Abstract
Although general mechanisms of RNA folding and catalysis have been elucidated, little is known about how ribozymes achieve structural stability at high temperature. A previous in vitro evolution experiment identified a small number of mutations that significantly increase the thermostability of the tertiary structure of the Tetrahymena ribozyme. Because we also determined the crystal structure of this thermostable ribozyme, we have for the first time the opportunity to compare the structural interactions and thermodynamic contributions of individual nucleotides in a ribozyme. We investigated the contribution of five mutations to thermostability by using temperature gradient gel electrophoresis. Unlike the case with several well-studied proteins, the effects of individual mutations on thermostability of this RNA were highly context dependent. The three most important mutations for thermostability were actually destabilizing in the wild-type background. A269G and A304G contributed to stability only when present as a pair, consistent with their proximity in the ribozyme structure. In an evolutionary context, this work supports and extends the idea that one advantage of protein enzyme systems over an RNA world is the ability of proteins to accumulate stabilizing single-site mutations, whereas RNA may often require much rarer double mutations to improve the stability of both its tertiary and secondary structures.
Keywords: structural stability, thermostability, ribozyme, temperature gradient gel electrophoresis, crystal structure, tertiary interactions
INTRODUCTION
Extensive studies have addressed the question of how proteins maintain structural stability and activity at high temperatures (Rees 2001; Vieille and Zeikus 2001; Yano and Poulos 2003; Korkegian et al. 2005). Several mechanisms, including strengthening salt bridges, hydrogen bonds, hydrophobic interactions, and secondary structures, are used by thermophilic proteins individually or in combination (Petsko 2001). In many examples, electrostatic interactions on the surface of a protein contribute to thermostability. Other proteins introduce a new metal ion binding site, or improve the interactions at the interface between subunits. These conclusions were elucidated by using several methods, including sequence and structural comparisons of mesophilic and thermophilic homologs, and directed evolution (Matsumura and Aiba 1985; Jaenicke and Bohm 2001; Martin et al. 2001; Rees 2001).
In contrast, we know relatively little about the rules governing the thermostability of tertiary structure of large RNA molecules. Increased G+C content and post-transcriptional modifications have been suggested to stabilize thermophilic tRNAs (Watanabe et al. 1980; Kowalak et al. 1994; Galtier and Lobry 1997). Large-scale comparison of thermophilic RNase P RNA sequences with their mesophilic homologs has revealed features such as the stabilized helices and shortened linkers between helices (Brown et al. 1993). Fang et al. (2001) have investigated a pair of catalytic domains from mesophilic and thermophilic RNase P RNAs by using thermodynamic methods. They have shown that the thermophilic RNA has a less structured folding intermediate and folds into the native structure more cooperatively. The correlation between a destabilized folding intermediate and increased folding cooperativity has been reinforced by the stepwise conversion of the mesophilic RNA to its thermophilic homolog (Fang et al. 2003).
We have been investigating the mechanism by which large ribozymes achieve thermostability by using directed evolution and X-ray crystallography. In a previous study, we designed a novel in vitro evolution procedure to select for mutants of the Tetrahymena ribozyme with improved stability at high temperature (Guo and Cech 2002). A small number of mutations (nine) were found to increase the melting temperature of the tertiary structure of the 393-nucleotide (nt) ribozyme by >10°C (Fig. 1A ▶). This work provided a pair of thermophilic and mesophilic homologs with minimal differences in sequence. Recently, a crystal structure of the catalytically active truncated form (247 nt) of this ribozyme, which contained five of these stabilizing mutations, was determined at 3.8 Å resolution (Fig. 1B ▶; Guo et al. 2004). The structure illustrated several features important for the structure and catalysis of the ribozyme: a series of interactions bridging the two domains (P4–P6 and P3–P9), a highly organized binding site for the guanosine substrate involving four layers of base triples, and a number of metal ions bound to the ribozyme.
FIGURE 1.
Thermostable mutations in the Tetrahymena ribozyme. (A) Stabilizing mutations portrayed on a secondary structure diagram. The regions that are not present in the crystal structure are represented as dashed lines. The five mutations analyzed here are indicated in red, and the other four (not present in the crystal structure) are drawn in gray. (B) A crystal structure of the Tetrahymena ribozyme core at 3.8 Å resolution. In both panels, the P4–P6 domain is drawn in blue; the P3–P9 domain is in green. The 2Fo-Fc composite omit electron density map around the residues of interest is contoured at 1 σ level. The hydrogen bonds are drawn as purple dashed lines. A possible interaction is drawn as an orange dashed line.
In the current study, we have used the crystal structure as a guide to dissect the origin of thermostability at the single-nucleotide level. The melting temperatures (Tm) of individual mutants were compared with that of the wild-type (WT) ribozyme by using temperature gradient gel electrophoresis (TGGE). Somewhat surprisingly, most stabilizing mutations did not act independently, but instead their contributions to thermostability were found to be highly context dependent. Some mutated nucleotides likely interacted directly with each other to improve structural stability. The importance of tertiary interactions was also demonstrated by a mutation that was stabilizing even though it eliminated a Watson-Crick base pair in a secondary structure. Therefore, we conclude that tertiary interactions in the molecular interior play a central role in structural stability of this and presumably other large RNA molecules.
RESULTS
Structural interactions involving thermostable mutations
Five thermostable mutations are present in our recent crystal structure (Guo et al. 2004) of a truncated but active version of the Tetrahymena ribozyme (Fig. 1B ▶). Although the resolution is relatively low (3.8 Å), it is still possible to infer the atomic interactions contributing to thermostability for some of the mutations. The A210G mutation is located in a bulge of P4 (paired region 4). The N2 amino group of this guanine (G) residue is 3.1 ± 0.4 Å from the O1P nonbridging oxygen atom of the phosphate of residue 302 (mean ± SD for the four independent molecules in the asymmetric unit), consistent with a hydrogen bond. The adenine (A) residue in the wild-type sequence lacks the N2 amino group and thus cannot make this hydrogen bond. In the case of A269G, the N2 amino group of the G is close enough to the base of U307 that an interaction is plausible. The A304G mutation is in the J8/7 region. This nucleotide forms a hydrogen bond with the base of residue 270, using its N2 amino group. Thus, both the A269G and A304G mutations introduce a N2 amino group for tertiary interactions, providing a plausible explanation for their stabilization effects.
The U277C mutation disrupts a Watson-Crick base-pairing interaction with A97 in the P3 helix (Fig. 1A ▶), which by itself would be destabilizing rather than stabilizing. Interestingly, the helical structure of P3 is not dramatically changed in the crystal structure. Although residues 97 and 277 are no longer within hydrogen bonding distance from each other, they maintain base stacking interactions with their neighboring residues (Fig. 1B ▶). The 97 base is also in position for a tertiary interaction with U300 using its Hoogsteen surface. We suggest that the disruption of the 97–277 base pair by the mutation allows this tertiary interaction to be strengthened, providing a net gain of tertiary structural stability.
The crystal structure does not provide a clear answer for the stability of the U259A mutation, which occurs in the J6/ 7 region. The A base of this residue stacks on U258 but is 3.7 ± 0.2 Å from C213, too far to indicate an H-bond. The 2′-hydroxyl of A259 is close enough to O6 of G108 to form an H-bond, but we do not know whether the 2′-OH of U259 in the wild-type ribozyme also forms that interaction. The electron density of the 259 residue is generally weaker than that of the others, possibly as a consequence of a flexible local structure.
Some thermostable mutations are by themselves destabilizing
We used site-directed mutagenesis to estimate the contribution of each thermostable mutation found in the crystal structure. Five mutations (A210G, U259A, A269G, U277C, and A304G) were individually introduced into the wild-type Tetrahymena L-21 ribozyme (393 nt), which was used in the previous in vitro evolution experiments (Guo and Cech 2002). The relative tertiary structure melting temperatures (ΔTm) of these mutants were estimated by using TGGE with the wild-type ribozyme as a reference (Fig. 2A ▶). TGGE employs a linear temperature gradient perpendicular to the direction of the electric field. It allows the melting curves of the tertiary structures of two or more structured RNAs to be displayed on a single gel (Riesner et al. 1989; Szewczak et al. 1998).
FIGURE 2.
TGGE was used to compare the melting temperatures of ribozyme variants. (A) TGGE with the Tetrahymena L-21 ribozyme WT, A269G and A304G mutants loaded sequentially. (B) TGGE with R14C and a single reverse mutant G269A loaded sequentially. Both gels were performed in 1× TH buffer and 0.4 mM MgCl2.
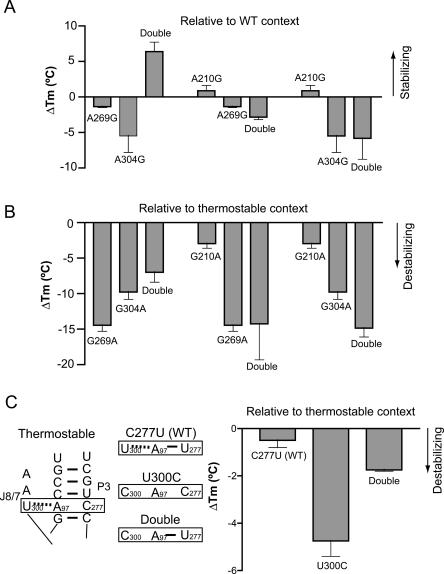
The results of this series of experiments turned out to be quite surprising. Only two of the individual mutations stabilized the structure, whereas the other three decreased the thermostability (Fig. 3A ▶). A210G and U277C slightly increased the melting temperature by 0.9°C and 1.6°C, respectively. In contrast, U259A and A304G destabilized the ribozyme by >5°C. A269G decreased the Tm by 1.4°C. Therefore, the contributions of these mutations to overall thermostability were far from equal and positive.
FIGURE 3.
Thermostability contributions of mutations are highly context dependent. (A) Five stabilizing mutations were introduced individually into the wild-type ribozyme and their ΔTm values relative to WT were determined by using TGGE in 0.4 mM MgCl2. (B) These five residues were reversed to the wild-type sequence in the thermo-stable background, and their ΔTm values relative to R14C were measured in 0.4 mM MgCl2. The ΔTm values are the mean of two to four independent measurements. The error bars encompass the range of measurements.
The contributions of individual mutations are context dependent
Are all the thermostable mutations important for the thermostability of the consensus mutant? To address this question, the five mutations mentioned above were reversed back to the wild-type sequence individually in the thermostable consensus background (R14C), and their melting temperatures relative to R14C were estimated by TGGE (Fig. 2B ▶). All the reverse mutants are less stable than the thermostable consensus (Fig. 3B ▶), indicating that the “forward” mutations all make positive contributions to the thermostability of R14C.
Comparison of the mutagenesis data in the wild-type and thermostable contexts indicated that the contributions to thermostability of the ribozyme by some mutations were highly context dependent (Fig. 3 ▶). Three mutations (positions 259, 269, and 304) were destabilizing individually in the wild-type context but became stabilizing in the presence of the other eight thermostable mutations. Only two mutations (positions 210 and 277) had the expected behavior of being stabilizing in both the wild-type and thermostable contexts, and even here the magnitudes of the effects were not equal. The context-dependent behavior was most dramatic for positions 259 and 304. U259A was destabilizing by 5.2°C in wild type and stabilizing by 16.5°C in the thermostable background. A304G destabilized the wild-type ribozyme structure by 5.5°C and increased the thermostability by 9.8°C in the R14C background. Interestingly, the most context-dependent mutations (positions 259, 269, and 304) are also the most important ones for structural stability in the thermostable mutant background (Fig. 3B ▶).
Cooperativity of A269G and A304G in enhancing stability
As has been mentioned, A304G is one of the most destabilizing mutations in the wild-type context and becomes highly stabilizing in the presence of all of the other mutations. The interactions observed in our crystal structure, which involve the N2 atom unique to the G residue, cannot explain the context-dependent behaviors of A304G and A269G unless the structures of the wide-type and the single mutants are different from that of the thermostable ribozyme. In the crystal structure, G304 and G269 are close to each other (~5 Å for the closest atom pairs), but no direct interaction is observed. The equivalent residues (positions 135 and 170) in the recent crystal structure of the Azoarcus group I intron (Adams et al. 2004) do directly interact with each other. Therefore, we explored the potential functional link between the two mutations.
A double mutant (A269G::A304G) in the context of the wild-type ribozyme was generated. In contrast to the single mutants, the double mutant was stabilized with a ΔTm of 6.4°C (Fig. 4A ▶), proving that the two mutations work together (synergistically) to improve thermostability of the ribozyme structure. These mutagenesis data are analogous to those obtained from conventional compensatory mutation analyses. These analyses have most often been used to demonstrate base-pairing interactions within the secondary structure of RNA molecules, but have also been used to establish tertiary interactions (Michel et al. 1990; Sampson et al. 1990; Murphy and Cech 1994). As a control, double mutants of A210G (which contacts neither residue 269 nor 304 in the crystal structure) in combination with either A269G (A210G::A269G) or A304G (A210G::A304G) did not have any stabilizing effect (Fig. 4A ▶). Therefore, the compensatory and synergistic effects are specific to the A269G and A304G mutations.
FIGURE 4.
Synergistic interactions of a pair of thermostable mutations, and a case where a tertiary interaction is improved at the expense of a base pair. (A) A269G and A304G are destabilizing individually and become stabilizing together, in the context of the wild-type ribozyme. As controls, the A210G mutation did not introduce extra stabilization when present in combination with either A269G or A304G. (B) Same stability patterns occur in the thermostable context. (C) U277C strengthens a tertiary interaction while disrupting a secondary structure. The local secondary structures of the R14C RNA and the three mutants are shown on the left. In all panels, the ΔTm values (in 0.4 mM MgCl2) are the mean of two to four independent measurements. The error bars encompass the range of values.
Similar mutational data in the context of the thermostable background also supported presence of compensation and synergism between residues 269 and 304 (Fig. 4B ▶). When residue 269 or 304 was reversed to the wild-type residue, the ribozyme structure was substantially destabilized, with ΔTm values of −14.4°C and −9.8°C, respectively. When both residues were reversed to A, the ribozyme was less destabilized with a ΔTm of −7.0°C. The consistent pattern of thermostability in both WT and R14C backgrounds is that when both 269 and 304 residues are G, a strong interaction is formed; when both residues are A, a weak interaction can be established; and when they are A and G in either order, the ribozyme is most destabilized due to disruption of the interaction.
A mutation improves thermostability by strengthening a tertiary interaction while disrupting a secondary structure
U277C disrupts an internal base pair in the P3 helix, yet this mutation made a small positive contribution to thermostability in both the wild-type and stabilized backgrounds (Fig. 3 ▶). Its base-pairing partner A97 formed a tertiary interaction with U300 in the crystal structure (Fig. 1B ▶; Guo et al. 2004). In a previous study, U300 was shown to form a base triple interaction with the A97–U277 base pair in the wild-type Tetrahymena ribozyme (Szewczak et al. 1999). The interaction between U300 and A97 found in our crystal structure was identical to the one proposed based on biochemical data. Here we consider the simplest possibility of dividing mutational effects on free energy change into those of tertiary interactions and secondary structures. We hypothesize that the tertiary interaction between A97 and U300 is important for thermostability, and disruption of the base pairing between U277 and A97 allows a more optimal tertiary interaction, possibly by releasing a conformational restraint. This hypothesis predicts the following thermostability properties of two mutants (both in R14C background): U300C, which disrupts the tertiary interaction, should most strongly decrease the ribozyme thermostability; U300C::C277U, which disrupts the tertiary interaction but restores the secondary structure, should be more stable than U300C alone. An alternative model is that the tertiary and secondary interactions are independent of each other, but disruption of the secondary structure is somehow favorable to thermostability. This latter model predicts that the U300C::C277U double mutant should be less stable than U300C in the R14C background.
We generated U300C and U300C::C277U mutants in the R14C background and measured their melting temperatures relative to R14C by using TGGE. The thermostability behaviors of these mutants were consistent with our first hypothesis (Fig. 4C ▶). U300C is the least thermostable with a ΔTm of −4.8°C; the U300C::C277U double mutant is more stable with a ΔTm of −1.8°C, likely due to the restored secondary structure; and C277U is almost as stable as R14C (ΔTm = −0.4°C).
Therefore, we present a case in which the overall structural stability of a large RNA can be improved by strengthening a tertiary interaction while disrupting a secondary structure. The interplay of secondary and tertiary interactions has been observed in the folding of the P5abc subdomain of the Tetrahymena ribozyme. Wu and Tinoco (1998) determined that a major rearrangement of secondary structure in the P5abc subdomain upon addition of magnesium ions, which is energetically unfavorable by itself, can be overcome by the free energy gained from new tertiary interactions (Silverman et al. 1999). Our experiments similarly indicate that secondary structures and tertiary interactions are not completely independent of each other in their contribution to thermostability of large RNA molecules. In the Tetrahymena ribozyme, the tertiary interactions, not secondary structures, appear to be the limiting factor for thermostability (Banerjee et al. 1993; Guo and Cech 2002; current study). It will be interesting to see whether similar conclusions are true for other large RNAs.
DISCUSSION
The origin of thermostability for large RNAs has not been investigated nearly as extensively as for proteins. The current study provides the first opportunity to compare how much individual nucleotides contribute to the stability of tertiary structure of a complex folded RNA for which the crystal structure has been determined. In fact, we can examine crystal structures of both the thermostable and native structures, although the resolutions are limited (3.8 and 5.0 Å, respectively) (Golden et al. 1998; Guo et al. 2004). We find that the contributions of individual mutations are context dependent and not additive but, instead, highly synergistic.
Comparison with RNase P RNA
In an earlier study, Fang et al. (2001) compared the thermodynamic properties of the catalytic domains (C-domains) of the RNase P RNAs of mesophilic and thermophilic bacteria. The thermophilic C-domain was shown to achieve increased stability by using a less structured intermediate and increased cooperativity. More recently, they converted the mesophilic ribozyme into the thermophilic homolog in a stepwise fashion (Fang et al. 2003). The two RNA molecules differ from each other at ~40 positions (out of ~260 nt), which were grouped into six motifs. Three motifs were shown to be important for the increased stability, and combining them resulted in a continuous increase in stability until achieving that of the thermophilic ribozyme with all three. This work also reinforced the correlation of stability and folding cooperativity found in the wild-type RNase P ribozymes.
Our present work shares some interesting features with the stepwise mutagenes is study on the C-domains of RNase P RNA. In both cases, a context-dependent effect of mutations on structural stability has been observed. In the RNase P RNA, two mutations in the P19 helix are modestly stabilizing in the mesophilic background, are neutral in the presence of the thermophilic P1 motif alone, and have a large stabilizing effect only in the presence of additional substitutions in the P1, J15/ J15.1, and J19/4 regions. Now by using single and double point-mutations, we show that individual mutations can be strongly destabilizing, and yet the combination of two or more of them can confer substantial stability to the RNA tertiary structure. These “compensatory” mutations are likely to replace existing interactions (involving A269 and A304) with stronger ones (involving A269G and A304G). The thermodynamic study by itself might suggest direct A269–A304 and G269–G304 interactions, but the crystal structure shows that they are instead cooperating via side-by-side interactions (Fig. 1B ▶).
Our in vitro selection using the Tetrahymena ribozyme indicated that a small number of mutations is capable of conferring thermostability to a large RNA molecule (nine mutations in this 393-nt RNA) (Guo and Cech 2002). Similarly, it was estimated that six mutations in the C-domain of 255-nt RNase P RNA contribute most of the stabilizing power as judged by the minimum magnesium concentration needed to achieve the native structure (Fang et al. 2003). Furthermore, we found that two stabilizing mutations (A269G and A304G) contribute a large part (6.4° of 10.5° total) of thermostability to the Tetrahymena R14C ribozyme. The similar observations from two independent types of RNA molecules, a group I intron and RNase P RNA, suggest that a small number of mutations might also be able to provide significant structural stability for other large RNA molecules. Indeed, three to four mutations in the GIR1 ribozyme were sufficient to significantly increase its site-specific hydrolysis activity and structural stability (Jabri and Cech 1998).
There are several critical differences between the two studies on the Tetrahymena group I intron and the RNase P RNA. We used TGGE to measure the melting transition temperatures of an RNA at one or two divalent metal ion concentrations (0.4 or 0.2 mM MgCl2) (Guo and Cech 2002). TGGE monitors the first unfolding transition that leads to a significant retardation in electrophoretic mobility, and it is possible that mutations could alter which structural element unfolds first. Furthermore, although TGGE was validated as an equilibrium method to monitor tertiary structure of the isolated P4–P6 domain of the ribozyme (Szewczak et al. 1998), the larger RNAs studied here may be more subject to kinetic traps in folding. The Tm value for an RNA undergoing cooperative unfolding is expected to approximate the temperature where the folded and unfolded states are equally stable (ΔG = 0), although they are not necessarily the same by definition. It has not been possible to obtain ΔG (37°C) values from our TGGE experiments. In contrast, the ΔG (37°C) values for RNase P RNA were extracted by monitoring the circular dichroism of the RNA while titrating with Mg2+ ions (or urea) (Fang et al. 1999). In the mutagenesis study, the RNase P RNAs from mesophilic and thermophilic bacteria were compared at 37°C, and their structural stability was represented by the magnesium ion concentrations in the middle of the last folding transition (from the last intermediate to native fold) (Fang et al. 2001, 2003). These methods are quite complementary and should be used together to allow them to be directly compared and contrasted in future studies.
Strategies used by proteins and RNAs to achieve thermostability
Because protein and RNA are very different biopolymers, it is not surprising that there are some fundamental differences in their folding. RNA has a polyanionic backbone, so metal ion binding is generally important for RNA folding (Draper et al. 2005). RNA has a hydrophilic rather than a hydrophobic core. In contrast, folding of protein is largely driven by hydrophobic interactions. Therefore, one would expect important differences in how RNA and protein achieve thermostability.
Many thermostable mutations in proteins are located on the surface (Zhao and Arnold 1999; Yano and Poulos 2003). They either introduce new electrostatic interactions or optimize existing ones (Chakravarty and Varadarajan 2002). We have shown that most stabilizing mutations selected in the Tetrahymena ribozyme are located in the molecular interior. They help to hold the secondary structure elements together, a function that is often accomplished in proteins by compact packing of the hydrophobic core. The core packing in proteins from mesophilic organisms appears to have already been optimized (Karshikoff and Ladenstein 1998), although exceptions to this general observation have been presented (Korkegian et al. 2005).
Some thermostable proteins increase their binding affinity for metal ions or create new metal ion binding sites. The thermostable versions of both the Tetrahymena ribozyme and the C-domain of the RNase P RNA fold at lower magnesium ion concentration than do the mesophilic ones (Fang et al. 2001; Guo and Cech 2002). Thus their affinity for metal ions is likely increased. However, this does not necessarily mean that any new metal-ion binding sites have been created, and in fact, none has been identified by comparison of the existing crystal structures.
Several thermodynamically stable proteins have been documented to achieve thermostability step-wise, with individual amino acid changes providing incremental, additive contributions (Serrano et al. 1993; Zhang et al. 1995; Zhao and Arnold 1999). Our RNA achieves thermostability in a very different way: Contributions by individual mutations are context dependent, nonadditive, but highly cooperative.
Correlation of thermostability data with crystal structures
The availability of two other group I intron crystal structures (Adams et al. 2004; Golden et al. 2005) allows us to gain further insights into how these stabilizing mutations work. Adams et al. (2004) have determined the three-dimensional structure of the group I tRNA intron (subgroup IC3) from the bacterium Azoarcus at 3.1 Å resolution. The intron was complexed with both exons and corresponded to the splicing intermediate before the exon ligation step. Although its host bacterium Azoarcus is not thermophilic, this group I intron has exceptional thermostability with peak splicing activity at 75°C, a property making it especially relevant to our current study (Tanner and Cech 1996). In addition, Golden and colleagues (2005) solved the structure of a group I intron from the bacteriophage Twort (subgroup IA2) at 3.6 Å resolution; this intron contained a paired P1 helix and corresponded to the product stage of a cleavage reaction. In contrast, our Tetrahymena ribozyme structure does not contain the P1 helix and corresponds to the catalytic stage prior to substrate binding (Guo et al. 2004). These three crystal structures align well with each other in the core region, but they differ in peripheral regions (Guo et al. 2004; Golden et al. 2005; Vicens and Cech 2006). Their similar catalytic core structures, despite differences in sequence, suggest that relatively small conformational changes are involved in substrate docking and catalysis.
A269G and A304G work synergistically to achieve thermostability in the Tetrahymena L-21 ribozyme. This observation is consistent with the proximity of the two nucleotides in the crystal structures. Intriguingly, in the thermostable Azoarcus intron, both positions are occupied by G residues (Tanner and Cech 1996). They directly interact through at least one hydrogen bond (between N7 of G269 and N1 of G304, Tetrahymena numbering here and hereafter) and possibly a second H-bond (between N7 of G269 and N2 of G304) (Adams et al. 2004). The crystal structure of the Azoarcus intron suggests that the two residues might interact directly with each other in the sequence context and solution conditions used in the current TGGE study. However, the hydrogen-bonding interactions between the 269 and 304 residues in the Azoarcus structure cannot explain clearly why it is so important to have a G at the 269 position for increased thermostability. Similar to the wild-type Tetrahymena ribozyme (subgroup IC1), the Twort ribozyme has A residues at both positions (Landthaler and Shub 1999). In the Twort ribozyme structure, these two residues are ~5 Å apart (for the closest atom pairs) (Golden et al. 2005). However, in the Twort ribozyme, A269 is right next to helices P7.1 and P7.2, which is characteristic of the IA2 subgroup of the group I intron family. It is possible that A269G and A304G have different modes of interactions (or lack of interactions) depending on local structural contexts and perhaps on specific catalytic stages.
U277C disrupts an internal base-pairing interaction and likely stabilizes the structure by strengthening the base triple interaction involving the first base (U300) in the J8/7 region. While the J8/7 region in the Tetrahymena ribozyme contains seven residues, the equivalent regions in the Azoarcus and Twort ribozymes are composed of six residues (Tanner and Cech 1996; Landthaler and Shub 1999). Therefore, it is not surprising that the base triple interaction (U300:U277–A97) is not present for them, as indicated by their crystal structures.
U259 has been proposed to interact with the second base pair of the P4 helix in the wild-type Tetrahymena intron (G108 and C213) (Michel et al. 1990). In both the Azoarcus and Twort crystal structures, the equivalent residues (G125 in Azoarcus and G117 in Twort) form a major groove contact as predicted (Adams et al. 2004; Golden et al. 2005). In all four molecules of the Tetrahymena crystal structure (Guo et al. 2004), the A residue at the 259 position stacks on U258 but does not seem to directly interact with the P4 helix. In the group I intron family, U and G are the dominantly abundant residues at this position (Cannone et al. 2002). It is possible that the U259A mutation improves the structural stability by improved stacking on U258, since A typically provides more stacking energy than does uracil (Sugimoto et al. 1987). As a separate consideration, it has been shown that the J6/7 region of the Tetrahymena ribozyme, in which residue 259 is located, is important for the folding of the P3–P9 domain (Zarrinkar and Williamson 1996). It is possible that U259A alters the folding pathway of the ribozyme, thus improving thermostability by changing the last folding intermediate. Indeed, we have observed additional bands for this mutant RNA on native gels that could correspond to these folding intermediates (data not shown).
Finally, the A210 residue is not conserved in the group I intron family and is present in neither the Azoarcus nor the Twort ribozyme.
Implications for evolution
If the context-dependent contribution of stabilizing mutations to the thermostability of a large ribozyme is general, it has important implications for molecular evolution. It would be easier to evolve a thermostable protein than a thermostable RNA, because many stabilizing mutations in proteins act independently (Serrano et al. 1993; Zhang et al. 1995). We have shown that in the Tetrahymena ribozyme, some of the most important stabilizing mutations synergize with each other. In this situation, achievement of stability requires simultaneous presence of two or three residues at specific positions. If the frequency of single-base changes is 10−5, the probability of a particular double-base change would be 10−10. The greatly reduced efficiency for RNA to develop stabilizing mutations might have been a serious disadvantage during evolution and have contributed to the eventual migration to our current world dominated by proteins and ribonucleoprotein (RNP) complexes. The idea that it takes compensatory double changes to improve structure is well known in the RNA field, but what is new in this study is that we see such effects in a freely evolving system and can compare the effects on stability with the crystal structure.
MATERIALS AND METHODS
RNA preparation
The wild-type and thermostable mutant (R14C) forms of the Tetrahymena L-21 ribozyme (containing nucleotides G22 through G414) were obtained from a previous study (Guo and Cech 2002). The R14C form contains the following nine mutations: A94G, A210G, U259A, A269G, U277C, A304G, U322G, U340A, and U377A. The 247-nt construct used for crystallization contains the following five mutations: A210G, U259A, A269G, U277C, and A304G. The transcription templates contained a T7 promoter, the ribozyme coding sequence, and an EarI site between the EcoRI and XbaI sites in a pUC19 plasmid. PCR methods were used to introduce the thermostable mutations individually into the wild-type ribozyme and to reverse individual mutations back to the native sequence in the R14C background. All mutagenized templates were then cloned and sequenced prior to transcription. The ribozymes were transcribed by T7 RNA polymerase at 37°C in the presence of [α-32P]CTP. The RNA was purified by using 6% denaturing polyacrylamide gels. The bands, visualized by autoradiography, were excised, gel-extracted in TEN buffer (10 mM Tris at pH 7.5, 1 mM EDTA, 0.25 M NaCl), and ethanol precipitated. The RNA was resuspended in TE buffer (10 mM Tris at pH 7.5, 1 mM EDTA) and stored at −20°C.
Temperature gradient gel electrophoresis
The experiments were performed essentially as described (Szewczak et al. 1998). Non-denaturing 6% polyacrylamide gels were run in a buffer (pH 7.4) containing 34 mM Tris and 66 mM NaHEPES and 0.4 mM MgCl2. The gel was typically prerun for 60 min and run with samples for ~2 h at 250 V. Running buffer was refreshed every hour. RNA samples in 10 mM sodium cacodylate (pH 6.5) were annealed by heating to 80°C for 3 min; moved to room temperature, where MgCl2 was added to a final concentration of 0.4 mM; and then allowed to cool slowly on the bench for an hour. An equal volume of 20% sucrose loading buffer with 0.02% each bromophenol blue and xylene cyanol was added just before loading on the gel. Samples were loaded 10–15 min apart.
Determination of Tm from TGGE
The horizontal temperature gradient established across the native gel was determined to be linear by in-gel measurements using a digital thermometer with a microprobe. The temperature at a given point was then calculated by measuring the distance of that point from the edge of the gel. Tm was defined as the temperature at which the band representing the folded form of the ribozyme decayed to half of its intensity at low temperature. See the Supplemental Figure (at http://www.doe-mbi.ucla.edu/People/Guo/supps/Guo_2006.pdf) for details.
Acknowledgments
We thank Dr. Quentin Vicens for valuable discussions and help on Figure 1B ▶. This work is supported by the Howard Hughes Medical Institute.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2198206.
REFERENCES
- Adams, P.L., Stahley, M.R., Kosek, A.B., Wang, J., and Strobel, S.A. 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50. [DOI] [PubMed] [Google Scholar]
- Banerjee, A.R., Jaeger, J.A., and Turner, D.H. 1993. Thermal unfolding of a group I ribozyme: The low-temperature transition is primarily disruption of tertiary structure. Biochemistry 32: 153–163. [DOI] [PubMed] [Google Scholar]
- Brown, J.W., Haas, E.S., and Pace, N.R. 1993. Characterization of ribonuclease P RNAs from thermophilic bacteria. Nucleic Acids Res. 21: 671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D’Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., et al. 2002. The Comparative RNA Web (CRW) Site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty, S. and Varadarajan, R. 2002. Elucidation of factors responsible for enhanced thermal stability of proteins: A structural genomics based study. Biochemistry 41: 8152–8161. [DOI] [PubMed] [Google Scholar]
- Draper, D.E., Grilley, D., and Soto, A.M. 2005. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 34: 221–243. [DOI] [PubMed] [Google Scholar]
- Fang, X., Pan, T., and Sosnick, T.R. 1999. A thermodynamic framework and cooperativity in the tertiary folding of a Mg2+-dependent ribozyme. Biochemistry 38: 16840–16846. [DOI] [PubMed] [Google Scholar]
- Fang, X.W., Golden, B.L., Littrell, K., Shelton, V., Thiyagarajan, P., Pan, T., and Sosnick, T.R. 2001. The thermodynamic origin of the stability of a thermophilic ribozyme. Proc. Natl. Acad. Sci. 98: 4355–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, X.W, Srividya, N., Golden, B.L., Sosnick, T.R., and Pan, T. 2003. Stepwise conversion of a mesophilic to a thermophilic ribozyme. J. Mol. Biol. 330: 177–183. [DOI] [PubMed] [Google Scholar]
- Galtier, N. and Lobry, J.R. 1997. Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J. Mol. Evol. 44: 632–636. [DOI] [PubMed] [Google Scholar]
- Golden, B.L., Gooding, A.R., Podell, E.R., and Cech, T.R. 1998. A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science 282: 259–264. [DOI] [PubMed] [Google Scholar]
- Golden, B.L., Kim, H., and Chase, E. 2005. Crystal structure of a phage Twort group I ribozyme-product complex. Nat. Struct. Mol. Biol. 12: 82–89. [DOI] [PubMed] [Google Scholar]
- Guo, F. and Cech, T.R. 2002. Evolution of Tetrahymena ribozyme mutants with increased structural stability. Nat. Struct. Biol. 9: 855–861. [DOI] [PubMed] [Google Scholar]
- Guo, F., Gooding, A.R., and Cech, T.R. 2004. Structure of the Tetrahymena ribozyme: Base triple sandwich and metal ion at the active site. Mol. Cell 16: 351–362. [DOI] [PubMed] [Google Scholar]
- Jabri, E. and Cech, T.R. 1998. In vitro selection of the Naegleria GIR1 ribozyme identifies three base changes that dramatically improve activity. RNA 4: 1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke, R. and Bohm, G. 2001. Thermostability of proteins from Thermotoga maritima. Methods Enzymol. 334: 438–469. [DOI] [PubMed] [Google Scholar]
- Karshikoff, A. and Ladenstein, R. 1998. Proteins from thermophilic and mesophilic organisms essentially do not differ in packing. Protein Eng. 11: 867–872. [DOI] [PubMed] [Google Scholar]
- Korkegian, A., Black, M.E., Baker, D., and Stoddard, B.L. 2005. Computational thermostabilization of an enzyme. Science 308: 857–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalak, J.A., Dalluge, J.J., McCloskey, J.A., and Stetter, K.O. 1994. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33: 7869– 7876. [DOI] [PubMed] [Google Scholar]
- Landthaler, M. and Shub, D.A. 1999. Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: Introns in multiple genes, a single gene with three introns, and exon skipping by group I ribozymes. Proc. Natl. Acad. Sci. 96: 7005–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, A., Sieber, V., and Schmid, F.X. 2001. In-vitro selection of highly stabilized protein variants with optimized surface. J. Mol. Biol. 309: 717–726. [DOI] [PubMed] [Google Scholar]
- Matsumura, M. and Aiba, S. 1985. Screening for thermostable mutant of kanamycin nucleotidyltransferase by the use of a transformation system for a thermophile, Bacillus stearothermophilus. J. Biol. Chem. 260: 15298–15303. [PubMed] [Google Scholar]
- Michel, F., Ellington, A.D., Couture, S., and Szostak, J.W. 1990. Phylogenetic and genetic evidence for base-triples in the catalytic domain of group I introns. Nature 347: 578–580. [DOI] [PubMed] [Google Scholar]
- Murphy, F.L. and Cech, T.R. 1994. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J. Mol. Biol. 236: 49–63. [DOI] [PubMed] [Google Scholar]
- Petsko, G.A. 2001. Structural basis of thermostability in hyperthermophilic proteins, or “there’s more than one way to skin a cat.” Methods Enzymol. 334: 469–478. [DOI] [PubMed] [Google Scholar]
- Rees, D.C. 2001. Crystallographic analyses of hyperthermophilic proteins. Methods Enzymol. 334: 423–437. [DOI] [PubMed] [Google Scholar]
- Riesner, D., Steger, G., Zimmat, R., Owens, R.A., Wagenhofer, M., Hillen, W., Vollbach, S., and Henco, K. 1989. Temperature-gradient gel electrophoresis of nucleic acids: Analysis of conformational transitions, sequence variations, and protein–nucleic acid interactions. Electrophoresis 10: 377–389. [DOI] [PubMed] [Google Scholar]
- Sampson, J.R., DiRenzo, A.B., Behlen, L.S., and Uhlenbeck, O.C. 1990. Role of the tertiary nucleotides in the interaction of yeast phenylalanine tRNA with its cognate synthetase. Biochemistry 29: 2523– 2532. [DOI] [PubMed] [Google Scholar]
- Serrano, L., Day, A.G., and Fersht, A.R. 1993. Step-wise mutation of barnase to binase. A procedure for engineering increased stability of proteins and an experimental analysis of the evolution of protein stability. J. Mol. Biol. 233: 305–312. [DOI] [PubMed] [Google Scholar]
- Silverman, S.K., Zheng, M., Wu, M., Tinoco Jr., I., and Cech, T.R. 1999. Quantifying the energetic interplay of RNA tertiary and secondary structure interactions. RNA 5: 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, N., Kierzek, R., and Turner, D.H. 1987. Sequence dependence for the energetics of dangling ends and terminal base pairs in ribonucleic acid. Biochemistry 26: 4554–4558. [DOI] [PubMed] [Google Scholar]
- Szewczak, A.A., Podell, E.R., Bevilacqua, P.C., and Cech, T.R. 1998. Thermodynamic stability of the P4–P6 domain RNA tertiary structure measured by temperature gradient gel electrophoresis. Biochemistry 37: 11162–11170. [DOI] [PubMed] [Google Scholar]
- Szewczak, A.A., Ortoleva-Donnelly, L., Zivarts, M.V., Oyelere, A.K., Kazantsev, A.V., and Strobel, S.A. 1999. An important base triple anchors the substrate helix recognition surface within the Tetrahymena ribozyme active site. Proc. Natl. Acad. Sci. 96: 11183–11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, M. and Cech, T. 1996. Activity and thermostability of the small self-splicing group I intron in the pre-tRNA(lle) of the purple bacterium Azoarcus. RNA 2: 74–83. [PMC free article] [PubMed] [Google Scholar]
- Vicens, Q. and Cech, T.R. 2006. Atomic-level architecture of group I introns revealed. Trends Biochem. Sci. (in press). [DOI] [PubMed]
- Vieille, C. and Zeikus, G.J. 2001. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65: 1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K., Oshima, T., Iijima, K., Yamaizumi, Z., and Nishimura, S. 1980. Purification and thermal stability of several amino acid– specific tRNAs from an extreme thermophile, Thermus thermophilus HB8. J. Biochem. 87: 1–13. [DOI] [PubMed] [Google Scholar]
- Wu, M. and Tinoco Jr., I. 1998. RNA folding causes secondary structure rearrangement. Proc. Natl. Acad. Sci. 95: 11555–11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, J.K. and Poulos, T.L. 2003. New understandings of thermostable and peizostable enzymes. Curr. Opin. Biotechnol. 14: 360–365. [DOI] [PubMed] [Google Scholar]
- Zarrinkar, P.P. and Williamson, J.R. 1996. The kinetic folding pathway of the Tetrahymena ribozyme reveals possible similarities between RNA and protein folding. Nat. Struct. Biol. 3: 432–438. [DOI] [PubMed] [Google Scholar]
- Zhang, X.J., Baase, W.A., Shoichet, B.K., Wilson, K.P., and Matthews, B.W. 1995. Enhancement of protein stability by the combination of pointmutations in T4 lysozyme is additive. Protein Eng. 8: 1017–1022. [DOI] [PubMed] [Google Scholar]
- Zhao, H. and Arnold, F.H. 1999. Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Eng. 12: 47–53. [DOI] [PubMed] [Google Scholar]