Abstract
The yeast Doa10 ubiquitin (Ub) ligase resides in the endoplasmic reticulum (ER)/nuclear envelope (NE), where it functions in ER-associated degradation (ERAD). Doa10 substrates include non-ER proteins such as the transcription factor Matα2. Here, we expand the range of Doa10 substrates to include a defective kinetochore component, a mutant NE membrane protein, and a substrate-regulated human ER enzyme. For all these substrates, Doa10 requires two Ub-conjugating enzymes, Ubc6 and Ubc7, as well as the Ubc7 cofactor Cue1. Based on a novel genomic screen of a comprehensive gene deletion library and other data, these four proteins appear to be the only nonessential and nonredundant factors generally required for Doa10-mediated ubiquitination. Notably, the Cdc48 ATPase facilitates degradation of membrane-embedded Doa10 substrates, but is not required for any tested soluble Doa10 substrates. This distinction is maintained even when comparing membrane and soluble proteins bearing the same degradation signal. Thus, while Doa10 ubiquitinates both membrane and soluble proteins, the mechanisms of subsequent proteasome targeting differ.
Keywords: Cdc48, Doa10, ERAD, proteasome, ubiquitin
Introduction
Protein degradation plays an essential role in many biological processes. In eukaryotes, the most common mechanism for degrading intracellular proteins is by the ubiquitin (Ub)–proteasome system (Hochstrasser, 1996; Pickart, 2001; Varshavsky, 2005). Polymers of Ub are ligated to the substrate protein, leading to the recognition and destruction of the substrate by the 26S proteasome. For Ub–protein conjugation, Ub is first activated in an energy-dependent reaction by Ub-activating enzyme (E1), followed by transfer of the Ub to a Ub-conjugating enzyme (E2). The E2, together with a third factor, a Ub–protein ligase or E3, transfers Ub to the target protein. E3s are enzymes that stimulate E2-dependent ubiquitination of substrates; because they usually bind directly to substrates, E3s are often the principal factors dictating substrate recognition (Pickart, 2001).
Substrates contain degradation signals or degrons that are recognized by an E3 or E3–E2 complex (Laney and Hochstrasser, 1999). Degrons vary greatly between substrates, and only a few have been characterized in detail. The Saccharomyces cerevisiae Matα2 transcription factor is a nuclear protein that is degraded by the Ub system. Two Ub pathways are required for normal rates of α2 degradation (Chen et al, 1993; Swanson et al, 2001). The first involves the E2s Ubc4 and Ubc5. The second pathway utilizes the endoplasmic reticulum (ER)-localized E2s Ubc6 and Ubc7 and the ER/nuclear envelope (NE) transmembrane E3 Doa10. The latter pathway recognizes a well-defined degron called Deg1 (Johnson et al, 1998).
Whereas α2 degradation serves a regulatory purpose (Laney and Hochstrasser, 2003), many misfolded or otherwise faulty proteins are detected by ‘quality-control' mechanisms and are also rapidly degraded by the Ub system. Failure of such quality control leads to the accumulation of misfolded proteins, which is associated with multiple human degenerative disorders (Ciechanover and Schwartz, 2004). A well-known site of protein quality control is the ER, where both luminal and membrane proteins, if not correctly folded or assembled, are degraded in a process called ER-associated degradation (ERAD). ERAD of membrane proteins involves their ubiquitination and retrotranslocation to the cytosol for degradation by the proteasome (Hampton, 2002; Hirsch et al, 2004).
Unexpectedly, some of the same Ub enzymes that participate in the proteolysis of soluble cytosolic or nuclear proteins also mediate membrane protein degradation at the ER. Matα2 was the first identified substrate for Ubc6, Ubc7, and Doa10, but these enzymes also have membrane protein substrates. Two mutant plasma-membrane transporters, Pma1-D378N and Ste6–166, are rapidly degraded in the ER by the Doa10 pathway, as is Ubc6 itself (Swanson et al, 2001; Wang and Chang, 2003; Huyer et al, 2004; Vashist and Ng, 2004). The other major yeast ERAD pathway involves the Hrd1/Der3 E3 and Ubc7 E2, which also targets both naturally short-lived and aberrant proteins, the latter including a mutant vacuolar carboxypeptidase Y called CPY* (Hampton, 2002; Hirsch et al, 2004). However, one apparent difference between yeast Doa10 and Hrd1 is that the Doa10 pathway recognizes both membrane proteins and soluble proteins of the cytoplasm/nucleus, whereas Hrd1 recognizes only membrane or luminal substrates.
The ability of the Doa10 pathway to recognize both membrane and nonmembrane proteins raises the question of whether both substrate classes have the same types of degrons. One difficulty in addressing this has been that all the known membrane substrates of Doa10 were aberrant proteins (except Ubc6, which is part of the Doa10 complex), while no soluble quality-control substrate had been identified. The recognition of regulatory and quality-control substrates might involve distinct features regardless of whether they are membrane or soluble proteins. To help untangle these factors, we sought and now report a number of new Doa10 targets, including a mutant nonmembrane protein subject to quality control and a functional membrane protein. Conversely, we have constructed a matched set of soluble and membrane proteins with the same degron and show that they are all efficient Doa10 substrates.
We were also interested in whether the proteolytic pathways for soluble and membrane Doa10 substrates showed any divergence. Our data indicate that membrane-embedded Doa10 substrates, but not soluble ones, require the Cdc48-Ufd1-Npl4 ATPase complex and the Rad23 and Dsk2 poly-Ub receptors for their degradation. The distinction is maintained even if the membrane and soluble proteins carry the same degron. This is the first identified substrate property that predicts the proteasomal targeting route among different substrates sharing the same Ub ligase. For both classes of substrate, the same four proteins—Doa10, Ubc6, Ubc7, and Cue1—are necessary for ubiquitination. A novel genome-wide screen with a Deg1-based substrate as well as further analysis of a previous selection that used a mutant nuclear substrate (Kopski and Huffaker, 1997) both yielded only these factors. Our results indicate that a diverse array of substrates, both membrane and nonmembrane, can be recognized by an ER-embedded Doa10 complex, and they also suggest that naturally short-lived regulatory proteins and aberrant quality control targets are likely to share related recognition determinants.
Results
As a way to find new Doa10 substrates, we scanned the literature for proteins whose degradation in yeast appeared to depend strongly on both Ubc6 and Ubc7 because previously identified substrates of Doa10 required both of these E2s. Three candidate proteins were identified: mutant versions of yeast Ndc10 and Mps2, and the human type II iodothyronine deiodinase. A series of synthetic degron fusions were also regarded as potential targets.
The Ndc10-2 kinetochore protein is a Doa10 pathway substrate
One potential substrate was the mutant Ndc10-2 protein (Kopski and Huffaker, 1997). Ndc10 is a subunit of the centromeric DNA-binding CBF3 complex and also associates with intranuclear mitotic spindle microtubules (Muller-Reichert et al, 2003). The temperature sensitivity of the ndc10-2 mutant is suppressed by loss of Ubc6 or Ubc7 (Kopski and Huffaker, 1997). This might reflect metabolic stabilization of a partially functional Ndc10-2 protein.
We combined the ndc10-2 allele with either cue1Δ (Cue1 anchors Ubc7 to the ER) or doa10Δ to determine if these Doa10 pathway components also suppressed ndc10-2 temperature sensitivity. This was indeed observed (not shown). To address whether suppression correlated with changes in the degradation of Ndc10-2, we analyzed its rate of disappearance after protein synthesis was blocked (Figure 1A). Minimal degradation of wild-type Ndc10 protein occurred during the 1 h chase, but mutant Ndc10-2 protein rapidly disappeared under these conditions. Most importantly, degradation of Ndc10-2 protein was strongly inhibited by the deletion of either CUE1 or DOA10.
Figure 1.
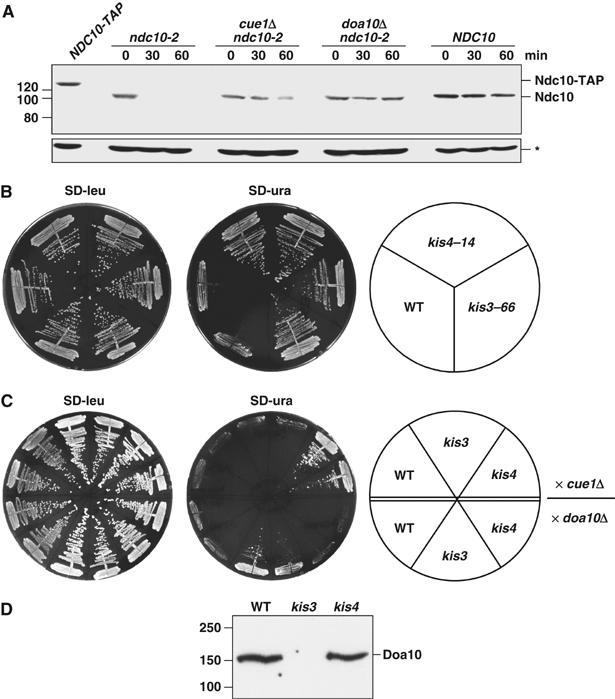
The Ndc10-2 kinetochore protein is a short-lived substrate of the Doa10 pathway. (A) Degradation of Ndc10-2 protein in doa10Δ and cue1Δ cells. Cycloheximide was added 15 min after shifting them to 37°C, and aliquots of cells were removed at the indicated times. Lysates were analyzed by anti-Ndc10 immunoblotting. A strain with the chromosomal NDC10 tagged with the TAP-tag coding sequence (lane 1) was used as a control for antibody specificity. Asterisk, a crossreacting protein that served as a loading control. (B) Two previously isolated suppressors of ndc10-2 are also defective for Deg1-mediated proteolysis. Strains carrying the suppressor mutations kis3–66 and kis4–14 were transformed with a LEU2 plasmid expressing the fusion Deg1-Ura3. Failure to degrade this protein rapidly allows growth on uracil. (C) Complementation analysis demonstrates that kis3–66 has a mutation in DOA10 and kis4–14 a mutation in CUE1. The strains on the upper half of the plate were mated to cue1Δ, while those on the bottom half were mated to doa10Δ cells. Two diploids from each cross were streaked on each plate. (D) Anti-Doa10 immunoblot analysis of kis mutants.
Extragenic ndc10-2 suppressor mutations in DOA10 and CUE1
Kopski and Huffaker (1997) found four complementation groups in their screen for suppressors of ndc10-2. Two of them, kis1 and kis2, had mutations in UBC7 and UBC6, respectively (Table I). The genes affected by the kis3 and kis4 mutations were not identified. To test if kis3 and kis4 strains also harbored mutations in known or unknown components of the Doa10 pathway, we transformed them with a plasmid encoding Deg1-Ura3, a convenient reporter for Doa10 pathway function. Wild-type cells rapidly degrade the reporter protein. Ura3 is required for uracil synthesis, and its rapid degradation prevents cells from growing on medium lacking uracil (SD-ura) (Chen et al, 1993). Doa10 pathway mutants are strongly impaired in Deg1-Ura3 turnover, allowing growth on SD-ura. As shown in Figure 1B, the kis3 and kis4 mutants also grew well under these conditions. We next tested whether the kis3 and kis4 strains might have mutations in Cue1 or Doa10. The Deg1-Ura3-based growth assay was used for complementation analysis. Growth on SD-ura was observed only for the crosses between doa10Δ and kis3–66 and between cue1Δ and kis4–14 (Figure 1C). These data suggested that kis3–66 is a mutant allele of DOA10 and kis4–14 is a mutation in CUE1 (Table I). In support of this, no Doa10 protein was detected by anti-Doa10 immunoblotting in kis3–66 cells, unlike wild-type or kis4–14 cells (Figure 1D), and sequencing of the CUE1 gene from the kis4–14 strain revealed a single C-to-T transition at nucleotide 250 in the ORF, changing Q84 to a stop codon.
Table 1.
Gene assignments for the ndc10-2 suppressors (Kopski and Huffaker, 1997)
| Complementation group | Alleles | Gene | Reference |
|---|---|---|---|
| kis1 | 10 | UBC7 | Kopski and Huffaker (1997) |
| kis2 | 3 | UBC6 | Kopski and Huffaker (1997) |
| kis3 | 10 | DOA10 | This study |
| kis4 | 1 | CUE1 | This study |
Synthetic degron-protein fusions are Doa10 substrates
Another group of short-lived proteins that were identified as Ubc6/Ubc7/Cue1-dependent substrates were the artificial SL17/CL degron fusions of Gilon et al (2000). Short peptides derived from a library of random yeast DNA inserts fused downstream of either the Escherichia coli gene for β-galactosidase (βgal) or yeast URA3 conferred rapid degradation on the encoded proteins. Degradation of a βgal-SL17 fusion was strongly impaired in doa10Δ cells (Figure 2A). Consistent with this, when wild-type and doa10Δ cells expressing a Ura3-SL17 fusion were compared for growth on SD-ura, robust growth was only seen in doa10Δ (Figure 2B). Growth analysis of six different Ura3-CL degron fusions gave similar results. Interestingly, these fusions with Ura3 localized to the cytoplasm and were apparently excluded from the nucleus (Figure 2C), in contrast to Deg1-protein fusions, which concentrate in the nucleus.
Figure 2.
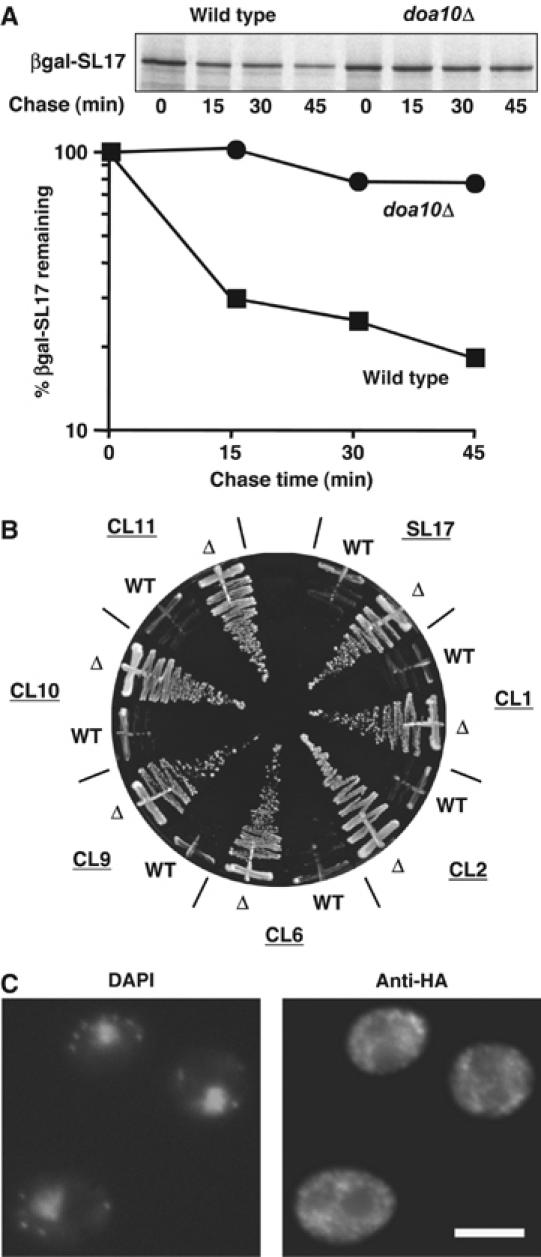
Doa10 substrates include a series of cytoplasmic synthetic degron fusions. (A) Degradation of a fusion of the SL17 degron with βgal measured by pulse-chase analysis at 30°C. (B) Growth on SD-ura of wild-type (WT) and doa10Δ (Δ) cells expressing fusions of the indicated degrons to Ura3. (C) Cytoplasmic localization of Ura3-HA-SL17 detected by anti-HA immunofluorescent staining in doa10Δ. Staining was similar in WT and doa10Δ cells, but much brighter in the latter; three fusions (SL17, CL1 and CL2) were tested and gave similar results. Scale bar, 5 μm.
Diverse membrane protein targets of Doa10
The preceding data are consistent with the idea that Doa10 is principally involved in recognizing degradation determinants on the cytosolic side of the ER membrane, while Hrd1 may mostly monitor luminally exposed structural features (Vashist and Ng, 2004). Relatively few proteins have been analyzed, however, so it is not certain that this will be generally true. Specificity for the Hrd1 and Doa10 E3s might arise through their recognition of substrate features on the same side of the membrane or the partitioning of E3s and substrates to different membrane subdomains. A broader range of membrane substrates for both E3s will be needed to determine the relative importance of these different mechanisms.
One candidate membrane substrate for Doa10 was the mutant Mps2-1 protein (McBratney and Winey, 2002). Mps2 is a single-pass membrane protein that helps tether the spindle pole body (SPB), the yeast equivalent of the vertebrate centrosome, to the NE. The thermolabile Mps2-1 point mutant is short-lived; when cells are shifted to high temperature, the protein disappears from SPBs (McBratney and Winey, 2002). The growth defect of mps2-1 cells at 36°C is suppressed by deleting CUE1, UBC6, or UBC7, but not HRD1, and the rapid degradation and SPB depletion of Mps2-1 protein is also strongly inhibited by cuelΔ. We deleted DOA10 from mps2-1 cells to determine if this would also suppress mps2-1 temperature sensitivity (Figure 3A). Four independent mps2-1 doa10Δ strains were tested. All showed strong suppression of the mps2-1 growth defect at 36°C, consistent with stabilization of a partially functional protein.
Figure 3.
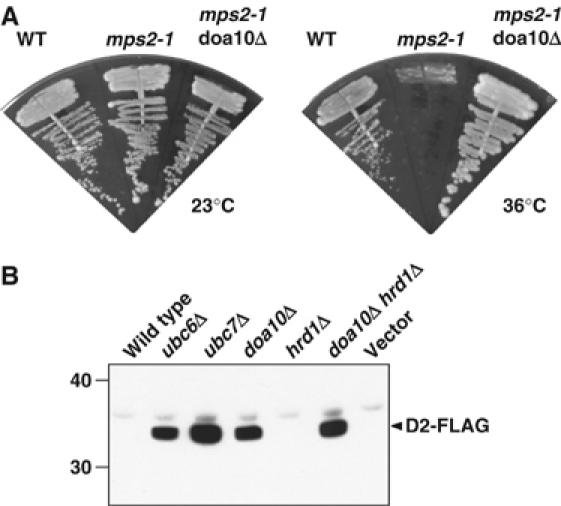
Degradation of a mutant transmembrane NE protein and a human ER-localized enzyme requires Doa10. (A) Mps2-1 is degraded by the Doa10 pathway. The DOA10 locus was deleted in an mps2-1 strain; the double mutant is shown alongside the original mps2-1 mutant and a wild-type (WT) control. Cells were grown on YPD for 3 days. (B) Degradation of the human ER-localized Dio2 requires Doa10 in yeast. Anti-FLAG immunoblotting was carried out for the indicated strains expressing the deiodinase D2-FLAG from a galactose-regulated promoter. The two selenocysteine codons in Dio2 were changed to cysteine for expression in yeast (Botero et al, 2002). Last lane, WT strain transformed with empty vector.
The human deiodinase D2 (Dio2) has its N-terminal end inserted in the ER membrane (Botero et al, 2002). This short-lived enzyme converts the thyroid hormone thyroxine (T4) to its active form; high levels of T4 increase the rate of Dio2 degradation. Dio2 proteolysis shows similar features when expressed in human and yeast cells. In particular, Ubc6 and Ubc7 have been implicated in Dio2 ubiquitination in both organisms, and T4 stimulates Dio2 degradation in yeast as well (Botero et al, 2002). We therefore asked whether Doa10 was important for Dio2 degradation (Figure 3B). As expected, deletion of either Ubc6 or Ubc7 caused much greater accumulation of Dio2 than in wild-type cells, where the protein was barely detectable. Similarly, high levels of Dio2 were observed in doa10Δ and doa10Δ hrd1Δ cells, but not in hrd1Δ. We conclude that the naturally short-lived Dio2 deiodinase is a substrate of Doa10 in yeast.
Together with earlier data on membrane protein substrates of Doa10, the new findings with Mps2-1 and Dio2 indicate that many structurally diverse membrane proteins can be ubiquitinated by Doa10. Some of these can be considered quality-control substrates, whereas others are wild-type proteins that are rapidly degraded as part of their normal regulation.
A genomic screen for genes required for Doa10 pathway function
Besides E2 and E3 enzymes and the proteasome, additional proteins generally are required for efficient degradation of ERAD substrates. These proteins participate in various stages of the proteolytic pathway, such as substrate recognition, retrotranslocation, and trafficking to the proteasome (Hirsch et al, 2004). Previous genetic screens for factors involved in the degradation of α2 identified the E2 and E3 enzymes of the Doa10 pathway as well as proteasome subunits and proteasome regulators. Only the Ubc6 and Ubc7 E2s (and the Ubc7 cofactor Cue1) and the Doa10 E3 were shown to be crucial for Deg1-mediated ubiquitination. To identify potential additional factors that contributed to the ubiquitination and degradation of Deg1-bearing substrates, we performed a high-throughput screen of a library of ∼4800 viable yeast gene deletion strains using synthetic genetic array (SGA) technology. Our proteolytic reporter was a fusion of Deg1 to Ura3 and a triplicated HA epitope (Deg1-Ura3-3HA). Rapid degradation of the fusion protein in wild-type cells prevents them from forming colonies on SD-ura, but mutations that hinder its degradation lead to uracil prototrophy.
The chromosomally integrated Deg1-URA3-3HA reporter was introduced into each of the single gene deletion yeast strains by the SGA method (Materials and methods). Over 99% of the 4753 deletion strains were successfully crossed and selected to carry both the reporter and the individual gene deletion. These were then pinned onto SD-ura plates. In all, 47 strains grew on the selective media. These strains, along with a negative control hrd1Δ strain, were transferred to a master plate and rescreened on SD-ura, leaving 17 mutants that consistently grew faster than controls (Table II; Supplementary Figure S1A). Four strains grew much faster than the rest: cue1Δ, doa10Δ, ubc7Δ, and nup120Δ. The first three inactivate known components of the Doa10 ubiquitination pathway (ubc6Δ was not in the library), while the fourth lacks the nuclear pore protein Nup120.
Table 2.
Mutants identified from genomic deletion screen with Deg1-Ura3-HA3
| Systematic name | Standard name | RGa | Functional description |
|---|---|---|---|
| Ubiquitin conjugation | |||
| YMR022W | UBC7 | 4 | Ubiquitin-conjugating enzyme |
| YMR264W | CUE1 | 4 | Recruits Ubc7 to the ER |
| YIL030C | DOA10 | 4 | E3 ubiquitin ligase |
| Proteasome | |||
| YDL020C | RPN4 | 2 | Transcriptional activator for proteasome genes |
| YBR173C | UMP1 | 3 | 20S proteasome maturation factor |
| YDR363W-A | SEM1 | 3 | 26S proteasome regulatory subunit |
| Chromosome/cell cycle/DNA replication | |||
| YGR188C | BUB1 | 3 | Protein kinase that functions in spindle checkpoint |
| YOR026W | BUB3 | 3 | Spindle checkpoint protein; binds Bub1 |
| YPL008W | CHL1 | 2 | Helicase, required for sister chromatid cohesion |
| YPR141C | KAR3 | 3 | Kinesin related, required for sister chromatid cohesion |
| YHR191C | CTF8 | 2 | Replication factor, required for sister chromatid cohesion |
| Telomerase | |||
| YLR233C | EST1 | 2 | Telomerase holoenzyme subunit |
| YLR318W | EST2 | 2 | Telomerase catalytic subunit |
| YIL009C-A | EST3 | 2 | Telomerase holoenzyme subunit |
| Other | |||
| YPR131C | NAT3 | 3 | Catalyzes acetylation of N-terminal methionine of proteins |
| YJL092W | SRS2 | 3 | DNA helicase, disassembles Rad51 filaments |
| aRelative growth value was based on three to four independent growth assays (scales 1–4, 4=fastest). | |||
Among the deletion strains that grew on SD-ura plates but more slowly were strains lacking subunits or regulators of the proteasome: the Sem1 regulatory subunit, the Ump1 maturation factor, and the Rpn4 proteasomal gene regulator. Most proteasome components are essential for viability and were therefore not present in the deletion library. Deletions of two nonessential proteasome subunits, Pre9 and Rpn10, do not impair Deg1-mediated proteolysis (Velichutina et al, 2004; Verma et al, 2004), explaining why they were not isolated in our screen. Some strains showed very weak growth enhancements on SD-ura, but intriguingly, the mutations affected proteins from a very limited number of cellular regulatory systems (Table II). Three telomerase subunits were identified, as were several proteins linked to the mitotic checkpoint and chromatid cohesion. Loss of the Nat3 Nα-acetyltransferase also enhanced growth. These findings suggest that these factors might modulate Doa10 pathway function, possibly indirectly.
For representative candidate strains, degradation of Ubc6-HA and Deg1-βgal test substrates was monitored (Supplementary Figure S1B). As predicted, degradation rates of the test substrates showed an inverse relationship with colony growth on SD-ura. Further analysis of the nup120Δ strain revealed that an unlinked lesion was responsible for the degradation defect (not shown). Based on complementation analysis and anti-Doa10 immunoblotting, the strain had a cryptic mutation in DOA10 (Supplementary Figure S1C).
In summary, a comprehensive screen of the nonessential genes in S. cerevisiae has identified a very small number that are required for Deg1-mediated degradation, and these are precisely the same genes previously shown to encode the Ub-ligation enzymes of the Doa10 pathway. Not insignificantly, the identical set of genes was identified in the selection for ndc10-2 suppressors (Table I).
The above analyses suggest that Doa10 pathway substrates require a common set of factors, including Doa10, Ubc6, Ubc7, and Cue1. Surprisingly, Cue1 was reported to have a much more limited role than Ubc6 or Ubc7 in degrading α2 itself (Lenk and Sommer, 2000). Cue1 is a transmembrane ER receptor for Ubc7. We re-examined the contribution of Cue1 to α2 degradation by deleting CUE1 or both CUE1 and UBC4 (Supplementary Figure S2). The half-life of α2 increased ∼2-fold in cue1Δ cells, similar to what was seen in ubc7Δ (Chen et al, 1993). Moreover, a synergistic impairment of degradation was observed in cue1Δ ubc4Δ cells, as expected, if both major α2 ubiquitination pathways were blocked. The ∼8-fold inhibition was similar to that previously observed in ubc4Δ ubc7Δ mutants. We conclude that Cue1 is required in the Doa10 pathway of α2 ubiquitination.
Membrane and soluble Doa10 substrates differ in their Cdc48 dependence
All of these genetic analyses suggested that Doa10, Ubc6, Ubc7, and Cue1 were the only nonessential and nonredundant factors encoded in the yeast genome that were required for efficient Doa10-dependent protein ubiquitination. However, it was possible that there were essential (or redundant) proteins—other than the proteasome—that also contributed to Doa10-dependent proteolysis. We examined the role of one such essential protein complex directly and also asked whether different classes of substrates shared all steps between Ub addition and proteolysis by the proteasome.
Different ubiquitinated proteins have distinct adaptors that bind to polyubiquitinated substrates and may ferry them to the 26S proteasome (Kim et al, 2004; Verma et al, 2004). Membrane substrates ubiquitinated by Hrd1 or Doa10 need the three-component Cdc48-Npl4-Ufd1 ATPase complex for their degradation (Bays et al, 2001; Wang and Chang, 2003; Huyer et al, 2004). The Cdc48 AAA ATPase can help extract proteins from the membrane (Ye et al, 2001), but the complex, which binds both poly-Ub and the proteasome, might also act more generally as a proteasome adaptor (Dai et al, 1998). Consistent with this latter idea, Cdc48 is also required for degradation of some nonmembrane proteins (Johnson et al, 1995; Cao et al, 2003).
To test the involvement of the Cdc48 complex in the degradation of nonmembrane Doa10 substrates, we measured the turnover of α2 and a series of short-lived Deg1 fusion proteins in various mutants. As expected, in cells lacking the Doa10 pathway (ubc6Δ), α2 was stabilized ∼2–3-fold relative to wild-type cells (Figure 4A). In contrast, mutations in CDC48, NPL4, or UFD1 had no detectable effect on its degradation. Similarly, mutations in these same Cdc48 complex components failed to alter the degradation of either Deg1-βgal or Deg1-Ura3 (Figure 4B and not shown). Simultaneous measurement of Deg1-βgal and CPY*-HA degradation in cdc48 cells showed that the latter was stabilized, as expected (Figure 4C).
Figure 4.
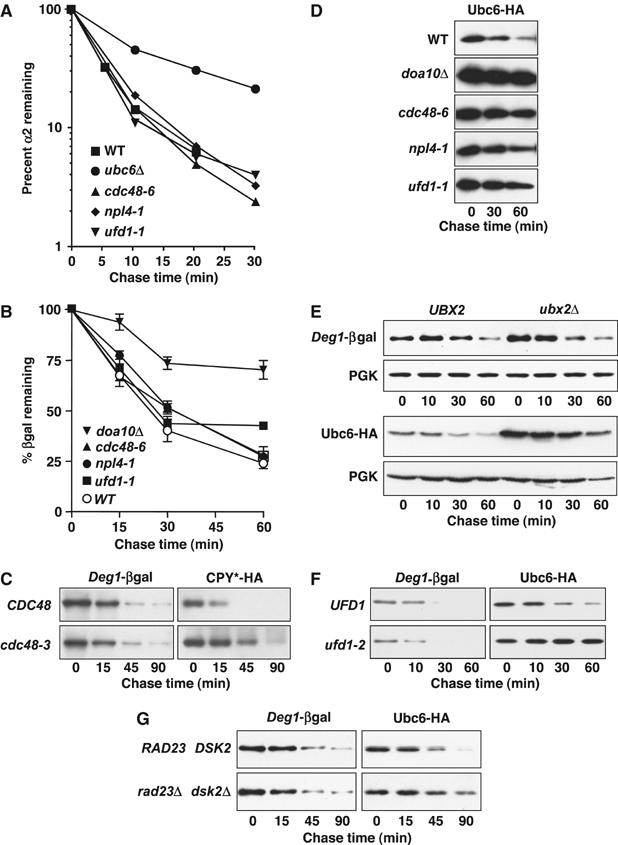
The Cdc48 ATPase complex is required for Doa10 membrane substrates, but not α2 and soluble Deg1-bearing substrates. (A) Pulse-chase analysis of α2 degradation in Cdc48 complex mutants. Proteins were precipitated with anti-α2 antibodies. (B) Loss of Deg1-βgal is not affected in mutants lacking specific Cdc48 complex components. Loss of βgal activity was followed after the addition of cycloheximide. Three transformants of each strain were assayed. Error bars are obscured by symbols at some data points. (C) Degradation of Deg1-βgal is not impaired in cdc48-3 cells, in contrast to that of CPY*-HA. Cells cotransformed with plasmids encoding the respective proteins were assayed by cycloheximide chase and immunoblotting with antibodies to βgal and HA. (D) Degradation of the Ubc6-HA membrane protein requires the Cdc48 complex. Degradation was assayed by cycloheximide chase and anti-HA immunoblotting. (E) Degradation of Ubc6-HA, but not Deg1-βgal, requires Ubx2, an ER receptor for Cdc48. Protein turnover was assayed as in (C). (F) Degradation of Deg1-βgal and Ubc6-HA in ufd1-2 cells. Strains cotransformed with expression plasmids for the two proteins were assayed as in (C). (G) Rad23 and Dsk2 contribute to Ubc6-HA, but not Deg1-βgal degradation. Strains cotransformed with expression plasmids for the two proteins were assayed at 30°C as in (C).
We also tested the degradation of fusions of the Deg1 degron to a single GFP, Deg1-GFP (Bays et al, 2001), or a tandem pair of GFPs, Deg1-GFP2 (Lenk and Sommer, 2000). Both proteins still required the Doa10 pathway for maximal degradation, although this dependence was notably weaker than for the above Deg1 reporters, presumably because an additional degradation pathway(s) can recognize the GFP constructs (Supplementary Figure S3). Although we consider these less than ideal test substrates, neither Deg1-GFP nor Deg1-GFP2 was degraded more slowly in any tested mutant of the Cdc48 complex. Others had previously tested the same GFP fusions in Cdc48 complex mutants. Our data agree with those of Bays et al (2001) and Medicherla et al (2004), but not Verma et al (2004) or Neuber et al (2005). The reason for the discrepancies is unknown, but might reflect effects on Doa10-independent pathways acting on these fusions.
Until recently, mutant versions of the Pma1 and Ste6 transporters were the only Doa10 membrane substrates tested for Cdc48-dependent degradation (Wang and Chang, 2003; Huyer et al, 2004), so it was possible that their Cdc48 dependence was unusual, perhaps reflecting their large size or large number of TM segments. We therefore tested Ubc6, a small single TM substrate of Doa10. Unlike the different soluble substrates we tested, this short-lived enzyme was strongly stabilized in the cdc48, npl4, ufd1, and ubx2 mutants (Figure 4E and F). This is consistent with a very recent report on the same substrate (Neuber et al, 2005). Ubx2, an ER membrane protein, helps dock Cdc48 to the ER (Schuberth and Buchberger, 2005; Neuber et al, 2005). Turnover of Deg1-βgal in cells lacking Ubx2 was not impaired (Figure 4E). Notably, Ubc6-HA was stabilized in ufd1-2 cells, whereas Deg1-βgal coexpressed in the same cells was unaffected (Figure 4F). Finally, loss of the Rad23 and Dsk2 proteins, which function as (partially redundant) adaptors between polyubiquitinated substrates and the proteasome, also specifically stabilized Ubc6-HA, but not coexpressed soluble Deg1-βgal (Figure 4G).
From the above data and from published results with mutant membrane transporters, it appears that the Cdc48 ATPase complex and Rad23/Dsk2 are crucial for the degradation of membrane protein substrates of Doa10, but not for the degradation of soluble substrates ubiquitinated by the same E3.
A Deg1 membrane substrate of Doa10 requires Cdc48 for its degradation
The preceding analyses used membrane substrates with different and largely uncharacterized degrons. To pinpoint a function for the Cdc48 complex that is specific for membrane substrates of the Doa10 pathway, one would ideally use a matched set of substrates in which exactly the same degron and Ub machinery are utilized, and the only difference is whether or not the substrate is inserted in the membrane. We therefore sought to create a Deg1-bearing substrate that inserts in the ER membrane and is degraded by the Doa10 pathway. Importantly, several Deg1 membrane protein fusions had been made previously, but none of them is degraded primarily by the Doa10 pathway (Mayer et al, 1998; Wilhovsky et al, 2000; our unpublished data).
We fused Deg1 to the N-terminus of Vma12, a resident ER protein with both termini facing the cytoplasm (Figure 5A) (Jackson and Stevens, 1997). A protein A (PrA) or GFP tag was present at the C-terminus (Materials and methods). With the GFP-tagged derivative, we observed typical cortical and perinuclear ER staining (Figure 5B). By cell fractionation, Deg1-Vma12-PrA (Deg1-VP) behaved as an integral membrane protein, only being solubilized by detergent (Figure 5C); both the N- and C-terminal domains faced the cytosol based on protease digestion of microsomes (not shown). In pulse-chase analyses, Deg1-VP was rapidly degraded (half-life 7–8 min) in a manner strictly dependent on Doa10, but not Hrd1 (Figure 5D). Degradation also required both Ubc6 and Ubc7 (not shown). Introduction of point mutations known to inactivate Deg1 (designated Deg1*), in soluble proteins (Johnson et al, 1998), also completely blocked Deg1-VP degradation (Figure 5E). Therefore, the degron and Ub enzyme requirements for Deg1-VP turnover are identical to those for soluble Deg1-bearing substrates.
Figure 5.
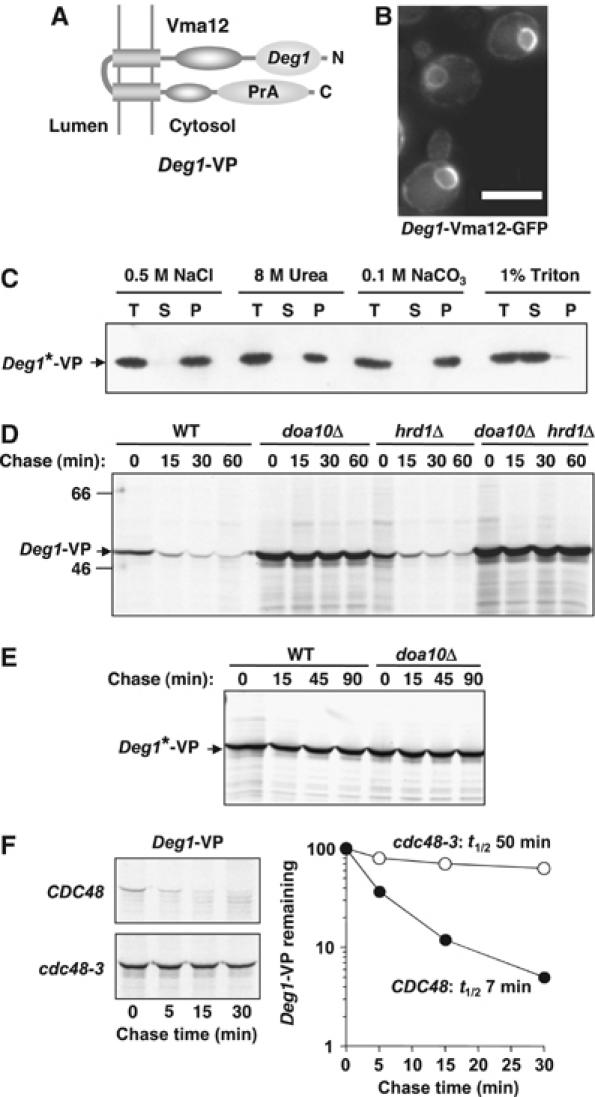
Degradation of a Deg1-Vma12 membrane protein requires both Doa10 and Cdc48. (A) Schematic of the predicted topology of Deg1-Vma12-PrA (Deg1-VP). (B) Deg1-Vma12-GFP localizes to the ER. The PrA moiety in Deg1-VP was replaced with GFP to allow fluorescence localization in live cells (doa10Δ). Scale bar, 5 μm. (C) Deg1*-VP is an integral membrane protein based on cell fractionation. The Deg1 degron was inactivated by mutation (F18S, I22T: Deg1*) to allow it to accumulate. (D) Deg1-VP is a short-lived substrate of the Doa10 pathway as measured by pulse-chase analysis. Proteins were precipitated with antibodies to α2. (E) Inactivation of the Deg1 degron (Deg1*) blocks Deg1*-VP degradation. Analysis was performed as in (D). (F) Degradation of Deg1-VP requires the Cdc48 complex. Analysis was carried out as in (D), except that cells grown overnight at 24°C were shifted to 30°C for 4 h, and then to 37°C for 1.5 h prior to pulse-chase analysis at 37°C; quantitation is shown on the right.
We then determined whether Deg1-VP degradation was dependent on Cdc48. By pulse-chase analysis in cdc48 cells, a striking stabilization of Deg1-VP was observed (Figure 5F). This contrasts to the lack of an observable defect in the degradation of soluble Deg1 substrates (Figure 4). Thus, the transplantation of Deg1 to a membrane protein created a new requirement for Cdc48 for Doa10-mediated degradation, as predicted from the previous correlative analyses. These data argue that the Cdc48 complex does not function as a general proteasome adaptor in the Doa10 pathway, but rather is likely to function primarily if not exclusively in substrate displacement from the membrane.
Discussion
The work presented here combines three related sets of results: (1) a genomics-based analysis that identified most or all nonredundant and nonessential factors necessary for general Doa10-dependent ubiquitination; (2) an expansion of known proteolytic substrates of Doa10 to include both soluble quality-control substrates and naturally short-lived membrane proteins; and (3) a demonstration that membrane and nonmembrane Doa10 substrates differ in a predictable manner in their requirements for subsequent proteasome targeting.
A novel genome-wide screen for Ub pathway components
Previously, classical genetic screens with Deg1-based reporters identified various genes that participate in the degradation of the α2 regulator (see Swanson et al, 2001). For Deg1-dependent protein ubiquitination, mutations in the E1 (Uba1), E2s (Ubc6 and Ubc7), E3 (Doa10), and an E2-anchoring factor (Cue1) were identified. However, because it has not yet been possible to reconstitute α2 or Deg1-reporter ubiquitination with purified components, it is not clear whether this represents the full list of Deg1-dependent ubiquitination factors. Moreover, it was still possible that these enzymes functioned indirectly and that other E2s and E3s were directly responsible for Deg1-mediated Ub ligation in vivo.
Our screen of ∼4800 viable yeast deletion strains for defects in Deg1-mediated proteolysis provides insight into these issues. We found only three mutants—doa10Δ, cue1Δ, and ubc7Δ—that caused strong stabilization of Deg1-based reporters. The ubc6Δ mutant was not in the library, but Ubc6 function in the degradation in Deg1-containing proteins is well established. Therefore, this screen suggests that Doa10, Ubc6, Ubc7, and Cue1 could well be the only nonessential and nonredundant factors strictly required for Deg1-dependent ubiquitination. Factors with more ancillary or substrate-specific roles may exist. The failure to find any additional E2 or E3 candidates in our genomic screen also provides an argument against the idea that Doa10 and Ubc6–Ubc7 only function indirectly in Deg1-mediated ubiquitination. We have now tested all 11 Ub E2s as well as the Rub1 E2, and only Ubc6 and Ubc7 emerged as relevant factors (Chen et al, 1993; this study).
Unexpected support for the idea that Doa10, Ubc6, Ubc7, and Cue1 are the primary proteins required for Doa10-dependent ubiquitination comes from the ndc10-2 suppressors (Kopski and Huffaker, 1997). The 24 mutants affected four genes, and our analysis now shows that these genes encode the four aforementioned Doa10 pathway proteins. This remarkable convergence of genetic results suggests first that these proteins represent the core components of the Doa10 ubiquitination machinery, and second that the Ndc10-2 quality-control substrate and the naturally short-lived α2 protein share a common mechanism of ubiquitination.
The genome-wide selection that we conducted can be compared to another recently described yeast genomic selection for proteolytic mutants, one in which mutants in the Hrd1 pathway were sought (Medicherla et al, 2004). In the earlier study, each of the ∼4800 deletion strains was transformed with a plasmid encoding a proteolysis reporter. This method is more laborious than the SGA method that we employed, which introduced the reporter into each strain by simple genetic crosses performed en masse. A chromosomal reporter also reduces the chance of copy-number amplification that can occur with plasmids when cells are placed under selective pressure for increased levels of a plasmid product.
Degrons in protein quality control and regulatory proteolysis
We now know of seven naturally derived substrates of the yeast Doa10 pathway and at least this number of synthetic degron-fusion substrates (Figure 6A). The substrates include membrane proteins, soluble proteins of the cytoplasm and nucleus, naturally short-lived regulators, and aberrant proteins subject to quality control. Several issues are raised by this unprecedented crosscompartment diversity of substrates: Are their degrons related? Do fundamental differences exist in the recognition of membrane and soluble substrates of Doa10 or of regulatory and quality-control substrates? What determines whether a membrane protein substrate is targeted by the Doa10 or Hrd1 pathway?
Figure 6.
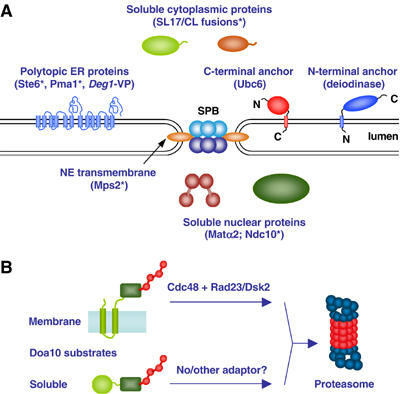
Different Doa10 substrates and routes to the proteasome. (A) Summary of Doa10 substrates. An asterisk by the protein name indicates a mutant protein (‘quality-control' substrate). See text for details. SPB, spindle-pole body. (B) Divergent pathways of proteasome targeting for Doa10 substrates.
Given the apparent diversity of Doa10 substrates, it would seem that they could not all share the same mechanism of recognition. In some cases, recognition might even be indirect. Mutant Pma1-D378N binds to the transmembrane ER disulfide isomerase Eps1, and this binding appears to occur prior to Doa10 action (Wang and Chang, 2003). If Eps1 also associates with Doa10, it could act as a substrate recognition subunit of the E3. Such ‘indirect recognition' is less likely for soluble substrates such as α2 and Ndc10-2 because only the four E2–E3 complex components emerged from our genome-wide screen and the ndc10-2 suppressor selection. Essential or redundant factors may yet be found (Swanson et al, 2001).
In our original model for Deg1 degron recognition, we suggested that the exposed hydrophobic face of an amphipathic helix was the key recognition determinant (Johnson et al, 1998). We propose here that Doa10 can directly bind such hydrophobic surfaces in various substrates at the ER membrane. The importance of a helical structure was inferred from the fact that Deg1 is largely helical, and the only nontolerated residues found on the hydrophilic face in the Deg1 determinant were the helix breakers Pro and Gly (Johnson et al, 1998). Amphipathic helices might be involved in many cases; however, the SL17/CL degrons all share regions of strong hydrophobicity, but only a few are obviously amphipathic (Gilon et al, 2000). In the present model, Doa10 recognition of degrons in naturally short-lived regulators and quality control substrates is based on similar principles. This is consistent with mutagenesis studies of the natural Deg1 degron and the artificial CL1 degron.
Importantly, the analysis of Deg1-VP degradation (Figure 5) formally demonstrates that the same Doa10-dependent degron can be recognized in the context of both soluble and membrane substrates. Previously, it was reasonable to argue that integral membrane substrates required a distinct type of degron (although not all Doa10 degrons need be the same). What is recognized in the substrates summarized in Figure 6A? The truncated C-terminal domain of the mutant Ste6-166 (Ste6*) protein is sufficient for Doa10-dependent degradation (Vashist and Ng, 2004). We suggest that loss of the C-terminal segment in Ste6* leads to the exposure of an amphipathic or hydrophobic helical segment recognized by Doa10; a prominent amphipathic sequence (residues 1123–1136) is present in this domain of Ste6. Similarly, both Mps2 and Ndc10 have amphipathic segments with the potential to form coiled coils, and these could serve as recognition determinants if the mutant proteins misfold. Doa10 might also recognize some membrane substrates through binding of a TM segment within the membrane or after partial dislocation to expose the hydrophobic TM helix (or both).
A final outstanding question is what determines whether a membrane ERAD substrate will be targeted by Doa10 or Hrd1 (or another E3). The new data remain consistent with Doa10 recognizing cytosolically exposed elements in its membrane substrates and Hrd1 recognizing luminal determinants, although this is probably an oversimplification. In apparent contradiction to this generalization, one Hrd1 target, the thermolabile Sec61-2 translocon subunit (Bordallo et al, 1998), has a lesion on the cytosolic side of the membrane (Nishikawa et al, 2001). However, the sec61-2 mutation immediately precedes an unusually short TM segment and might destabilize it, causing a structural change recognized by the Hrd1 complex on the luminal side of (or within) the membrane.
Function of the Cdc48 complex in the Doa10 pathway
For efficient targeting to the proteasome, polyubiquitinated substrates utilize a variety of different poly-Ub-binding factors, of which some can be regarded as proteasome adaptors and others as transfer factors acting further upstream (Hirsch et al, 2004; Kim et al, 2004; Verma et al, 2004). No clear rules for linking particular substrates to particular factors had emerged from previous studies, even for proteins ubiquitinated by the same E2–E3 pathway (Verma et al, 2004). In contrast, our data suggest that a substrate ubiquitinated by Doa10 will require certain poly-Ub-binding factors for routing to the proteasome only if it is a membrane-embedded protein. Specifically, the Cdc48-Npl4-Ufd1 complex (and the Rad23/Dsk2 adaptors) is required for the degradation of all Doa10 membrane substrates tested to date, but none of the soluble substrates (Figure 6B). This suggests that the exclusive role of the Cdc48 complex in the degradation of Doa10 substrates is in the extraction or release of those substrates that are inserted in the membrane, a well-documented biochemical activity of this ATPase (Ye et al, 2001). Comparison of the membrane Deg1-VP substrate to soluble Deg1 substrates demonstrates that even for proteins with the same degron and the same Ub enzyme requirements, only the membrane Doa10 substrate requires Cdc48. Our results also support the suggestion of some kind of mechanistic coupling between Cdc48 and the Rad23 and Dsk2 poly-Ub-binding adaptors (Medicherla et al, 2004; Richly et al, 2005).
Cdc48 is required for the proteolysis of certain soluble substrates, but its exact role is less well established in these cases (Johnson et al, 1995; Dai et al, 1998; Cao et al, 2003; Richly et al, 2005). In the Doa10 pathway, Cdc48 might use its ATPase activity to help dissociate polyubiquitinated soluble substrates from the membrane E3 complex. However, our data indicate that either such an activity is not absolutely required for soluble Doa10 substrates or there are dissociation factors that act redundantly with Cdc48. The latter idea would imply that the Cdc48 complex acts at multiple steps in the degradation of membrane-bound Doa10 substrates, but only the membrane extraction step strictly requires it.
With the substrates described in this work, we now know of 14 proteins (not counting the different Deg1 fusion variants) that are degraded by the yeast Doa10 pathway. These offer a rich set of membrane and nonmembrane targets for further analysis of protein quality control and regulatory proteolysis.
Materials and methods
Yeast and bacterial methods
Yeast rich (YPD) and minimal (SD) plates were prepared as described, and standard methods were used for genetic manipulation of yeast (Guthrie and Fink, 1991). Standard techniques were used for recombinant DNA work.
Plasmids
Plasmids encoding fusions of the Deg1 degron to various reporters have been described previously (Hochstrasser and Varshavsky, 1990; Chen et al, 1993; Lenk and Sommer, 2000; Wilhovsky et al, 2000), as have plasmids expressing Ubc6-HA (Walter et al, 2001), human Dio2 (Botero et al, 2002), and CPY*-HA (Huyer et al, 2004). pRS305-Deg1-URA3-3HA was constructed by a two-step PCR-based method. The final insert included the entire intergenic region between MATα1 and MATα2, and extended through the transcriptional terminator after the URA3 and 3HA segments. The p414MET25-Deg1-Vma12-PrA plasmid was made by recombination in yeast between the PCR-amplified VMA12 ORF and a gapped p414MET25 plasmid containing the Deg1-FLAG-Sec62-PrA coding sequence (Mayer et al, 1998). This resulted in an exact exchange of the VMA12 and SEC62 ORFs. The PrA segment was exchanged by a similar procedure with a GFP coding sequence (W Greenblatt, unpublished). All ORF inserts were checked by DNA sequencing.
Yeast strains
Strains used in this study are listed in Supplementary Table I, except those from the Saccharomyces Genome Deletion Project (http://sequence-www.stanford.edu/group/yeast_deletion_project/), which were purchased from Open Biosystems. Strains from this library were used for the genome-wide doa screen and for the analysis of ubx2. MHY1687 was a segregant from a cross between congenic cue1Δ and ubc4Δ strains. MHY3185 was created by PCR-mediated replacement of DOA10 with a doa10Δ::HIS3 allele in MHY1824. MHY3272 was derived from a cross between MHY3188 and the cue1Δ strain from Open Biosystems, while MHY3273 was from a cross between MHY3188 and MHY1685 (the a version of MHY1631). To generate MHY3528, the ufd1-1 allele from PM373 (Johnson et al, 1995) was crossed into the MHY500 background followed by an additional backcross. For the genomic screen for doa mutants, a bait strain with a chromosomal Deg1-URA3-3HA reporter was constructed. Yeast strain Y5563 (from C Boone) was transformed with pRS305-Deg1-URA3-3HA, which had been cleaved with Tth111 I to direct it to the leu2Δ0 locus, generating MHY3011. Leu+ transformants were checked by colony PCR to verify the integration site and by anti-HA immunoblotting.
Genomic screen of yeast deletion library
The Deg1-Ura3-3HA substrate-expressing strain MHY3011 was crossed to all the strains from the Saccharomyces Genome Deletion Project. Selection for diploids, sporulation, and selection for the desired a-type haploids were carried out similarly to published protocols (Tong et al, 2001). Details are provided in Supplementary Table SII. All matings and cell transfers were carried out in a 96-well plate format using 96-pin replicators. Haploid cells that had both the reporter gene and the specific library gene deletion were plated on SD-ura plates and colony growth at 30°C was monitored. Colonies that grew on SD-ura were re-plated on a single plate to allow a retest of growth and better comparison of relative growth rates.
Pulse-chase and immunoblot analyses
Pulse-chase analysis was carried out as described (Chen et al, 1993). For immunoblot/cycloheximide chases, several methods were used for cell extraction. Culture aliquots from each time point were added to sodium azide (0.1% final) on ice. Extractions of soluble substrates were usually carried out by incubating cells with 0.1 N NaOH for 5 min at 23°C, spinning down the cells, and dissolving the pellet in sample buffer (62.5 mM Tris–HCl, pH 6.8, 20% glycerol, 1.5% SDS, 8 M urea, 50 mg/ml DTT, bromphenol blue). After boiling for 5 min, samples were loaded onto gels. For ER proteins, a previously described method was used for lysis (Huyer et al, 2004). When soluble and ER proteins were monitored in the same cells, lysis was carried out by the second of the two methods. Antibodies used were anti-α2 antiserum (Hochstrasser and Varshavsky, 1990), affinity-purified anti-Ndc10 antibody (Muller-Reichert et al, 2003), 16B12 anti-HA and 9E10 anti-myc antibodies (Covance), affinity-purified anti-Doa10 (unpublished), and anti-FLAG M2 antibody (Sigma). Proteins were visualized by ECL (Amersham).
To measure βgal activity of Deg1-βgal in various mutants, cells were transformed with a 2μ-based Deg1-lacZ plasmid, and cycloheximide was added to 0.5 mg/ml at time zero to block protein synthesis. Cell lysis and activity assays were carried out as described (Plemper et al, 1997). Loss of βgal activity has been shown to mirror the loss of Deg1-βgal protein as measured by immunoblotting (M Deng and M Hochstrasser, unpublished).
For analysis of Cdc48 complex components, partial loss-of-function mutants (Richly et al, 2005) were grown at 24°C and shifted to 37°C for 30 min prior to pulse-chase, immunoblot/cycloheximide-chase, or βgal activity-chase analyses. Radiolabeling and chase were carried out at 37°C. This protocol was followed for all but the α2 pulse-chases, where cultures were instead shifted to 30°C for 1.5 h prior to labeling, and pulse-chase analysis was carried out at 32°C.
Supplementary Material
Supplementary Table I
Supplementary Table II
Supplementary Figure Legends
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Acknowledgments
We thank R Swanson for the data in Figure 2A, W Greenblatt for the Deg1-Vma12-GFP construct, and E McCargo for help with the genomic screen. We are grateful to A Buchberger, B Gereben, T Huffaker, G Huyer, S Jentsch, R Kulka, T Sommer, R Verma, and M Winey for strains or plasmids and to A Desai for Ndc10 antibody. This work was supported by NIH Grant GM46904.
References
- Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY (2001) HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell 12: 4114–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH (1998) Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell 9: 209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botero D, Gereben B, Goncalves C, De Jesus LA, Harney JW, Bianco AC (2002) Ubc6p and Ubc7p are required for normal and substrate-induced endoplasmic reticulum-associated degradation of the human selenoprotein type 2 iodothyronine monodeiodinase. Mol Endocrinol 16: 1999–2007 [DOI] [PubMed] [Google Scholar]
- Cao K, Nakajima R, Meyer HH, Zheng Y (2003) The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 115: 355–367 [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M (1993) Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell 74: 357–369 [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Schwartz AL (2004) The ubiquitin system: pathogenesis of human diseases and drug targeting. Biochim Biophys Acta 1695: 3–17 [DOI] [PubMed] [Google Scholar]
- Dai RM, Chen E, Longo DL, Gorbea CM, Li CC (1998) Involvement of valosin-containing protein, an ATPase co-purified with IκBα and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IκBα. J Biol Chem 273: 3562–3573 [DOI] [PubMed] [Google Scholar]
- Gilon T, Chomsky O, Kulka RG (2000) Degradation signals recognized by the Ubc6p–Ubc7p ubiquitin-conjugating enzyme pair. Mol Cell Biol 20: 7214–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR (1991) Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego [Google Scholar]
- Hampton RY (2002) ER-associated degradation in protein quality control and cellular regulation. Curr Opin Cell Biol 14: 476–482 [DOI] [PubMed] [Google Scholar]
- Hirsch C, Jarosch E, Sommer T, Wolf DH (2004) Endoplasmic reticulum-associated protein degradation—one model fits all? Biochim Biophys Acta 1695: 215–223 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M (1996) Ubiquitin-dependent protein degradation. Annu Rev Genet 30: 405–439 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Varshavsky A (1990) In vivo degradation of a transcriptional regulator: the yeast α2 repressor. Cell 61: 697–708 [DOI] [PubMed] [Google Scholar]
- Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S (2004) Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem 279: 38369–38378 [DOI] [PubMed] [Google Scholar]
- Jackson DD, Stevens TH (1997) VMA12 encodes a yeast endoplasmic reticulum protein required for vacuolar H+-ATPase assembly. J Biol Chem 272: 25928–25934 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Ma PCM, Ota IM, Varshavsky A (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270: 17442–17456 [DOI] [PubMed] [Google Scholar]
- Johnson PR, Swanson R, Rakhilina L, Hochstrasser M (1998) Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin–proteasome pathway. Cell 94: 217–227 [DOI] [PubMed] [Google Scholar]
- Kim I, Mi K, Rao H (2004) Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell 15: 3357–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopski KM, Huffaker TC (1997) Suppressors of the ndc10-2 mutation: a role for the ubiquitin system in Saccharomyces cerevisiae kinetochore function. Genetics 147: 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney JD, Hochstrasser M (1999) Substrate targeting in the ubiquitin system. Cell 97: 427–430 [DOI] [PubMed] [Google Scholar]
- Laney JD, Hochstrasser M (2003) Ubiquitin-dependent degradation of the yeast Matα2 repressor enables a switch in developmental state. Genes Dev 17: 2259–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenk U, Sommer T (2000) Ubiquitin-mediated proteolysis of a short-lived regulatory protein depends on its cellular localization. J Biol Chem 275: 39403–39410 [DOI] [PubMed] [Google Scholar]
- Mayer TU, Braun T, Jentsch S (1998) Role of the proteasome in membrane extraction of a short-lived ER-transmembrane protein. EMBO J 17: 3251–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBratney S, Winey M (2002) Mutant membrane protein of the budding yeast spindle pole body is targeted to the endoplasmic reticulum degradation pathway. Genetics 162: 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B, Kostova Z, Schaefer A, Wolf DH (2004) A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep 5: 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Reichert T, Sassoon I, O'Toole E, Romao M, Ashford AJ, Hyman AA, Antony C (2003) Analysis of the distribution of the kinetochore protein Ndc10p in Saccharomyces cerevisiae using 3-D modeling of mitotic spindles. Chromosoma 111: 417–428 [DOI] [PubMed] [Google Scholar]
- Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T (2005) Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol 7: 993–998 [DOI] [PubMed] [Google Scholar]
- Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T (2001) Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol 153: 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Ann Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH (1997) Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388: 891–895 [DOI] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S (2005) A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120: 73–84 [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A (2005) Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol 7: 999–1006 [DOI] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M (2001) A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev 15: 2660–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Varshavsky A (2005) Regulated protein degradation. Trends Biochem Sci 30: 283–286 [DOI] [PubMed] [Google Scholar]
- Vashist S, Ng DT (2004) Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol 165: 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velichutina I, Connerly PL, Arendt CS, Li X, Hochstrasser M (2004) Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. EMBO J 23: 500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin–proteasome system. Cell 118: 99–110 [DOI] [PubMed] [Google Scholar]
- Walter J, Urban J, Volkwein C, Sommer T (2001) Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J 20: 3124–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Chang A (2003) Substrate recognition in ER-associated degradation mediated by Eps1, a member of the protein disulfide isomerase family. EMBO J 22: 3792–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhovsky S, Gardner R, Hampton R (2000) HRD gene dependence of endoplasmic reticulum-associated degradation. Mol Biol Cell 11: 1697–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414: 652–656 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table I
Supplementary Table II
Supplementary Figure Legends
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3


