google Scholar]
- 18.Ward R. D., Zemlak T. S., Innes B. H., Last P. R., Hebert P. D. N. Philos. Trans. R. Soc. London B. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillis D. M., Dixon M. T. Q. Rev. Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- 20.Hajibabaei M., Janzen D.H., Burns J.M., Hallwachs W., Hebert P.D.N. Proc. Natl. Acad. Sci. USA. 2006 doi: 10.1073/pnas.0510466103. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janzen D. H. J. Appl. Ecol. 2004;41:181–187. [Google Scholar]
- 22.Hajibabaei M., deWaard J. R., Ivanova N. V., Ratnasingham S., Dooh R. T., Kirk S. L., Mackie P. M., Hebert P. D. N. Philos. Trans. R. Soc. London B. 2005;360:1959–1967. doi: 10.1098/rstb.2005.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall T. A. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 24.Kimura M. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 25.Saitou N., Nei M. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Kumar S. K., Tamura K., Nei M. Brief. Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 27.Ji Y.-J., Zhang D.-X., He L.-J. Mol. Ecol. Notes. 2003;3:581–585. [Google Scholar]
- 28.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Wheeler D. L. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.