Abstract
The Arabidopsis genes FT and TERMINAL FLOWER1 (TFL1) encode related proteins with similarity to human Raf kinase inhibitor protein. FT, and likely also TFL1, is recruited to the promoters of floral genes through interaction with FD, a bZIP transcription factor. FT, however, induces flowering, while TFL1 represses flowering. Residues responsible for the opposite activities of FT and TFL1 were mapped by examining plants that overexpress chimeric proteins. A region important in vivo localizes to a 14-amino-acid segment that evolves very rapidly in TFL1 orthologs, but is almost invariant in FT orthologs. Crystal structures show that this segment forms an external loop of variable conformation. The only residue unambiguously distinguishing the FT and TFL1 loops makes a hydrogen bond with a residue near the entrance of a potential ligand-binding pocket in TFL1, but not in FT. This pocket is contacted by a C-terminal peptide, which also contributes to the opposite FT and TFL1 activities. In combination, these results identify a molecular surface likely to be recognized by FT- and/or TFL1-specific interactors.
Keywords: florigen, flowering, PEBP, Raf kinase, RKIP
Introduction
The timing of flowering is fine-tuned by a balance between inducing and inhibitory signals. Genes that act as floral inducers in Arabidopsis thaliana and many other plants include FT and its immediate activator, CONSTANS (CO) (Kardailsky et al, 1999; Kobayashi et al, 1999; Samach et al, 2000; Searle and Coupland, 2004; Wigge et al, 2005), as well as LEAFY (LFY), which acts redundantly with FT in specifying floral fate (Weigel et al, 1992; Ruiz-García et al, 1997). The most prominent floral inhibitor is encoded by FLOWERING LOCUS C (FLC), likely to be another direct regulator of FT (Michaels and Amasino, 1999; Sheldon et al, 1999; Samach et al, 2000). Flowering is also potently repressed by the FT homolog TERMINAL FLOWER1 (TFL1), which in turn is negatively regulated by LFY (Shannon and Meeks-Wagner, 1991; Alvarez et al, 1992; Simon and Coupland, 1996; Bradley et al, 1997; Ratcliffe et al, 1998; Parcy et al, 2002).
In contrast to proteins such as CO, FLC or LFY, which encode transcription factors (Parcy et al, 1998; Michaels and Amasino, 1999; Sheldon et al, 1999; Samach et al, 2000), FT and TFL1 encode small proteins of about 175 amino acids. The founding member of the gene family was identified in mammals as phosphatidyl ethanolamine-binding protein (PEBP) (Schoentgen et al, 1987). Separately, it was noted that PEBP can give rise to a small peptide that stimulates acetylcholine synthesis in brain explants (Tohdoh et al, 1995). Subsequently, it was shown that PEBP proteins inhibit the activity of various enzymes, including Raf kinase and thrombin in mammals, and carboxypeptidase Y and Ras GTPase-activating protein in yeast (Bruun et al, 1998; Yeung et al, 1999, 2001; Hengst et al, 2001; Chautard et al, 2004). The most extensively studied protein among the PEBPs is Raf kinase inhibitor protein (RKIP) (Yeung et al, 1999, 2001; Trakul and Rosner, 2005).
PEBPs might also act more generally as either scaffolds for or regulators of signaling complexes, as demonstrated by the finding that SELF PRUNING (SP), a tomato homolog of TFL1, can interact with a range of diverse proteins (Pnueli et al, 2001). In Arabidopsis, FT interacts with the bZIP transcription factor FD (Abe et al, 2005; Wigge et al, 2005). FD, in turn, recruits FT to the promoter of florally expressed genes such as APETALA1 (Wigge et al, 2005). Importantly, most of the SP/FT/TFL1 interactors bind in a similar manner to both SP/TFL1 and FT, suggesting that the partners mediating the opposite activities of TFL1 and FT have not yet been identified (Pnueli et al, 1998; Wigge et al, 2005).
The structures of several FT- and TFL1-related proteins have been determined. Analyses of human and bovine PEBPs, and their complexes with the ligands phosphoethanolamine and cacodylate, identified a distinctive and highly conserved anion-binding site, which may serve to recognize phosphorylated residues in interacting partners (Banfield et al, 1998; Serre et al, 1998; Simister et al, 2002). In addition to human and bovine PEBP/RKIP, crystal structures have been determined for a bacterial PEBP homolog (Serre et al, 2001), and the TFL1 ortholog CENTRORADIALIS (CEN) from snapdragon (Banfield and Brady, 2000). The folds are all similar and dominated by a large, central β-sheet, with the anion-binding site located at one end, but with variation in the placement of their connecting loop regions.
To begin to understand the structural basis for the opposite actions of FT and TFL1, we have used overexpression of chimeric proteins to map regions that confer FT- or TFL1-like activity in vivo, similar to the study by Hanzawa et al (2005), who exchanged individual residues. Together with the analysis of FT and TFL1 crystal structures, the in vivo experiments identify a molecular surface likely to be recognized by yet-to-be-identified FT- and TFL1-specific interactors.
Results
Expression of chimeric genes in transgenic plants
The predicted polypeptides encoded by the Arabidopsis FT and TFL1 genes are 175 and 177 amino acids long, respectively, with only 39 residues involving nonconservative changes, including substitutions and insertions/deletions (Figure 1A). To map the regions responsible for their distinct in vivo activities, we constructed chimeric genes, in which individual exons were swapped (Figure 2, left). Since each gene has four exons, with conserved exon/intron boundaries, there are 14 possible chimeras in addition to the wild-type versions of FT and TFL1. All were expressed from the constitutively active cauliflower mosaic virus 35S promoter in ft tfl1 double mutants, to avoid confounding dominant-negative effects. At least 10 independent transformants were analyzed for each construct. In subsequent experiments, 13 smaller-scale chimeras were generated (Figure 2, right).
Figure 1.
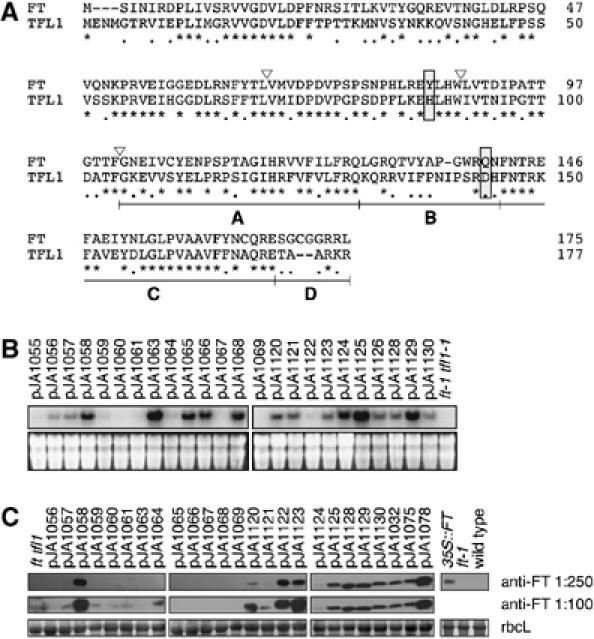
Sequence and expression of chimeras. (A) Amino-acid sequences of the parental FT and TFL1 proteins. Asterisks indicate identical residues, dots residues with similar biochemical properties, and dashes gaps introduced to optimize the sequence alignment. Triangles show the exon boundaries of FT and TFL1. The four segments used to generate the segmental chimeras within the fourth exon are shown as A, B, C and D. The Tyr85/His88 and Gln140/Asp144 residues, which form a hydrogen bond in TFL1, but not FT, and which are likely the most critical residues for distinguishing FT and TFL1 activity, are boxed. (B) RNA accumulation in transgenic plants. Blots were probed with a mixture of FT and TFL1 probes. The bottom panels show ethidium bromide-stained gels as loading controls. (C) Protein blot analysis using anti-FT antibody. The bottom panels show Ponceau S-stained blots (major band is large subunit of Rubisco).
Figure 2.
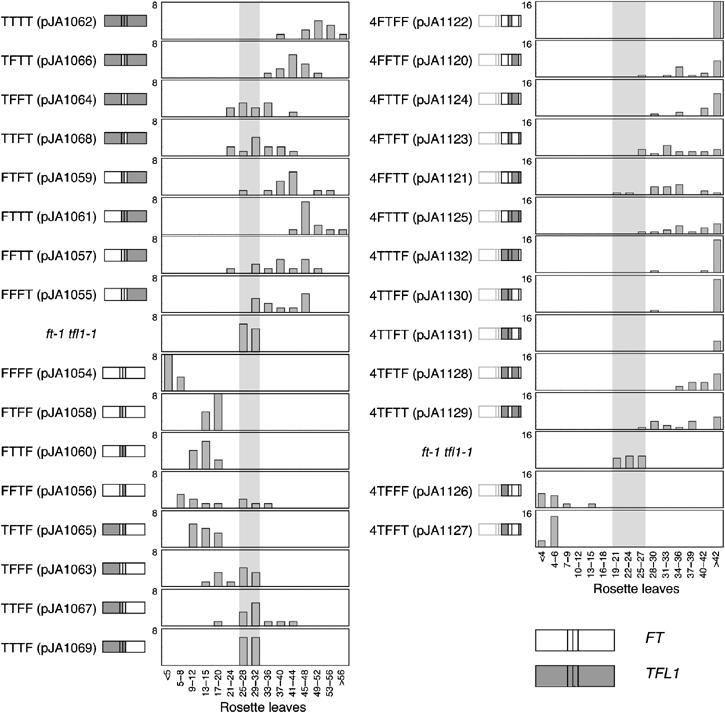
Flowering times of Arabidopsis plants expressing FT/TFL1 chimeras. The structure of each chimeric gene is shown next to the name of each construct. Sequences of FT and TFL1 are shown as open and gray boxes, respectively. Left, exon chimeras. Most transgenes that contain the fourth exon of TFL1 flower (top) cause later flowering than what is seen in the nontransgenic ft tfl1 control. Chimeras that contain the fourth exon of FT cause mostly late flowering (bottom). Right, swaps of segments in the fourth exon, with exons one to three from FT. Chimeras that cause later flowering are shown above the nontransgenic ft tfl1 control. In this experiment, the control plants flowered earlier than in the exon-swapping experiment (22.9±1.1 versus 28.6±0.8 rosette leaves), due to changed growth conditions.
Many transgenic constructs affected flowering time, which suggested that cosuppression or instability of chimeric proteins was not a major problem. In addition, we used RNA blot analysis to ascertain that chimeric genes were transcribed. Using a mixture of FT and TFL1 probes, the endogenous genes were below the limit of detection in RNA blots with material from untransformed ft tfl1 plants. In contrast, although variable in level, most transgenic constructs produced RNA blot signals (Figure 1B). Similarly, an anti-FT antiserum recognized a protein in several lines, but without a clear correlation between RNA blot and protein blot signals (Figure 1C). The lower efficiency of detecting chimeric proteins compared to chimeric messages is likely due to the fact that we used a mixed FT/TFL1 probe for the RNA blots, but had only an anti-FT antibody available for the protein blots. Importantly, many of the lines in which we could not detect a chimeric RNA or protein produced nevertheless a phenotype, indicating that the chimeric genes were active.
The fourth exon has a critical role
The in vivo activity of chimeric genes was analyzed by counting the number of rosette leaves in the T1 generation. Stem (cauline) leaves were excluded, because TFL1 overexpression can convert flowers into shoot-like structures and thus result in plants that produce only leaves (Ratcliffe et al, 1998). The phenotypes observed in the T1 generation were heritable, as shown by carrying several lines to the T2 generation (Supplementary Table S1).
As expected, overexpression of FT (pJA1054) in an ft tfl1 background caused early flowering, while overexpression of TFL1 (pJA1062) delayed flowering (Figure 2, left), similar to what had been reported for overexpression in wild-type and single-mutant backgrounds (Ratcliffe et al, 1998; Kardailsky et al, 1999; Kobayashi et al, 1999). Therefore, early and late flowering of transgenic plants was subsequently taken as an indication of FT- or TFL1-like activity, respectively. None of the chimeric genes was as effective in accelerating or delaying flowering as authentic FT or TFL1 (Figure 2, left).
To facilitate discussion of the chimeras, we will use an annotation that indicates the origin of each of the four exons; for example, FTFF indicates that exons one, three and four are from FT, and exon two from TFL1. Six chimeras caused predominantly late flowering, five early flowering, and three had no clear effect. The first exon accounts for 40% of the protein, but it did not affect FT- versus TFL1-like activity. For instance, FTTT (pJA1061) plants were almost as late as those overexpressing wild-type TFL1 (pJA1062). Although the effect of the opposite construct, TFFF (pJA1063), was not as strong as that of regular FT, it was still slightly early, indicating at least partial FT activity. Similar observations were made for swaps of the second and third exons (Figure 2).
In contrast, a striking change in activity was observed when the fourth exon of FT, which is similarly long as the first, was substituted for the fourth TFL1 exon. FFFT (pJA1055) plants flowered much later than the controls, although not quite as late as TFL1 overexpressers. Although the opposite construct, TTTF (pJA1069), had no clear effect on flowering time, most combinations of the fourth exon with other exons of FT did cause early flowering (Figure 2). The TTTF lines did not produce protein that reacted with the anti-FT antibody, which may reflect either that this antibody recognizes mostly epitopes encoded by other exons, or that this particular chimeric protein is unstable.
FT- and TFL1-specific activity map to a divergent segment of the fourth exon
Owing to the strong effect of the fourth exon, we divided it into four segments (Figure 1A), and generated 13 additional chimeras in the backbone of the FFFT (pJA1055) construct (Figure 2, right). Segments A and C, which together comprise 26 residues, are very similar between FT and TFL1, while segments B and D are rather different. We are using a similar annotation as for the exon swaps; for example, 4FFFT denotes a chimera in which segments A through C are from FT, and segment D from TFL1.
Most of the fourth-exon chimeras were late flowering, the exceptions being 4TFFF (pJA1126) and 4TFFT (pJA1127), in which segments B and C were from FT. Both constructs were almost as effective in accelerating flowering as regular 35S:FT. Segments B and C alone in 4TFTT (pJA1129) and 4TTFT (pJA1131) by themselves were not sufficient to impart FT-like activity. Similarly, combinations of segments B or C with segments A or D did not cause early flowering. Instead, these chimeras caused late flowering, ruling out a trivial explanation for segments B and C from FT alone having no effects, namely that these chimeras were simply nonfunctional.
While substituting segments B and C from FT alone was not sufficient to impart FT-like activity on TFL1, either segment B or C from TFL1, when placed in an FT background, could delay flowering (constructs 4FTFF (pJA1122) and 4FFTF (pJA1120)). The 4FTFF chimeric protein, which showed the strongest effect and caused plants to flower as late as authentic TFL1, differs from FT in only 12 amino-acid residues.
In conclusion, segments B and C together are required for FT-like activity, but either segment B or C is sufficient for TFL1-like activity.
Conserved function of segment B in Antirrhinum CEN
Overexpression of the TFL1 ortholog CEN from snapdragon delays flowering in tobacco, but Arabidopsis TFL1 is ineffective in tobacco (Amaya et al, 1999). Tobacco is in the Asteridae, and thus more closely related to snapdragon than Arabidopsis, which is in the Rosidae. Segment B sequences are not only very different between FT and TFL1, but also evolve very rapidly between TFL1 orthologs: CEN and TFL1 share only five identical residues in this 17-amino-acid segment.
We replaced segment B of the fourth exon of TFL1 with the corresponding sequence of CEN (pJA1147; Figure 3), and overexpressed the chimera in tobacco. In the R0 generation, tobacco plants transformed with the wild-type TFL1 construct flowered 11 nodes earlier than the TFL1/CEN chimeric plants. Delayed flowering conferred by segment B of CEN was confirmed in the R1 generation (Figure 3).
Figure 3.
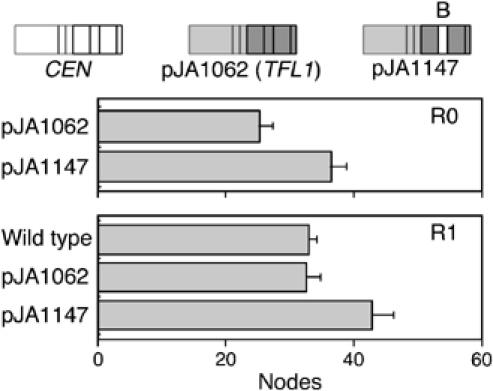
Flowering times of tobacco plants with CEN/TFL1 segment B swap. Primary transformants (R0) and progeny (R1) are shown. Only plants that overexpress a chimeric gene with segment B of the fourth exon of CEN in the TFL1 background are late flowering.
Crystal structures of FT and TFL1
To complement the in vivo studies, we solved the crystal structures of FT and TFL1 (Figure 4A). Both structures show a fold very similar to those previously reported for snapdragon CEN (Banfield and Brady, 2000) and a range of mammalian and bacterial homologs (Banfield et al, 1998; Serre et al, 1998, 2001; Simister et al, 2002). The final models include residues 6–167 of the FT sequence, and 7–171 of TFL1. There are two molecules in the asymmetric unit of TFL1 crystals, and four molecules in that of FT crystals. Nonetheless, there are only small variations between each form; root mean square (r.m.s.) deviations for the 162 equivalent Cα positions for the two TFL1 molecules are 0.91 Å overall, and for the four FT molecules 0.51 Å. These differences are smaller than those that distinguish FT and TFL1 (1.22 Å r.m.s. on 156 equivalent Cα positions), or FT-TFL1 and CEN (2.2–2.5 Å) (Banfield and Brady, 2000), or FT-TFL1 and hPEBP (3.6–4.1 Å) (Banfield et al, 1998).
Figure 4.
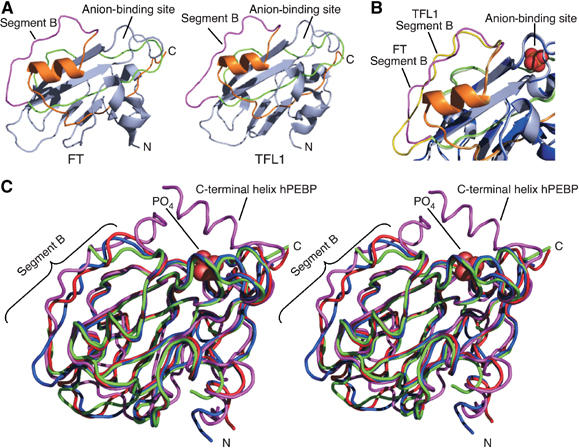
Crystal structures of FT and TFL1. (A) Cartoon diagrams of FT and TFL1. For residues encoded by the fourth exon, segment A is colored green, segment B magenta, and segment C orange. Segment D forms the C-terminus of each protein, but is disordered in both crystal structures. Amino (N) and carboxy (C) termini are labeled. (B) Close-up showing an overlay of segment B and surrounding regions of FT and TFL1. A phosphate ion, from the hPEBP structure, is modeled as orange and red spheres in the anion-binding site. (C) Stereo view of an overlay of FT (red), TFL1 (blue), CEN (green), and hPEBP (magenta) structures as ribbon traces. A phosphate ion bound within the anion ligand-binding site of hPEBP is shown as red spheres. The C-terminal helix in hPEBP, not present in the FT and TFL1 structures, is indicated.
In both TFL1 and FT, there is a large central β-sheet, which is flanked on one side by a smaller β-sheet and on the other by an α-helix. The pocket comprising the anion-binding site found in other members of the family is present in both FT and TFL1, although the electron density suggests that only water molecules are bound in this site by FT and TFL1, despite the inclusion of cacodylate as a ligand analog in the crystallization buffer for TFL1.
In the mammalian PEBPs, Asp70, His86, Tyr120, and the main-chain amino group of Gly110 (numbering as per hPEBP) all contribute to complexes formed with anion ligands (Figure 5A and B) (Banfield et al, 1998; Serre et al, 1998; Simister et al, 2002). The conformation of this pocket is very similar in FT and TFL1. The TFL1-binding site is identical to the equivalent site in CEN, the main difference with hPEBP being a phenylalanine rather than tyrosine at position 120 (Figure 5B). There are several other potential hydrogen bond donors that may compensate for the loss of the hydroxyl group in the side chain at position 120. Notably, the FT-binding pocket instead has a valine side chain at position 120, creating a larger volume in the potential binding cavity (Figure 5B).
Figure 5.
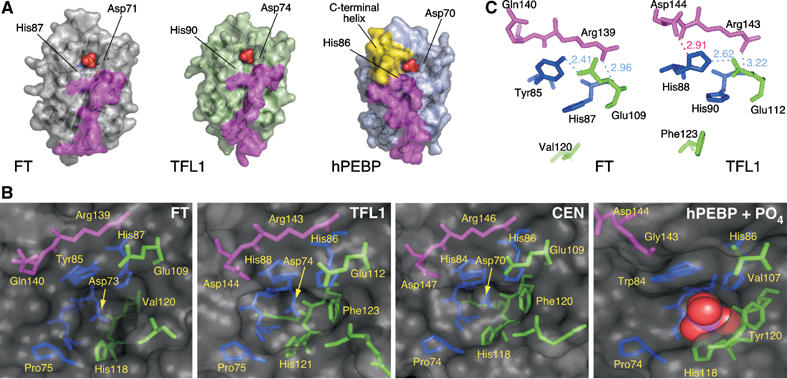
Details of the ligand-binding sites and external loop. (A) Surface representation of FT, TFL1 and hPEBP in similar orientation. Phosphate ligands (red and orange spheres, from hPEBP structure) are shown bound within the anion-binding site of each structure. The surfaces formed by segment B are colored magenta. The surface formed by the C-terminal helix in hPEBP (yellow) makes the ligand-binding site less accessible than in FT and TFL1. The two key conserved residues that bind the ligand (histidine and aspartic acid) are labeled. (B) Details of the amino acids lining the ligand-binding site. The pocket of hPEBP includes a bound phosphate ion (red and magenta spheres). Amino acids are colored by segment: residues in segment A (or equivalent) are colored green, residues in segment B magenta, and residues that precede segment A blue. (C) Interactions of segment B residues with amino acids at the entrance to the binding pocket. Hydrogen bonds are shown as light dashed lines (distances marked in Å), with amino acids colored by segment as in (B). Residues His87 and Val120 in FT and His90 and Phe123 in TFL1 are shown for orientation (see panel B). Only TFL1 shows an interaction between Asp144 and His88 (red line), while the corresponding residues in FT, Gln140 and Tyr85 do not interact.
A major difference between hPEBP and the three plant proteins in this region is that the periphery of the cavity in hPEBP is extended by a C-terminal helix (residues 175–184), together with an adjacent loop (residues 139–144) (Figure 5A). The helix may potentially regulate ligand access (Banfield et al, 1998). In the plant proteins, both features are absent, leading to a more accessible binding pocket (Figure 5A). Although no anion ligands are found in FT or TFL1 crystals, the arrangement of amino acids is otherwise consistent with the ability of FT and TFL1 to bind anions in this site. Importantly, the conformation of the potential ligand-binding pocket does not readily explain differences in FT and TFL1 activity.
His88 in TFL1 and the corresponding Tyr85 in FT have been found to be important for the opposite activities of FT and TFL1 (Hanzawa et al, 2005). These residues lie at the entrance to the ligand-binding pocket, with the aromatic rings of their side chains in the same orientation (Figure 5C). It seems likely that His88 and Tyr85 would contact any partner protein that associates with TFL1/FT via the potential ligand-binding pocket.
Structural differences of segment B encoded by the fourth exons of FT and TFL1
The most substantial differences between FT and TFL1 are in the external loop (residues 128–145), which corresponds to segment B identified as critical for FT versus TFL1 activity in vivo. Equivalent Cα atom positions are displaced by up to 3 Å. The temperature factors of this segment are much higher (around 25 Å2 for Cα atoms) than for the remainder of the molecules (around 16 Å2). Although this implies some flexibility, possibly to accommodate a binding partner, the external loop is still well ordered. This same region was not ordered in crystals of CEN protein (Banfield and Brady, 2000). The external loop is in close proximity to, and ultimately leads directly to, the residues that form part of the potential ligand-binding site (Figures 4A and 5B and C).
To identify specific residues that might be critical for activity differences, we aligned segments B and C of FT and TFL1 homologs of different plant species. Of 23 TFL1 and 13 FT homologs analyzed, at least eight and three, respectively, behave like their Arabidopsis counterparts in vivo (Supplementary Table S2). FT homologs stand out, as 11 of the 14 residues of segment B are invariant (Figure 6A). In contrast, segment B of TFL1 homologs is variable in length, with 14–17 residues, and only four residues are invariant. Of these, three are shared with FT, leaving only a single residue that unambiguously distinguishes FT and TFL1 homologs: Gln140 in FT versus Asp144 in TFL1. In each case, the side chain of this residue is located at the entrance to the ligand-binding pocket, and potentially available to interact with a protein ligand (Figure 5B and C). These amino acids are also immediately adjacent to the functionally critical residues Tyr85/His88 identified by Hanzawa et al (2005). In the TFL1 structure, the Asp144 side chain forms a charged hydrogen bond (2.9 Å) with the ND1 nitrogen of the His88 imidazolium ring, an interaction that is replicated in the CEN structure (Banfield and Brady, 2000). In contrast, in FT no equivalent contacts are observed between the phenol ring of Tyr85 and the side chain of Gln140. As with Gln140 and Asp144, the interacting residues Tyr85 and His88 in FT and TFL1, respectively, are absolutely conserved (see sequences in Supplementary Table S2) (Hanzawa et al, 2005).
Figure 6.
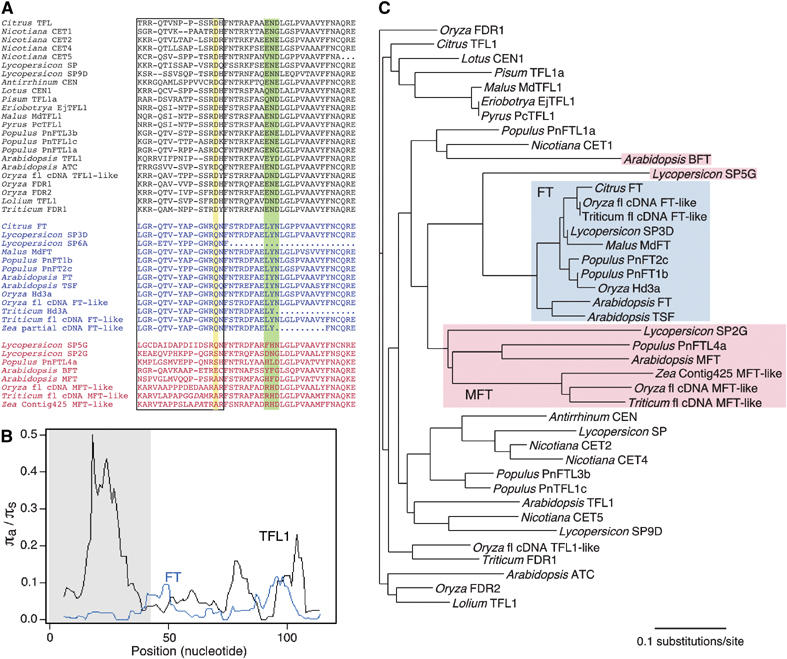
Sequence comparison of FT/TFL1 family members from flowering plants. (A) Alignments of segments B and C encoded by the fourth exons of TFL1-like genes (top, black), FT-like genes (middle, blue) and other members of the family (bottom, red). Dashes denote gaps, dots missing data. Additional indels in the two lower most sequences are indicated by italics. Segment B is boxed; the Asp144/Gln140 residue distinguishing all FT and TFL1 members is highlighted in yellow. The triad in segment C, which is also more conserved in FT than TFL1 proteins, is highlighted in green. (B) Average pairwise ratio of nonsynonymous (πa) to synonymous substitutions (πs) (sliding window, window size 12, step 1) (see also Supplementary Table S3). TFL1 genes show an excess of nonsynonymous substitutions in segment B. (C) Phylogenetic neighbor-joining tree. FT orthologs form a well-resolved clade, with the somewhat more divergent MFT-like proteins as sister clade. TFL1 sequences do not appear monophyletic because of their rapid divergence.
The interacting pairs, His88–Asp144 and Tyr85–Gln140, are not conserved in other members of the plant PEBP family, with the exception of SP5G from tomato, which is closest in sequence to the FT group (Figure 6). Segment B in non-FT homologs is variable in length, with only two identical residues shared with TFL1 and FT (Figure 6A). Among them is Arg139/143, immediately preceding Gln140/Asp144, which forms charged hydrogen bonds with the main chain carbonyl oxygen of Ser107/Ser110 (2.7/2.6 Å) and with His87/His90 (3.0/3.2 Å) (Figure 5B). Residues in this region form one wall of the ligand-binding pocket, and their strong conservation is therefore not surprising.
To evaluate the sequence diversity in FT and TFL1 homologs more closely, we performed a sliding window analysis of rates of pairwise nonsynonymous (πa) and synonymous (πs) substitutions. While segment B of FT proteins has almost no amino-acid substitutions, for a πa/πs ratio close to zero (0.03), indicative of strong purifying selection, segment B of TFL1 has a many-fold higher πa/πs ratio (0.38), suggesting more relaxed selection (Figure 6B and Supplementary Table S3; note that absolute πs is very similar for both). The sequence divergence of segment B is also reflected in a neighbor-joining tree of segments B and C from FT/TFL1-like proteins: FT proteins form a well-resolved single clade with short branch lengths and Lycopersicon SP5G as an outgroup. In contrast, TFL1 proteins do not appear as monophyletic because of their excessive divergence with much longer branches indicative of accelerated evolution (Figure 6C).
The experiments with chimeric proteins had shown that either segment B or the adjacent segment C, which forms a surface helix before entering the core of the protein to complete the central β-sheet, alone is sufficient to confer at least some TFL1-like activity, but that segment B together with the adjacent segment C is required for FT-like activity. While segment C is more similar between TFL1 and FT homologs than segment B, there is a triad that is essentially invariable in FT, but variable in TFL1 (Figure 6A). This triad starts in FT with residue 150, which is leucine or isoleucine, followed by tyrosine and asparagine. This triad is immediately adjacent to and makes contact with the segment B external loop, in the region where there is the greatest diversity between TFL1 and FT structures. The equivalent positions are occupied by a range of residues in TFL1 proteins, but residue 150 is never a leucine or isoleucine. The same triad distinguishes FT from other members of the family (Figure 6A). In mammalian RKIP, these residues are adjacent to a regulatory site, Ser153, phosphorylation of which leads to disruption of the Raf-1/RKIP complex (Corbit et al, 2003). However, the same mechanism of regulation is not expected for the plant proteins, where the equivalent residues are Glu146 in FT and Lys150 in TFL1.
Discussion
A central role in making the decision when to flower is played by the floral integrator FT and its antagonist TFL1 (Ratcliffe et al, 1998; Kardailsky et al, 1999; Kobayashi et al, 1999). We have combined in vivo analyses with structural studies to define an external loop along with an adjacent peptide, both of which play an important role in determining the opposite functions of FT and TFL1. Our work complements the identification of a residue that is similarly critical for FT versus TFL1 activity (Hanzawa et al, 2005). We have shown here that this residue, which is located near the potential ligand-binding site, interacts with the external loop.
Interactions between different exons
The majority of FT/TFL1 chimeric proteins we assayed had at least some activity when overexpressed in plants. Although this suggests that there is relatively little coevolution between different parts of the protein, none of the exon swap chimeras showed as strong effects as the native proteins (Figure 2, left), indicating that there is at least some interaction between residues from different exons. In support of this conclusion, substitution of His88 in the second TFL1 exon, which interacts with Asp144 in exon four, with the equivalent residue from FT, Tyr85, confers flower-promoting activity on TFL1 when expressed in a ft tfl1 background (Hanzawa et al, 2005). However, the activity appears to be much weaker than that of wild-type FT, since TFL1 (His88Tyr) had no effect on flowering in a wild-type background, and occasionally even led to conversion of flowers into inflorescence shoots, a phenotype typical of TFL1 overexpressers (Hanzawa et al, 2005). The converse mutation in FT (Tyr85His) also showed a paradoxical behavior, strongly delaying flowering of wild type, but partially promoting flowering in an ft tfl1 background. Visual inspection of the crystal structures suggests that Gln140 of FT could make a similar hydrogen bond with His85, possibly explaining why the FT (Tyr85His) mutant has at least some TFL1-like activity. In our set of chimeras, substituting the entire second exon of TFL1 with that of FT attenuated TFL1 function, but the corresponding plants were still later than the ft tfl1 parents (Figure 2). In contrast, substituting the entire second exon of FT with that of TFL1 had a similar effect as the Tyr85His single amino-acid exchange in FT reported by Hanzawa et al (2005), in that it attenuated FT activity (Figure 2). In addition, while a swap of FT exon four alone into TFL1 was ineffective, FT exons two and four together conferred strong flower-promoting activity on a TFL1 backbone, further supporting a synergistic effect of residues encoded by these two proteins.
Despite these complex effects, it appears that both the ligand-binding pocket, notably His88 in TFL1 and the corresponding Tyr85 in FT, and the external loop contribute to functional specificity of the proteins. In the crystal structures, His88/Tyr85 are in very similar locations. An overlay of the structures indicates that the aromatic rings of the histidine and tyrosine side chains lie in the same orientation, implying that the effect of swapping these residues may reflect altered ability to form a complex with an associated protein.
Structure of the peptide encoded by the fourth exon
We have shown that residues encoded by the fourth exons of FT and TFL1 play a critical role in conferring biological specificity on the two proteins. Within the fourth exon, segment B, which is highly variable in TFL1 orthologs but almost invariant in FT orthologs, has particularly strong effects, together with the adjacent segment C. The crystal structures of FT and TFL1 show that segment B forms an external loop, which would be easily accessible to interactors. Although a range of proteins that interact with FT, TFL1 and its orthologs have been identified (Pnueli et al, 2001; Abe et al, 2005; Wigge et al, 2005), the identity of the interactors that distinguish between FT and TFL1 is unknown. Nonetheless, the structural analysis of FT and TFL1 provides some insight into the nature of this interaction. Firstly, both FT and TFL1 are fully folded species, making it very unlikely that one protein simply passively interferes with the action of the other protein. Secondly, the overall structures of both FT and TFL1 are highly similar. This similarity is maintained in the potential anion-binding site that has previously been proposed as a key determinant in binding to phosphorylated protein interactors (Banfield and Brady, 2000), indicating that differential interaction with a ligand is unlikely to be the sole reason for opposite activities of FT and TFL1.
Another possibility is that the adjacent external loop contributes to the periphery of a protein–protein interface centered on the anion-binding pocket. In this case, exchange of segment B might sufficiently destabilize this interaction. This hypothesis is supported by the observation that the His88/Asp144 pair at the entrance to the binding pocket in TFL1 plays a crucial role in determining protein specificity (this work; Hanzawa et al, 2005). An alternative possibility is that the anion-binding site and segment B represent separate interfaces for two interactors. Conservation of the potential ligand-binding pocket might, therefore, leave both FT and TFL1 competent to fulfill the functions of one part of a signaling cascade, with segment B acting as a switch between two downstream possibilities.
In support of the loop providing a molecular surface that may act independently of the anion ligand-binding site, sequence and structural differences in this loop are most prevalent in the region distant from the binding pocket (132–139; TFL1 numbering), whereas the external loop conformation adjacent to the binding pocket is much more similar between the three plant proteins FT, TFL1 and CEN. The variable part of this loop is also adjacent to the 60–66 loop, another region of variability between FT and TFL1 (Figure 4). The surface formed by the confluence of these loops is at the opposite end of the FT/TFL1 molecules from the ligand-binding pocket, and close to the reported regulation site of Ser153 for RKIP (Corbit et al, 2003). The varied conformations observed in FT, TFL1 and CEN are consistent with the external loop contributing to a protein–protein interface, possibly leading to stabilization of its structure.
Effects of chimeric proteins and implications for FT and TFL1 action
Although FT and TFL1 have broadly antagonistic functions, the phenotypes resulting from loss of function and overexpression are not exact mirror images of each other, even when redundancy with recent duplicates such as TSF is accounted for (Koornneef et al, 1994; Ratcliffe et al, 1998, 1999; Kardailsky et al, 1999; Kobayashi et al, 1999; Michaels et al, 2005; Yamaguchi et al, 2005). Independent activities of FT and TFL1 can also be deduced from the fact that both 35S:FT tfl1 and ft 35S:TFL1 plants show additive phenotypes, as do ft tfl1 double mutants (Kardailsky et al, 1999; Kobayashi et al, 1999). On the other hand, overexpression of FT strongly suppresses 35S:TFL1 phenotypes. These observations are consistent with a model in which FT and TFL1 compete for common interacting partner(s), which have some intermediate level of activity in the absence of FT or TFL1. Such a model is supported by the recent identification of the FD bZIP transcription factor, which can interact with both FT and TFL1 (Abe et al, 2005; Wigge et al, 2005). FD in turn recruits FT to the promoter of the floral identity gene AP1, which is activated by the FD/FT complex (Wigge et al, 2005). FD, however, is not completely dependent on FT activity, as fd ft double mutants are more extreme than either single mutant (P Wigge, personal communication).
In one scenario, FD is a weak activator that is converted into a strong activator by FT, but into a repressor by TFL1 (Figure 7). Segments B and C in the fourth exon together might be required for FT to recruit a coactivator. TFL1, through segment B, might instead recruit a corepressor and therefore not only interfere with FD activity, but also cause an FD/TFL1 complex to have dominant-negative activity compared to FD alone. A positive role of segment B in TFL1 and orthologs is supported by the finding that segment B from the ortholog CEN can endow TFL1 with the ability to delay flowering in tobacco. While segment C from TFL1 on its own might not be sufficient to recruit the corepressor, it might nevertheless be sufficient to prevent interaction of the chimeric protein with a coactivator normally recruited by FT (Figure 7, bottom). Binding of this chimeric protein to FD would then render FD inactive, explaining the delay in flowering conferred by TFL1 segment C alone. A formally equivalent model is one in which FT and TFL1 actively switch the activity of a common interactor from transcriptional repression to activation. A chimeric protein that cannot bind such a hypothetical interactor might still reduce FD activity and delay flowering in an ft tfl1 background. Genetic tests of this model await the identification of FT- and TFL1-specific interactors.
Figure 7.
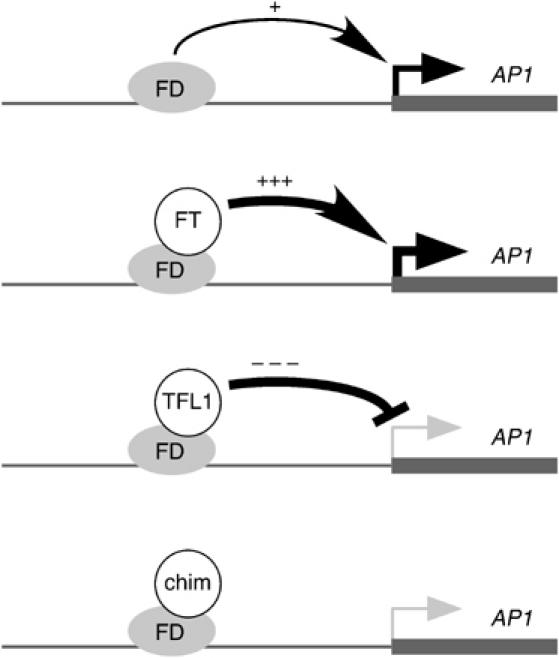
Model for interaction of FD with FT, TFL1 and certain chimeras. FD, which binds to the AP1 promoter (Wigge et al, 2005), is proposed to be a weak activator on its own. FT converts FD into a strong activator, while TFL1 converts FD into a strong repressor, explaining why TFL1 overexpressers are not merely late-flowering, like ft mutants, but also show floral defects (Ratcliffe et al, 1998). Certain chimeras (chim), such 4FFTF, might not have true TFL1 function, but still cause moderate late flowering, because they interfere with the weak activator function of FD by itself.
Materials and methods
Plant material and analysis
The ft tfl1 double mutant was derived from tfl1-1 (Columbia) and ft-1 (Columbia), which had been obtained by backcrossing ft-1 (Ler) seven times to wild-type Columbia.
Transgenic plants were generated using the floral dip method of Agrobacterium transformation (Weigel and Glazebrook, 2002). After selection for 8 days on kanamycin-supplemented MS medium, plants were transferred to soil at 23°C under long day conditions (16 h light/8 h dark). The ft tfl1 controls were grown on MS solid medium, without antibiotics, for 8 days as well.
Nicotiana tabacum cv. Xanthi was used for tobacco transformation. Transgenes were introduced via Agrobacterium-mediated transformation of leaf disks (Horsch et al, 1985). Regenerated kanamycin-resistant shoots were transferred to rooting media to induce root formation. Regenerated plantlets were transplanted to pots containing vermiculite and grown in a greenhouse under sunlight.
Chimeric genes
Primers were designed to contain both FT and TFL1 sequences, such that the first half (approximately 20 nucleotides) of each oligonucleotide contained the end sequence of an exon of FT or TFL1, whereas the second half contained the starting sequence of an adjacent exon of TFL1 or FT, respectively. After amplification of the appropriate fragments of FT and TFL1 cDNAs in a first round of PCR, gel-purified fragments were mixed and used as template to obtain the entire chimeric gene. Sequence-verified chimeras were placed behind the cauliflower mosaic virus 35S promoter in the pCHF3 vector (Jarvis et al, 1998). Oligonucleotide sequences and further details are provided in Supplementary Methods and Supplementary Table S4.
RNA and protein blot analysis
RNA and protein blot analyses were performed according to standard protocols (Ausubel et al, 1991), with minor modifications. Details are provided in Supplementary data.
Expression and crystallization of FT and TFL1
TFL1 and FT coding sequences were cloned into pET28a (Novagen) for expression in Escherichia coli BL21 (DE3) with a thrombin-cleavable N-terminal 6-histidine tag. The vector introduced three additional residues at the N-terminus (Gly-Ser-His). For FT, three cysteine residues in the native sequence were altered to promote crystallization. Cys107 and Cys164 were mutated to serines, and Cys170 was removed by deletion of the seven C-terminal residues. Details of protein expression and purification are given in Supplementary data.
Diffraction quality crystals of TFL1 (15 mg/ml) were grown by the hanging drop method, with 0.3 M (NH4)2SO4 and 22% w/v PEG 5K MME, buffered with 0.1 M cacodylate (pH 5.0), as the precipitant. Crystals were cryopreserved by the addition of 15% glycerol to this buffer. Crystals of FT (10 mg/ml) were grown from 0.14 M (NH4)2SO4 and 38% PEG 5K MME, buffered with 0.1 M MES (pH 7.1).
Structure determination
Diffraction data were collected at 100 K using the ADSC Quantum 4R CCD detector at stations PX14.1 and PX14.2, respectively, of the Daresbury Synchrotron Radiation Source. Data were processed using the HKL suite of programs (Otwinowski and Minor, 1997). Key data for both crystals are summarized in Table I.
Table 1.
Data processing and refinement statistics for TFL1 and FT
| TFL1 | FT | |
|---|---|---|
| Unit-cell parameters (Å) | a=109.3, b=109.3, c=64.7, α=β=90°, γ=120° | a=54.0, b=61.5, c=66.8, α=75.2°, β=72.2°, γ=70.1° |
| Space group | P321 | P1 |
| Wavelength (Å) | 1.488 | 1.488 |
| Resolution range (Å) | 50–1.8 | 20–2.6 |
| No. of unique reflections | 41 659 (5116) | 21 870 (1944) |
| Redundancy | 6.8 | 2.24 |
| Completeness (%) | 100 (99.7) | 93.2 (66.2) |
| Rmerge (%) | 10.9 (21.0) | 7.6 (17.0) |
| I/(I) | 20.11 (5.51) | 12.05 (3.79) |
| Refinement | ||
| Resolution range (Å) | 42–1.8 (1.85–1.8) | 20–2.6 (2.66–2.6) |
| Rcryst (%) | 18.4 (19.7) | 18.1 (22.4) |
| Rfree (%) | 20.7 (25.8) | 22.6 (29.1) |
| R.m.s.d., bond lengths (Å) | 0.016 | 0.012 |
| R.m.s.d., bond angles (deg) | 1.539 | 1.400 |
| No. of non-H protein atoms | 2682 | 5217 |
| No. of solvent molecules | 402 | 195 |
| Average B value (Å2) | ||
| Main chain | 16.8 | 21.2 |
| Side chain | 18.2 | 21.9 |
| Solvent molecules | 39.4 | 37.4 |
| Values shown in parenthesis are for the highest resolution shell. | ||
The crystal structures of FT and TFL1 were solved by molecular replacement, using the coordinates of snapdragon CEN as a search model (Banfield and Brady, 2000) (PDB code 1qou) for TFL1, and subsequently one molecule of TFL1 for FT. Rotation and translation searches using AMoRE (Navaza, 1994) identified the expected two clear solutions for TFL1, and the four monomers in the asymmetric unit of FT. The models were manually mutated to the FT and TFL1 sequences using O (Jones et al, 1991) and refined with REFMAC5. Noncrystallographic symmetry (NCS) restraints were maintained throughout the refinement on NCS-related monomers, with the exception of the variable regions, and removed in the final refinement cycles. The quality of the resulting models (summarized in Table I) was monitored with PROCHECK (Laskowski et al, 1993). A Ramachandran plot analysis using PROCHECK (Laskowski et al, 1993) showed that 89% of the nonglycine residues in the FT structure fall within the most favored regions, with a further 11% in the additionally allowed regions. No residues are in the generously allowed or disallowed regions. Similarly, the TFL1 model comprises 87.2% of nonglycine residues in the most favored regions and 12.8% in additionally allowed regions. Coordinates of the final models and structure factors have been deposited in the Protein Data Bank (Accession codes: 1WKP (FT), 1WKO (TFL1)). Software programs used for structure solution and refinement were those implemented in the CCP4 suite (Collaborative, 1994).
Phylogenetic analyses
Sequences of FT/TFL1 family members were identified by BLAST searches of GenBank (Viridiaeplantae) and manually aligned. Sequences (see Supplementary Table S2) were manually aligned. πa/πs ratios were calculated with DnaSP v4.10 (http://www.ub.es/dnasp) (Rozas and Rozas, 1999). Phylogenetic trees of complete sequences were generated using ClustalW (http://www.ebi.ac.uk/clustalw/index.html) (Chenna et al, 2003).
Supplementary Material
Supplementary Methods
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Acknowledgments
We are grateful to NY Kwon, C Vu, D Hyunh, T Dabi, H Trinh, A Alex and A Medina for growing plants; to R Conners and the staff at the Daresbury SRS synchrotron for access to and assistance with the data collection facilities; to P Wigge for communicating unpublished results; and to the diligent anonymous reviewers for comments. SYY and JHL were supported by the BK21 program in CLB, Korea University; DM by a studentship award from the ORS scheme; and VJW by a studentship from the BBSRC. This work was supported by grants from the Plant Signaling Network Research Center and CFGC (Crop Functional Genomics Center) to JHA; and by a grant from the Human Frontier Science Program Organization to RLB and DW, who is a director of the Max Planck Institute.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Alvarez J, Guli CL, Yu X-H, Smyth DR (1992) Terminal flower: a gene affecting inflorescence development in Arabidopsis thaliana. Plant J 2: 103–116 [Google Scholar]
- Amaya I, Ratcliffe OJ, Bradley DJ (1999) Expression of CENTRORADIALIS (CEN) and CEN-like genes in tobacco reveals a conserved mechanism controlling phase change in diverse species. Plant Cell 11: 1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RZ, Moore DD, Seideman JG, Smith JA, Struhl K (1991) Current Protocols in Molecular Biology. New York, NY: Wiley [Google Scholar]
- Banfield MJ, Barker JJ, Perry AC, Brady RL (1998) Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure 6: 1245–1254 [DOI] [PubMed] [Google Scholar]
- Banfield MJ, Brady RL (2000) The structure of Antirrhinum CENTRORADIALIS protein (CEN) suggests a role as a kinase regulator. J Mol Biol 297: 1159–1170 [DOI] [PubMed] [Google Scholar]
- Bradley DJ, Ratcliffe OJ, Vincent C, Carpenter R, Coen ES (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Bruun AW, Svendsen I, Sorensen SO, Kielland-Brandt MC, Winther JR (1998) A high-affinity inhibitor of yeast carboxypeptidase Y is encoded by TFS1 and shows homology to a family of lipid binding proteins. Biochemistry 37: 3351–3357 [DOI] [PubMed] [Google Scholar]
- Chautard H, Jacquet M, Schoentgen F, Bureaud N, Benedetti H (2004) Tfs1p, a member of the PEBP family, inhibits the Ira2p but not the Ira1p Ras GTPase-activating protein in Saccharomyces cerevisiae. Eukaryot Cell 3: 459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Corbit KC, Trakul N, Eves EM, Diaz B, Marshall M, Rosner MR (2003) Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J Biol Chem 278: 13061–13068 [DOI] [PubMed] [Google Scholar]
- Hanzawa Y, Money T, Bradley D (2005) A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA 102: 7748–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst U, Albrecht H, Hess D, Monard D (2001) The phosphatidylethanolamine-binding protein is the prototype of a novel family of serine protease inhibitors. J Biol Chem 276: 535–540 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Jarvis P, Chen LJ, Li H, Peto CA, Fankhauser C, Chory J (1998) An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282: 100–103 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Part 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla V, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Blankestijn-de Vries H, Hanhart C, Soppe W, Peeters T (1994) The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta phenotype. Plant J 6: 911–919 [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol 137: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr A 50: 157–163 [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology, Carter CW, Sweet RM (eds). Vol. 276, pp 307–326. New York: Academic Press [DOI] [PubMed] [Google Scholar]
- Parcy F, Bomblies K, Weigel D (2002) Interaction of LEAFY, AGAMOUS and TERMINAL FLOWER1 in maintaining floral meristem identity in Arabidopsis. Development 129: 2519–2527 [DOI] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D (1998) A genetic framework for floral patterning. Nature 395: 561–566 [DOI] [PubMed] [Google Scholar]
- Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E (1998) The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125: 1979–1989 [DOI] [PubMed] [Google Scholar]
- Pnueli L, Gutfinger T, Hareven D, Ben-Naim O, Ron N, Adir N, Lifschitz E (2001) Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 13: 2687–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O, Amaya I, Vincent C, Rothstein S, Carpenter R, Coen E, Bradley D (1998) A common mechanism controls the life cycle and architecture of plants. Development 125: 1609–1615 [DOI] [PubMed] [Google Scholar]
- Ratcliffe OJ, Bradley DJ, Coen ES (1999) Separation of shoot and floral identity in Arabidopsis. Development 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175 [DOI] [PubMed] [Google Scholar]
- Ruiz-García L, Madueño F, Wilkinson M, Haughn G, Salinas J, Martínez-Zapater JM (1997) Different roles of flowering time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9: 1921–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schoentgen F, Saccoccio F, Jolles J, Bernier I, Jolles P (1987) Complete amino acid sequence of a basic 21-kDa protein from bovine brain cytosol. Eur J Biochem 166: 333–338 [DOI] [PubMed] [Google Scholar]
- Searle I, Coupland G (2004) Induction of flowering by seasonal changes in photoperiod. EMBO J 23: 1217–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre L, Pereira de Jesus K, Zelwer C, Bureaud N, Schoentgen F, Benedetti H (2001) Crystal structures of YBHB and YBCL from Escherichia coli, two bacterial homologues to a Raf kinase inhibitor protein. J Mol Biol 310: 617–634 [DOI] [PubMed] [Google Scholar]
- Serre L, Vallee B, Bureaud N, Schoentgen F, Zelwer C (1998) Crystal structure of the phosphatidylethanolamine-binding protein from bovine brain: a novel structural class of phospholipid-binding proteins. Structure 6: 1255–1265 [DOI] [PubMed] [Google Scholar]
- Shannon S, Meeks-Wagner DR (1991) A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3: 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simister PC, Banfield MJ, Brady RL (2002) The crystal structure of PEBP-2, a homologue of the PEBP/RKIP family. Acta Crystallogr D 58: 1077–1080 [DOI] [PubMed] [Google Scholar]
- Simon R, Coupland G (1996) Arabidopsis genes that regulate flowering time in response to day-length. Sem Cell Dev Biol 7: 419–425 [Google Scholar]
- Tohdoh N, Tojo S, Agui H, Ojika K (1995) Sequence homology of rat and human HCNP precursor proteins, bovine phosphatidylethanolamine-binding protein and rat 23-kDa protein associated with the opioid-binding protein. Brain Res Mol Brain Res 30: 381–384 [DOI] [PubMed] [Google Scholar]
- Trakul N, Rosner MR (2005) Modulation of the MAP kinase signaling cascade by Raf kinase inhibitory protein. Cell Res 15: 19–23 [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T (2005) TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol 46: 1175–1189 [DOI] [PubMed] [Google Scholar]
- Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, Sedivy JM, Kolch W (1999) Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 401: 173–177 [DOI] [PubMed] [Google Scholar]
- Yeung KC, Rose DW, Dhillon AS, Yaros D, Gustafsson M, Chatterjee D, McFerran B, Wyche J, Kolch W, Sedivy JM (2001) Raf kinase inhibitor protein interacts with NF-κB-inducing kinase and TAK1 and inhibits NF-κB activation. Mol Cell Biol 21: 7207–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4


