Abstract
The m7GpppN cap structure of eukaryotic mRNA is formed by the sequential action of RNA triphosphatase, guanylyltransferase, and (guanine N-7) methyltransferase. In trypanosomatid protozoa, the m7GpppN is further modified by seven methylation steps within the first four transcribed nucleosides to form the cap 4 structure. The RNA triphosphatase and guanylyltransferase components have been characterized in Trypanosoma brucei. Here we describe the identification and characterization of a T. brucei (guanine N-7) methyltransferase (TbCmt1). Sequence alignment of the 324–amino acid TbCmt1 with the corresponding enzymes from human (Hcm1), fungal (Abd1), and microsporidian (Ecm1) revealed the presence of conserved residues known to be essential for methyltransferase activity. Purified recombinant TbCmt1 catalyzes the transfer of a methyl group from S-adenosylmethionine to the N-7 position of the cap guanine in GpppN-terminated RNA to form the m7GpppN cap. TbCmt1 also methylates GpppG and GpppA but not GTP or dGTP. Mutational analysis of individual residues of TbCmt1 that were predicted—on the basis of the crystal structure of Ecm1—to be located at or near the active site identified six conserved residues in the putative AdoMet- or cap-binding pocket that caused significant reductions in TbCmt1 methyltransferase activity. We also report the identification of a second T. brucei RNA (guanine N-7) cap methyltransferase (named TbCgm1). The 1050–amino acid TbCgm1 consists of a C-terminal (guanine N-7) methyltransferase domain, which is homologous with TbCmt1, and an N-terminal guanylyltransferase domain, which contains signature motifs found in the nucleotidyl transferase superfamily.
Keywords: mRNA capping, m7G methyltransferase, Trypanosoma brucei
INTRODUCTION
The earliest event in the modification of the 5′ terminus of eukaryotic mRNA is the formation of the m7GpppN cap structure, which is required for mRNA stability and efficient translation. Capping entails three enzymatic reactions in which the 5′-triphosphate terminus of the nascent RNA is cleaved to a diphosphate by RNA triphosphatase, capped with GMP by RNA guanylyltransferase, and methylated at the N-7 position of the cap guanosine by RNA (guanine N-7) methyltransferase (hereafter referred to as m7G methyl-transferase) (Shuman 2000). Consistent with the fundamental role of the cap in eukaryotic gene expression, all three activities are essential for viability in budding yeast (Schwer and Shuman 1994; Schwer et al. 2001). Studies in yeast and mammals suggest that the specificity of capping pol II transcripts is achieved by the direct interaction of capping enzyme components with the elongating form of pol II (Cho et al. 1997; McCracken et al. 1997; Yue et al. 1997; Pillutla et al. 1998; Ho and Shuman 1999).
In kinetoplastid protozoa, the mechanism by which the mRNA acquires a cap is different from other eukaryotes. The cap is formed on the spliced leader transcript (SL RNA), then transferred to the 5′ end upstream of the start site of individual precursor mRNAs by trans-splicing to form mature monocistronic mRNAs (Agabian 1990; Adams et al. 1996; Liang et al. 2003). The SL RNA cap is unique among eukaryotes in that the first four nucleotides following the cap are hypermethylated and is referred to as cap 4. Cap 4 consists of a standard m7GpppN with 2′-O methylations on the first four ribose sugars (AmAmCmUm), and additional base methylations on the first adenine (m6,6A) and the fourth uracil (m3U) (Bangs et al. 1992; Mair et al. 2000). Analysis of Trypanosoma brucei SL RNA biosynthesis using cap methyltransferase inhibitors suggests that the cap is formed cotranscriptionally and that the methylation steps that lead to cap 4 are essential for the trans-splicing reaction (Ullu and Tschudi 1991; McNally and Agabian 1992; Mair et al. 2000). It has been suggested that cap methyltransferases are promising drug targets against kinetoplastid parasites, because the mechanism of cap formation and the structure of the cap are fundamentally different from those of the host (Tschudi and Ullu 2002). However, only the RNA triphosphatase and the RNA guanylyltransferase have been characterized biochemically (Silva et al. 1998; Ho and Shuman 2001).
T. brucei RNA triphosphatase (TbCet1) is a 253–amino acid monomeric protein that catalyzes the removal of the γ phosphate from triphosphate-terminated RNA in the presence of divalent cations (Ho and Shuman 2001). TbCet1 is mechanistically similar to the RNA triphosphatases found in fungi, microsporidia, and other protozoa but is unrelated to the cysteine phosphatase-type RNA triphosphatases found in metazoans and plants. The T. brucei guanylyltransferase (TbCe1) catalyzes the transfer of GMP from GTP to the 5′-diphosphate terminus of the RNA to form GpppN (Silva et al. 1998). The primary structure and mechanism of TbCe1 resemble those of RNA guanylyltransferases from other eukaryotes, except that TbCe1 contains an N-terminal extension of 250 amino acids of unknown function that is dispensable for guanylyltransferase activity. Neither TbCet1 nor TbCe1 appear to possess any cap methyltransferase activity.
For synthesis of the m7GpppN cap, kinetoplastids must encode a separate m7G methyltransferase. We searched the T. brucei proteome for a polypeptide resembling human and yeast m7G methyltransferases, and identified two candidate m7G methyltransferases: a 324–amino acid polypeptide, which we refer to as TbCmt1 (T. brucei m7G cap methyl-transferase 1), and a 1050–amino acid polypeptide, which we refer to as TbCgm1 (T. brucei cap guanylyltransferase-methyltransferase 1). Here we show that recombinant TbCmt1 has intrinsic m7G methyltransferase activity; it catalyzes the transfer of a methyl group from S-adenosylmethionine (AdoMet) to a cap-terminated RNA (GpppRNA) to form methylated capped RNA (m7GpppRNA) and S-adenosylhomocysteine (AdoHcy). Mechanistic conservation among the m7G methyltransferases is suggested by structure-based mutational analysis of the putative AdoMet- and cap-binding pockets in TbCmt1, with reference to the crystal structures of the microsporidian parasite Encephalitozoon cuniculi m7G methyltransferase (Fabrega et al. 2004; Hausmann et al. 2005).
RESULTS
Identification of a T. brucei m7G methyltransferase
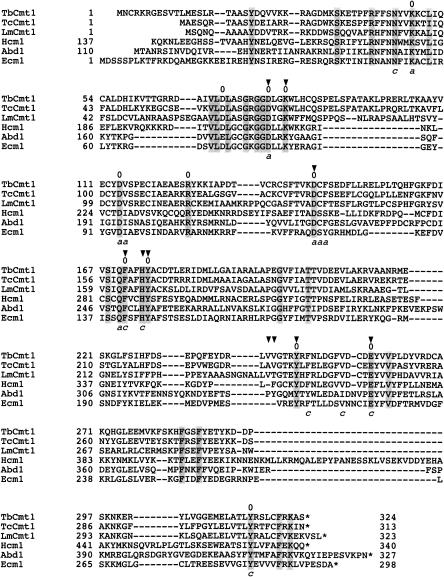
We searched for candidate T. brucei cap methyltransferase enzymes by querying the Sanger Database for proteins related to known RNA (guanine N-7) cap methyltransferases. We identified an open reading frame on T. brucei chromosome 10 with extensive homology with the catalytic domain of human (Hcm1, blast score 199) and Saccharomyces cerevisiae (Abd1, blast score 152) m7G methyltransferases. The alignment of the primary structures of TbCmt1, Abd1, Hcm1, and E. cuniculi m7G methyltransferases (Ecm1) is shown in Figure 1 ▶. TbCmt1 displays conservation at 76 of 324 positions among these enzymes and contains a well-characterized AdoMet binding motif (70-VLDLASGRGGD LGK-83). Additionally, residues that contact AdoMet and the cap, as observed in the co-crystal structures of Ecm1 complexed with AdoMet, AdoHcy, and the m7GpppA cap analog are present in TbCmt1 (Fabrega et al. 2004). All 12 residues that were found to be critical for m7G methyltransferase activity in Ecm1 are present in TbCmt1 (indicated by “0” in Fig. 1 ▶), leading to the prediction that this gene product encodes a m7G cap methyltransferase component of the T. brucei capping apparatus (Mao et al. 1995, 1996; Wang and Shuman 1997; Fabrega et al. 2004; Hausmann et al. 2005).
FIGURE 1.
Sequence similarity among the protozoan, the fungal, and the mammalian RNA m7G methyltransferases. The amino acid sequences of m7G methyltransferases encoded by T. brucei (TbCmt1: Sanger Institute TV10.v4.004), T. cruzi (TcCMT1: XP_811819.1), and L. major (LmCmt1: CAJ08963.1) are aligned to the sequences of H. sapiens (Hcm1), S. cerevisiae (Abd1), and E. cuniculi (Ecm1) m7G methyltransferases. Gaps in the alignment are indicated by dashes. Side-chain identity in all proteins is highlighted by shaded boxes. Ecm1 residues that interact with AdoMet and the cap analog are indicated below as “a” and “c,” respectively. Conserved residues in Ecm1 that were found by mutational analysis to be essential for function are indicated by “0.” TbCmt1 residues that were mutated to alanine in this study are denoted by arrows.
To determine if other kinetoplastid protozoa encode m7G methyltransferases, we performed a reciprocal search using the TbCmt1 sequence and found TbCmt1 homologs in Trypanosoma cruzi (TcCmt1: 66% identity and 80% similarity) and Leishmania major (LmCmt1: 45% identity and 58% similarity) from the National Center for Biotechnology Information database (Fig. 1 ▶). The T. cruzi (313 amino acids) and L. major (323 amino acids) proteins are similar in size to the T. brucei protein. The kinetoplastid proteins are slightly larger than the “minimal” cap methyl-transferase Ecm1 (298 amino acids) but are smaller than Abd1 (549 amino acids) and Hcm1 (520 amino acids) (Pillutla et al. 1998; Saha et al. 1999; Hausmann et al. 2002). Note that kinetoplastid m7G methyltransferases lack a counterpart to the N-terminal ~150–amino acid segment present in the budding yeast and human enzymes. These N-terminal segments were dispensable for their methyltransferase activities in vitro and in vivo (Mao et al. 1996; Saha et al. 1999).
Expression of TbCmt1 in bacteria
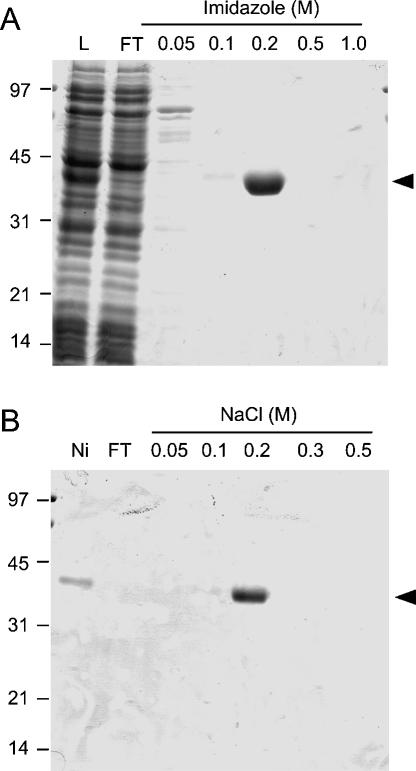
To evaluate the m7G methyltransferase activity, the recombinant TbCmt1 protein was produced in bacteria. The TbCMT1 coding sequence was amplified from T. brucei genomic DNA and inserted into an inducible T7 RNA polymerase-based pET vector such that 10 tandem histidines (His-tag) were fused N-terminal to the TbCmt1 protein. The pET-TbCMT1 expression plasmid was introduced into Escherichia coli BL21(DE3)Gold, a strain that contains the T7 RNA polymerase gene under the control of a lacUV5 promoter. A 39-kDa protein corresponding to His-tagged TbCmt1 was detectable by SDS-PAGE in soluble extracts of IPTG-induced bacteria (Fig. 2A ▶, lane L). The TbCmt1 protein was purified from soluble extract by nickel-agarose, followed by phosphocellulose chromatography. The recombinant protein adsorbed to the phosphocellulose and was recovered in a 0.2 M NaCl fraction following step elution (Fig. 2B ▶). The phosphocellulose preparation was virtually homogeneous with respect to the 39-kDa protein.
FIGURE 2.
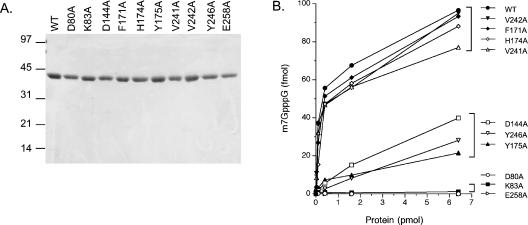
Expression and purification of TbCmt1. The protein compositions of the recombinant TbCmt1 protein at sequential stages of purification were analyzed by SDS-PAGE. (A) Ni-NTA Aga-rose fractions: Soluble lysate of IPTG-induced BL21(DE3)Gold/pET-TbCMT1 (lane L), Ni-NTA agarose flowthrough (FT), and indicated imidazole eluates. (B) Phosphocellulose fractions: Aliquots of the 0.2-M imidazole eluate from Ni-Agarose chromatography (Ni), phosphocellulose flowthrough (FT), and indicated NaCl eluates. The gels were fixed and stained with Coomassie blue dye. The positions and sizes (in kilodaltons) of the coelectrophoresed marker proteins are shown to the left. The position of TbCmt1 is indicated by an arrow to the right.
Characterization of TbCmt1 activity
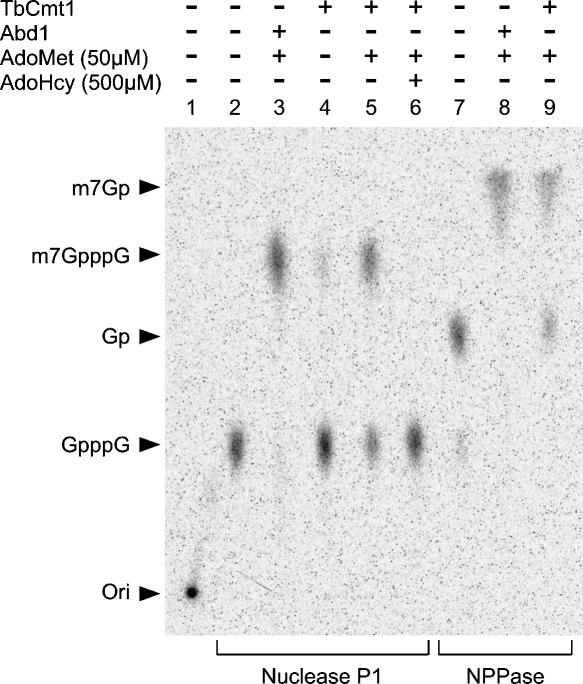
RNA m7G cap methyltransferases catalyze the transfer of a methyl group from AdoMet to GpppRNA to form m7GpppRNA and AdoHcy. To determine if TbCmt1 is capable of catalyzing this reaction, purified TbCmt1 was incubated with 32P-GpppRNA53 (bold type indicates the labeled phosphorus) in the presence and the absence of AdoMet. The substrate for this assay, a 53-mer 32P-GpppRNA53 was prepared from a triphosphate-terminated runoff transcript synthesized in vitro by T7 RNA polymerase, then cap-labeled with [α-32P]GTP using the mammalian capping enzyme (see Materials and Methods). The reaction products were digested to cap dinucleotides with nuclease P1 and analyzed by polyethyleneimine-cellulose (TLC) plate (Fig. 3 ▶). In the absence of nuclease P1 digestion, all 32P-labeled material remained at the origin in polynucleotide form (Fig. 3 ▶, lane 1). Treatment of the reaction products with nuclease P1 resulted in liberation of a labeled species that co-migrated with m7GpppG, generated in a parallel reaction mixture containing purified yeast Abd1 (Fig. 3 ▶, lanes 3,5). A trace amount (~10%) of methylation was observed in the absence of AdoMet (Fig. 3 ▶, lane 4), suggesting that some AdoMet may have been bound to the enzyme and co-purified during the TbCmt1 preparation. One of the products, AdoHcy (500 μM), was inhibitory in a reaction containing 50 μM AdoMet (Fig. 3 ▶, lane 6). We further verified that methylation occurs at the terminal guanosine nucleoside by digesting the reaction products with nucleotide pyrophosphatase (NPPase), a nuclease that cleaves between the γ and the β phosphates in the capped structure. Both the TbCmt1 and Abd1 reaction products liberated m7Gp, which migrated near the top of the TLC (Fig. 3 ▶, lanes 7–9). These results indicate that TbCmt1 catalyzes methylation at the N-7 position of the terminal cap guanosine.
FIGURE 3.
Analysis of the methyltransferase reaction product. Methyltransferase reaction mixture (10 μL) contained 100 fmol of 32P-GpppRNA53 with the following components: 2 pmol Abd1 + 50 μM AdoMet (lanes 3,8), 2 pmol TbCmt1 (lane 4), 2 pmol TbCmt1 + 50 μM AdoMet (lanes 5,9), 2 pmol TbCmt1 + 50 μM AdoMet and 500 μM AdoHcy (lane 6). Control reactions in the absence of enzyme are indicated (lanes 1,2,7). After incubation at 27°C for 30 min, samples were incubated with either 140 ng of nuclease P1 (lanes 2–6) or 0.1 unit of nucleotide pyrophosphatase (NPPase) (lanes 7–9) for 60 min at 37°C. The digests were then spotted on polyethyleneimine-cellulose TLC plates and developed with 0.45 M (NH4)2SO4. A PhosphorImager scan of the chromatogram is shown. The chromatographic origin (ori) and positions of m7Gp, m7GpppG, GMP, and GpppG are noted.
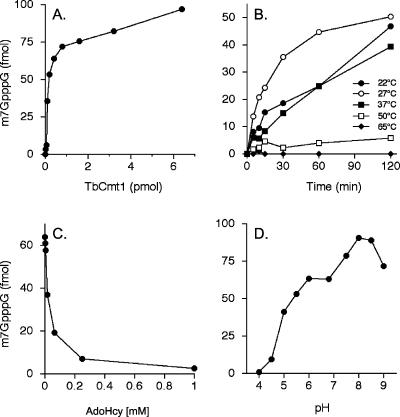
The amount of m7GpppG formed during a 30-min reaction in the presence of 50 μM AdoMet was proportional to the amount of added TbCmt1 (Fig. 4A ▶). At saturating enzyme, input 32P-GpppRNA53 substrate was quantitatively methylated to form m7GpppRNA. Kinetic analysis of methyl transfer indicated that the reaction was completed within 120 min at 27°C. The yield of methylated product varied with AdoMet concentration. Half-maximal activity was observed at ~1 μM AdoMet and plateau at 10 μM (data not shown). TbCmt1 formed ~70 fmol of methylated capped ends per 50 fmol of enzyme in 30 min. This value corresponds to a turnover number of 0.047 min−1, which is similar to the turnover number for recombinant Hcm1 (0.037 min−1) (Saha et al. 1999). The activity was measured at temperatures ranging from 22° to 65°C. The extent of m7GpppG formation was optimal at 27°C, whereas activity was ~50% of the optimum value at either 22° or 37°C, and the enzyme was completely inactive at 65°C. Methylation was optimal in 50 mM Tris-HCl buffer between pH 8.0 and 8.5 and declined sharply at and below neutral pH (Fig. 4D ▶). The standard methyltransferase reaction mixture contained 10 mM NaCl from the enzyme preparation. The effect of increasing ionic strength on methyltransferase activity was measured by supplementing the reactions with NaCl. Activity in the presence of 60 mM NaCl was reduced to ~25% of that of 10 mM NaCl and was virtually null at 125 mM NaCl (data not shown).
FIGURE 4.
Cap methylation activity of recombinant TbCmt1. (A) Protein titration. Methyltransferase reaction mixtures (10 μL) containing 100 fmol of 32P-GpppRNA53, 50 μM AdoMet, and TbCmt1 as specified were incubated for 30 min at 27°C. The extent of m7GpppG formed was plotted as a function of input protein. (B) Kinetics. Reaction mixtures (10 μL) contained 100 fmol of 32P-GpppRNA53, 50 μM AdoMet, and 2 pmol TbCmt1. The reaction was initiated by enzyme addition and was incubated at the indicated temperature. Aliquots (2 μL) were withdrawn at the times indicated, and the reaction was quenched immediately by addition of sodium acetate to pH 5.5 and heating for 5 min at 80°C. Products were then digested with nuclease P1. The extent of m7GpppG formed is plotted as a function of incubation time. (C) Inhibition by AdoHcy. Reaction mixtures (10 μL) contained 100 fmol of 32P-GpppRNA53, 50 μM AdoMet, 2 pmol TbCmt1, and AdoHcy as indicated. The yield of m7GpppG is plotted as a function of AdoHcy concentration. (D) pH dependence. Reaction mixtures (10 μL) containing 50 mM Tris buffer (either Tris-acetate at pH 4.0, 4.5, 5.0, 5.5, 6.0, 6.8, and 7.0 or Tris-HCl at pH 7.5, 8.0, 8.5, and 9.0), 5 mM dithiothreitol, 100 fmol of 32P-GpppRNA53, 50 μM AdoMet, and 2 pmol TbCmt1 were incubated for 30 min at 27°C. The extent of m7GpppG formed is plotted as a function of pH.
Structure-based mutagenesis of TbCmt1
Crystal structures of Ecm1 have been solved complexed with AdoMet, AdoHcy, m7GpppG, and more recently with aza-AdoMet (Fabrega et al. 2004; Hausmann et al. 2005). The active site of Ecm1 is composed of two separate ligand-binding pockets: one for AdoMet and one for the GpppN cap. Fabrega and colleagues (Fabrega et al. 2004) suggest that cap guanine N-7 methylation is accomplished predominantly by optimally orienting the AdoMet and GpppN cap in close proximity, facilitating the methyl transfer reaction. Alanine mutations of the residues that contact the AdoMet ribose (Ecm1: Asp94), methionine (Ecm1: Lys54, Asp78), or adenine (Ecm1: Ile95, Asp122, Ser123, Tyr124), as well as cap ribose (Ecm1: Tyr284, Asn50) and cap guanine (Ecm1: Phe141, Tyr284) determined that they were all critical for Ecm1 m7G methyltransferase function. The majority of these residues are present in TbCmt1 and are conserved among other m7G methyltransferases (Fig. 1 ▶).
If TbCmt1 is a true member of the m7G methyltransferase family, then removal of side chains that make contacts to the substrates should elicit a significant loss in activity. To illuminate the structure–function relationship for TbCmt1, 10 residues in TbCmt1 were selected for alaninemutagenesis. These residues were selected based on the active site structure of Ecm1 and their conservation in m7G methyltransferase family members (denoted by arrows in Fig.1). These include residues in TbCmt1 that may comprise the putative AdoMet-binding pocket (Asp80, Lys83, Asp144) and cap-binding pocket (Phe171, His174, Tyr175, Tyr246, Glu258). Alanine substitutions were also introduced at nonconservative residues (Val241, Val242). The D80A, K83A, D144A, F171A, H174A, Y175A, V241A, V242A, Y246A, and E258A mutations were introduced into the TbCMT1 gene, and the TbCmt1-Ala proteins were expressed as His10-tagged derivatives in E. coli in parallel with wild-type TbCmt1. In every case, the 39-kDa TbCmt1 protein was the predominant poly-peptide. The recombinant mutant TbCmt1 proteins were purified from soluble bacterial extracts by nickel-agarose and phosphocellulose chromatography as described for wild-type TbCmt1. SDS-PAGE analysis of the polypeptide compositions of the phosphocellulose protein preparations revealed that a similar extent of purification was achieved for each mutant as for the wild-type protein (Fig. 5A ▶).
FIGURE 5.
Effect of alanine mutations on m7G methyltransferase activity of TbCmt1. (A) Protein purification. Aliquots (3 μg) of phospho-cellulose fractions of wild-type (WT) TbCmt1 and the indicated Ala-mutants were analyzed by SDS-PAGE. Proteins were visualized by staining with Coomassie blue. The positions and sizes (in kilodaltons) of marker proteins are indicated to the left. (B) Methylation of capped RNA. Reaction mixtures (10 μL) contained 100 fmol of 32P-GpppRNA53, 50 μM AdoMet, and wild-type or mutant enzymes as specified. The extent of m7GpppG formed is plotted as a function of input enzyme.
Effect of alanine mutations on TbCmt1 m7G methyltransferase activity
Cap methyltransferase activity of the wild-type and mutant TbCmt1 proteins was assayed in the presence of 50 μM AdoMet and 10 nM cap-terminated RNA. The specific activities of the TbCmt1-Ala mutants were quantified from protein titrations in the linear range of enzyme dependency (Fig. 5B ▶). The D80A, K83A, and E258A mutants were unable to support methylation at levels of input protein sufficient to convert all the input substrate to the methylated form by wild-type TbCmt1. We calculated that the specific activities of D80A, K83A, and E258A mutants were <0.1% of the wild-type activity. Severe mutational effects were also observed for D144A (5% of wild-type activity), Y175A (3% of wild-type activity), and Y246A (2% of wild-type activity). In contrast, the F171, H174A, V241A, and V242A mutants displayed near-wild-type activities and thus were not essential for the activity of TbCmt1. With the exception of the F171A mutant, these results are in agreement with the mutational analyses reported for Ecm1 and Abd1 (Mao et al. 1996; Wang and Shuman 1997; Fabrega et al. 2004; Hausmann et al. 2005). It is thus likely that TbCmt1 adopts a similar active site as that of Ecm1.
Methyl acceptor specificity
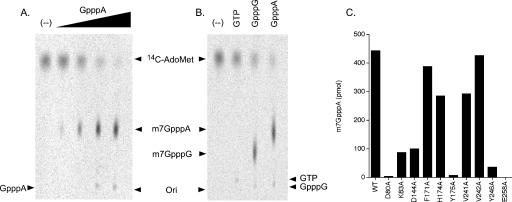
To determine the minimum substrate for TbCmt1 methyl-transferase activity, we tested whether nucleotides and cap analogs can serve as methyl acceptors for TbCmt1. Recombinant TbCmt1 was incubated with GpppA and GpppG, and various nucleotides in the presence of [14C-CH3]AdoMet, and the individual products were resolved by TLC. The positions of unlabeled standards were identified by UV shadowing. Incubation of increasing amounts of GpppA with 50 μM [14C-CH3]AdoMet and TbCmt1 resulted in the formation of [14C-CH3]-labeled product that co-migrated with the m7GpppA standard (Fig. 6A ▶). Approximately 90% of input label was converted to the m7GpppA product in the presence of 4 mM GpppA. TbCmt1 also catalyzed near-quantitative methyltransfer to a GpppG acceptor to form m7GpppG (Fig. 6B ▶). In contrast, GTP was a poor methyl acceptor even at the highest concentration sufficient to methylate cap analogs (4 mM). Neither UTP, CTP, ATP, nor dGTP was detectably methylated under these conditions (data not shown). The TbCmt1 reaction was identical in all respects to methylation of GpppA and GpppG cap analogs by the Abd1 enzyme as analyzed in parallel reactions (data not shown). These results suggest that TbCmt1 has a higher affinity to the cap dinucleotides than GTP, and that the RNA component is not strictly essential for catalysis.
FIGURE 6.
Methylation of cap analog by TbCmt1. (A) GpppA titration. Reaction mixtures (10 μL) containing 50 mM Tris-HCl (pH 8.0), 5 mM dithiothreitol, 50 μM [14C-CH3]AdoMet, 8 pmol TbCmt1, and increasing concentration of GpppA (0.004, 0.04, 0.4, and 4 mM, proceeding from left to right in the titration series) were incubated for 30 min at 27°C. A control reaction lacking GpppA is indicated by (--). An aliquot (2 μL) of the mixture was applied to a PEI-cellulose TLC plate, which was developed with 0.4 M LiCl and visualized by PhosphorImager. (B) Methyl-acceptor specificity. Reaction mixtures (10 μL) containing 50 μM [14C-CH3]AdoMet, 8 pmol TbCmt1, and either 4 mM GTP, GpppG, GpppA or no methyl-acceptor as indicated by (–). Reaction products were analyzed by TLC and visualized by PhosphorImager. The positions of GTP, GpppA, and GpppG were located by UV illumination as indicated. (C) Mutational analysis. Reaction mixtures (10 μL) contained 50 μM [14C-CH3] AdoMet, 4 mM GpppA and 8 pmol of wild-type or mutant TbCmt1. Reaction products were analyzed by PEI-cellulose TLC. The extent of [14C-CH3] m7GpppA formation, quantitated by PhosphorImager, is plotted for each enzyme.
The effects of mutations on TbCmt1’s ability to transfer a methyl group from AdoMet to the GpppA acceptor were measured under the reaction conditions that convert 90% of input AdoMet to m7GpppA by wild-type TbCmt1 (Fig. 6C ▶). All six mutations that were essential for methylation of capped RNA also elicited severe defects in m7GpppA formation. The activities of the D80A, Y175A, and E258A mutants were <1% of the wild type, whereas the Y246A mutant formed trace amounts of methylated product (4% of wild-type level). The K83A and D144A mutations had a less drastic effect on GpppA methylation than on methylation of cap-terminated RNA (~20% that of the wild-type value). The F171A, H174A, V241A, and V242A mutants had modest (within twofold) or no effect on m7GpppA formation. These results further support the conclusion that these essential residues comprise the TbCmt1 active site.
DISCUSSION
Kinetoplastid protozoan mRNAs acquire their 5′ caps via trans-splicing of an SL RNA sequence containing the hyper-methylated cap 4 structure (Bangs et al. 1992; Mair et al. 2000). While the RNA triphosphatase (TbCet1) and guany-lyltransferase (TbCe1) have been characterized (Silva et al. 1998; Ho and Shuman 2001), none of the cap-methylating enzymes have been identified in kinetoplastid protozoa. Here we report the first identification of a cap RNA methyltransferase in T. brucei. Recombinant TbCmt1 transfers a methyl group from AdoMet to the N-7 position of the GpppN cap to form the conventional m7GpppN structure present in all eukaryotes. The catalytic properties of the trypanosome enzyme are similar to other cellular m7G methyltransferases with respect to AdoMet-dependency and product inhibition by AdoHcy. TbCmt1 also methylates cap analogs. However, GTP was not an effective methyl acceptor for TbCmt1. Thus, the trypanosome enzyme is capable of discriminating cap analogs from GTP, ensuring that methylation takes place only on capped substrates.
We can make reasonable predictions about the roles of TbCmt1 residues by comparison with Ecm1 (Fabrega et al. 2004; Hausmann et al. 2005). Based on structural homology, TbCmt1 Asp80, Lys83, and Asp144 likely coordinate AdoMet in the active site, as alanine substitutions of any of these residues either abolished or diminished the enzyme’s ability to methylate GpppA or cap-terminated RNA. TbCmt1 Lys83 may interact with Asp80, which in turn may make a water-mediated contact to the AdoMet amine as in Ecm1 (Fabrega et al. 2004; Hausmann et al. 2005). Alanine substitution of the corresponding Asp and Lys in Ecm1 failed to support growth in an ABD1-deficient yeast strain. TbCmt1 Asp144 is the putative equivalent of Asp122 in Ecm1 and is thought to form a hydrogen bond to the adenine N6 atom of AdoMet. Ecm1 Asp122 is not strictly essential for catalysis but elicits a temperature sensitive growth defect in yeast (Fabrega et al. 2004). Based on these results, we predict that Asp80, Lys83, and Asp144 participate in forming the AdoMet binding pocket in TbCmt1.
Of the five TbCmt1 mutations introduced in the putative cap guanine binding pocket, replacement of Tyr175, Tyr246, and Glu258 by alanine eliminated methyltransferase activity. With reference to the Ecm1 crystal structure complexed with AdoMet or m7GpppG, Tyr175 (equivalent of Ecm1 Tyr145) may bridge the two substrates in the active site by making a water-mediated contact with the cap guanine O6 atom as well as van der Waals contacts with AdoMet (Fabrega et al. 2004). Hausmann et al. (2005) showed that Ecm1 Tyr145 can be replaced either by phenylalanine or by leucine, suggesting that the dominant role of tyrosine at this position is to provide aliphatic contacts with AdoMet. Consistent with our finding that Tyr175 is essential for the TbCmt1 activity, the corresponding tyrosines in fungal (Abd1: Tyr254), metazoan (Hcm1: Tyr289), and viral (vaccinia: Tyr683) m7G methyltransferases were indispensable for their activities (Mao and Shuman 1996; Mao et al. 1996; Saha et al. 1999, 2003). TbCmt1 Glu258 (Ecm1: Glu225) may form hydrogen bonds with the N1 atom and may also contact the exocyclic 2-NH2 of the guanine. The hydroxyl of Tyr246 (Ecm1: Tyr212) may assist this interaction by forming a hydrogen bond to the Glu225 carboxylate. Ecm1 Y212A and E225A mutants both elicit temperature-sensitive phenotypes in yeast strains lacking the ABD1 gene (Fabrega et al. 2004), consistent with the requirement for each residue in TbCmt1. Alanine substitution at the conserved His174 did not affect TbCmt1 activity. While the Ecm1 structure suggests that the corresponding His144 acts as a hydrogen donor to the guanine O6 and, thereby, provides additional guanine specificity, this residue does not appear to have a significant effect on catalysis. In support of this view, Ecm1 His144 was dispensable for methyltransferase activity (Hausmann et al. 2005). However, Phe171 was dispensable for TbCmt1 activity, whereas the equivalent Phe141 of Ecm1 is absolutely essential for methyltransferase activity in vitro and the F141A mutant failed to complement the ABD1-deficient yeast strain (Hausmann et al. 2005). In the Ecm1-cap structure, Phe141 makes multiple van der Waals contacts with the cap guanine and ribose (Fabrega et al. 2004). It is conceivable that an alternate phenylalanine or other aliphatic residues in TbCmt1 may contribute to this essential interaction. Further mutagenesis studies should clarify this apparent discrepancy.
We explored a yeast genetic complementation approach to address whether the trypanosome m7G methyltransferase can function in yeast cells. We transformed a plasmid containing TbCMT1 under a constitutive yeast TPI promoter into an S. cerevisiae strain with a deletion of the chromosomal copy of ABD1 (Mao et al. 1995). We found that expression of TbCMT1 on either CEN or 2-μ plasmid did not sustain growth in the Abd1 deletion strain (data not shown). A plausible explanation for the failure of TbCMT1 to function in yeast could be that the trypanosome protein fails to interact with the yeast RNA polymerase II. In fungi, plants, and metazoans, capping enzyme components interact with the carboxyl-terminated domain (CTD) of the large subunit of RNA polymerase II, which is composed of YSPTSPS heptapeptide repeats. The phosphorylated CTD acts as a landing pad and targets the capping apparatus to the transcription elongation complex (Cho et al. 1997; McCracken et al. 1997; Ho and Shuman 1999; Fabrega et al. 2003). It is not clear how the trypanosome capping apparatus is recruited to sites of transcription as the large subunit of T. brucei RNA polymerase II lacks the conventional CTD element, although it does appear to be phosphorylated (Evers et al. 1989; Smith et al. 1989; Chapman and Agabian 1994).
A further search of the NCBI database using the TbCmt1 sequence uncovered a second m7G methyltransferase ORF encoded in all three kinetoplastid protozoa (Fig. 7 ▶). The 1050–amino acid T. brucei protein (designated TbCgm1) is significantly larger than TbCmt1, and homology with the m7G methyltransferases is restricted to the C-terminal 350–amino acid segment (Fig. 7A ▶). The most significant difference between TbCmt1 and TbCgm1 is that TbCgm1 contains motifs characteristic of RNA guanylyltransferases (Fig. 7B ▶). The N-terminal 650–amino acid segment of TbCgm1 has all six conserved nucleotidyl transferase motifs (I, III, IIIa, IV, V, and VI) involved in GTP-binding and catalysis in the standard order and spacing, except that the 139–amino acid interval between motifs I and III is slightly longer than that of known RNA guanylyltransferases (Hakansson et al. 1997; Wang et al. 1997). Motif I (127-KADGTR-132) contains a conserved lysine residue to which GMP becomes covalently linked via a phosphoamide bond to the ɛ-amino group of the enzyme (Schwer and Shuman 1994; Wang et al. 1997). All of the amino acid residues that are essential for the function of the S. cerevisiae RNA guanylyltransferase Ceg1 are conserved in TbCgm1, as are the residues that make direct contacts with the GTP substrate in the Chlorella virus and Candida albicans RNA guanylyltransferases (Schwer and Shuman 1994; Hakansson et al. 1997; Wang et al. 1997; Hakansson and Wigley 1998; Fabrega et al. 2003). These similarities strongly suggest that TbCgm1 possesses RNA guanylyltransferase activity in addition to the putative m7G methyltransferase activity.
FIGURE 7.
Trypanosoma and Leishmania encode a second m7G methyltransferase with an associated guanylyltransferase domain. (A) m7G methyltransferase domains. Amino acid sequence alignment of the C-terminal methyltransferase domains of T. brucei Cgm1 (TbCgm1, accession no. XP_824771), T. cruzi Cgm1 (TcCgm1, XP_820423), and L. major Cgm1 (LmCgm1, CAJ03800.1). Side-chain identity and similarity in all proteins are indicated by asterisks and dots, respectively. Equivalent residues that interact with AdoMet and the cap analog in the Ecm1 co-crystal structure are indicated below the alignment as “a” and “c,” respectively. (B) Conserved sequence elements in mRNA guanylyltransferases. Alignment of six colinear guanylyltransferase motifs (I, III, IIIa, IV, V, and VI) from Chlorella virus PBCV-1 (ChV), S. cerevisiae (Sce), Schizosaccharomyces pombe (Spo), C. albicans (Cal), Caenorhabditis elegans (Cel), Plasmodium falciparum (Pfa), T. brucei Ce1 (TbCe1), T. brucei Cgm1 (TbCgm1), T. cruzi (TcCgm1), and L. major (LmCgm1). A number of amino acid residues separating the motifs are indicated.
It is noteworthy that two distinct RNA guanylyltransferase polypeptides have been detected in T. brucei and the related parasite Crithidia fasciculata. Silva et al. (1998) showed that 68-kDa and ~116-kDa polypeptides reacted with GTP to form covalent GMP catalytic intermediates in T. brucei whole-cell extract, as gauged by SDS-PAGE analysis. While the 68-kDa polypeptide was purified, cloned, and characterized as TbCe1, the identity of the 116-kDa polypeptide remained in question. Based on the calculated molecular weight of the TbCgm1 polypeptide, we predict that the 115-kDa protein-GMP complex corresponds to the TbCGM1 gene product.
To examine whether TbCgm1 indeed possesses m7G cap methyltransferase activity, the C-terminal TbCgm1 methyltransferase domain, TbCgm1 (717–1050), was expressed in bacteria and purified. Preliminary experiments indicate that TbCgm1 (717–1050) is capable of transferring a methyl group from AdoMet to a GpppN cap terminated poly(A) to form an m7GpppA structure (Y. Takagi, M.P. Hall, and C.K. Ho, unpubl.).
We speculate that a bifunctional TbCgm1 guanylyltransferase-m7G methyltransferase and TbCet1 triphosphatase are likely to act together to form the cap 0 structure. TbCet1 may also function together with TbCe1 guanylyltransferase and TbCmt1 m7G methyltransferase, unless TbCe1 has an intrinsic 5′-processing activity that can act as a triphosphatase (Silva et al. 1998). Alternatively, an additional RNA triphosphatase enzyme may be encoded by T. brucei. However, we have been unable to identify a homolog that resembles either TbCET1 or a cysteine phosphatase-type RNA triphosphatase from the trypanosome genome.
The identification of TbCMT1 and TbCGM1 raises the interesting question of why two separate m7GpppN cap-forming activities are encoded in trypanosomes. One plausible explanation is that two separate sets of capping enzymes act on different RNA substrates. Tschudi and Ullu (2002) proposed that m7GpppN formation on SL RNA and snRNA utilizes fundamentally different mechanisms. In most eukaryotes, a single set of capping enzymes is sufficient to cap mRNA and snRNA as they are both transcribed by pol II and the cap is added co-transcriptionally to all pol II transcripts. In trypanosomes, while the SL RNA is transcribed by pol II and capped co-transcriptionally, the trimethylguanosine-capped U2, U3, and U4 snRNAs appear to be transcribed by pol III (Tschudi et al. 1986; Mottram et al. 1989; Fantoni et al. 1994; Nakaar et al. 1994). It is not clear whether U snRNAs are capped co-transcriptionally, as other pol III transcripts, such as tRNA, 5S RNA, and 7SL RNA, lack a cap structure. We speculate that one set of capping enzymes is responsible for SL RNA capping and another set is designated for snRNA capping. It is also conceivable that additional capping enzymes may be required for capping polycistronic pre-mRNAs. Alternatively, both sets of capping enzymes may act on SL RNA and regulate the level of trans-splicing-competent capped SL RNA. Further biochemical and genetic characterization of the T. brucei capping apparatus should clarify the role of individual capping enzyme components in trypanosome cap biosynthesis.
MATERIALS AND METHODS
T7-based vector for expression of T. brucei RNA m7G cap methyltransferase
Preliminary sequence data for TbCMT1 were obtained from the Sanger Institute T. brucei Genomes Web site (http://www.sanger.ac.uk/Projects/T_brucei). A DNA fragment containing the TbCMT1 ORF on chromosome 10 was amplified by PCR from total T. brucei brucei genomic DNA (a gift from Laurie Read, SUNY Buffalo) with the use of oligonucleotide primers designed to introduce an NdeI restriction site at the predicted translation start codon and a BamHI site 3′ of the predicted stop codon. The ~1.0-kbp PCR product was digested with NdeI and BamHI and inserted into the T7 RNA polymerase-based expression vector pET16b (Novagen) to generate the plasmid pET-TbCMT1. The nucleotide sequence of the trypanosome DNA insert was determined. The predicted amino acid sequence of the 324–amino acid TbCmt1 protein encoded by this plasmid is shown in Figure 1 ▶.
Expression and purification of T. brucei RNA m7G cap methyltransferase
The pET-TbCmt1 plasmid was transformed into E. coli BL21-Gold(DE3). A 2-L culture amplified from a single transformant was grown at 37°C in LB medium containing 0.1 mg/mL ampicillin until the OD600 reached 0.4. The culture was adjusted to 0.4 mM IPTG and incubated at 17°C for 18 h. Cells were harvested by centrifugation, and the pellet was stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 100 mL of buffer A (50 mM Tris·HCl at pH 7.5, 10% sucrose) containing 0.25 M NaCl. Cell lysis was achieved by the addition of lysozyme and Triton X-100 to final concentrations of 100 μg/mL and 0.1%, respectively. The lysate was sonicated to reduce viscosity, and insoluble material was removed by centrifugation for 45 min at 14,000 rpm in a Beckman TA14–50 rotor. The soluble extract (450 mg of protein) was applied to a 5-mL column of Ni-NTA-agarose resin (Qiagen) that had been equilibrated with buffer A containing 0.1% Triton X-100. The column was washed with 50 mL of the same buffer and then eluted stepwise with 15-mL aliquots of buffer B (50 mM Tris-HCl at pH 8.0, 0.25 M NaCl, 10% glycerol) containing 0.05, 0.1, 0.2, 0.3, and 0.5 M imidazole. The protein composition of the column fractions was monitored by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). The recombinant trypanosome protein was retained on the column and recovered in the 0.2 M imidazole eluate. This fraction was adjusted to 0.025 M NaCl by addition of 135 mL of buffer B, then applied to a 5-mL column of phosphocellulose pre-equilibrated in buffer B with 0.025 M NaCl. The column was washed with 50 mL of the same buffer and then eluted stepwise with 15 mL of buffer B containing 0.05, 0.1, 0.2, 0.3, 0.5, and 1.0 M NaCl. The wild-type and mutant trypanosome proteins were retained on the column and recovered predominantly in the 0.2 M NaCl fraction (~5 mg). The phosphocellulose preparation was concentrated by centrifugal ultrafiltration (Millipore), and the final protein concentrations were determined with the Bio-Rad dye reagent using bovine serum albumin as the standard.
Alanine mutants of TbCmt1
Alanine-substitution mutations were introduced into the TbCMT1 gene by polymerase chain reaction using the two-stage overlap extension method. The residues targeted for amino acid substitution were Asp80, Lys83, Asp144, Phe171, His174, Tyr175, Val241, Val242, Tyr246, and Glu258. The pET-TbCMT1 plasmid was the template for the first-stage amplifications. The DNA products of the second-stage amplification were digested with NdeI and BamHI and inserted into pET16b. The presence of the desired mutations was confirmed by DNA sequencing; the inserted restriction fragment was sequenced completely to exclude the acquisition of unwanted mutations during amplification and cloning. The His-tagged mutant proteins were purified from soluble bacterial lysates by nickel-agarose and phosphocellulose chromatography steps as described for the wild-type TbCmt1 protein. Protein concentration was determined by using the Bio-Rad dye reagent with bovine serum albumin as the standard.
Preparation of RNA substrates
Triphosphate-terminated RNA, pppRNA53: 5′-ppp(GpApGpUp Up)10GAG, was synthesized by in vitro transcription from plasmid pTZ18a (a gift of Paul Gollnick, SUNY Buffalo) (Elliott et al. 2001) using T7 RNA polymerase. The 53-mer pppRNA53 was freed of protein and nucleotides by electrophoresis through a nondenaturing 8% polyacrylamide gel. The pppRNA53 was localized by UV illumination, excised, and recovered by soaking the gel slice for 12 h in Tris-HCl (pH 8.0) and 1 mM EDTA. The 32P-labeled capped RNA (GpppRNA53) was prepared in a reaction mixture (200 μL) containing 50 mM Tris-HCl (pH 8.0), 5 mM dithiothreitol, 2.5 mM MgCl2, 20 pmol of pppRNA53, 0.33 μM [α-32P]GTP, and 40 pmol of purified recombinant mammalian capping enzyme Mce1 (Ho et al. 1998). After incubation at 37°C for 20 min, the product was resolved by electrophoresis through a native 8% polyacryl-amide gel. The 32P-cap labeled GpppRNA53 was located by autoradiography of the wet gel, eluted from an excised gel slice in 10 mM Tris-HCl (pH 8.0), 1 mM EDTA and precipitated by ethanol. Recovery of labeled RNA was assessed by scintillation counting. The molar concentration of cap-labeled RNA was calculated according to the specific activity of the input [α-32P]GTP donor in the capping reaction.
Methyltransferase assay
Standard reaction mixtures (10 μL) containing 50 mM Tris-HCl (pH 8.0), 5 mM dithiothreitol, 100 fmol of 32P-GpppRNA53, and TbCmt1 as specified were incubated for 30 min at 27°C. The reaction mixtures were adjusted to 50 mM sodium acetate (pH 5.5) and samples were incubated with 140 ng of nuclease P1 (US Biologicals) or 0.1 unit of nucleotide pyrophosphatase (Sigma) for 60 min at 37°C. Aliquots (2 μL) were spotted onto a PEI cellulose thin-layer chromatography (TLC) plate (EMD Bioscience), which was developed with 0.45 M ammonium sulfate. The extent of methylation of the cap (m7GpppG /[m7GpppG + GpppG]) was quantitated by scanning the TLC plate with a Storm 860 PhosphorImager.
Materials
[14C-CH3]AdoMet was purchased from GE Healthcare—Amersham Bioscience. AdoMet and AdoHcy were purchased from Sigma. Cap analogs GpppG, GpppA, m7GpppA and m7GpppA were purchased from Ambion.
Acknowledgments
We thank Stewart Shuman (Sloan-Kettering Institute) for providing useful comments, plasmids, and bacterial strains; Laurie Read (SUNY Buffalo) for T. brucei genomic DNA; Paul Gollnick (SUNY Buffalo) for the pTZ18a plasmid; and Ed Niles (SUNY Buffalo) for helpful discussions.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2250606.
REFERENCES
- Adams, M.D., Rudner, D.Z., and Rio, D.C. 1996. Biochemistry and regulation of pre-mRNA splicing. Curr. Opin. Cell Biol. 8: 331–339. [DOI] [PubMed] [Google Scholar]
- Agabian, N. 1990. Trans splicing of nuclear pre-mRNAs. Cell 61: 1157–1160. [DOI] [PubMed] [Google Scholar]
- Bangs, J.D., Crain, P.F., Hashizume, T., McCloskey, J.A., and Boothroyd, J.C. 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267: 9805–9815. [PubMed] [Google Scholar]
- Chapman, A.B. and Agabian, N. 1994. Trypanosoma brucei RNA polymerase II is phosphorylated in the absence of carboxyl-terminal domain heptapeptide repeats. J. Biol. Chem. 269: 4754–4760. [PubMed] [Google Scholar]
- Cho, E.J., Takagi, T., Moore, C.R., and Buratowski, S. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 11: 3319–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, M.B., Gottlieb, P.A., and Gollnick, P. 2001. The mechanism of RNA binding to TRAP: Initiation and cooperative interactions. RNA 7: 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers, R., Hammer, A., and Cornelissen, A.W. 1989. Unusual C-terminal domain of the largest subunit of RNA polymerase II of Crithidia fasciculata. Nucleic Acids Res. 17: 3403–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrega, C., Shen, V., Shuman, S., and Lima, C.D. 2003. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol. Cell 11: 1549–1561. [DOI] [PubMed] [Google Scholar]
- Fabrega, C., Hausmann, S., Shen, V., Shuman, S., and Lima, C.D. 2004. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol. Cell 13: 77–89. [DOI] [PubMed] [Google Scholar]
- Fantoni, A., Dare, A.O., and Tschudi, C. 1994. RNA polymerase III-mediated transcription of the trypanosome U2 small nuclear RNA gene is controlled by both intragenic and extragenic regulatory elements. Mol. Cell. Biol. 14: 2021–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson, K. and Wigley, D.B. 1998. Structure of a complex between a cap analogue and mRNA guanylyl transferase demonstrates the structural chemistry of RNA capping. Proc. Natl. Acad. Sci. 95: 1505–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson, K., Doherty, A.J., Shuman, S., and Wigley, D.B. 1997. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell 89: 545–553. [DOI] [PubMed] [Google Scholar]
- Hausmann, S., Vivares, C.P., and Shuman, S. 2002. Characterization of the mRNA capping apparatus of the microsporidian parasite Encephalitozoon cuniculi. J. Biol. Chem. 277: 96–103. [DOI] [PubMed] [Google Scholar]
- Hausmann, S., Zheng, S., Fabrega, C., Schneller, S.W., Lima, C.D., and Shuman, S. 2005. Encephalitozoon cuniculi mRNA cap (guanine N-7) methyltransferase: Methyl acceptor specificity, inhibition BY S-adenosylmethionine analogs, and structure-guided mutational analysis. J. Biol. Chem. 280: 20404–20412. [DOI] [PubMed] [Google Scholar]
- Ho, C.K. and Shuman, S. 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3: 405–411. [DOI] [PubMed] [Google Scholar]
- ———. 2001. Trypanosoma brucei RNA triphosphatase. Antiprotozoal drug target and guide to eukaryotic phylogeny. J. Biol. Chem. 276: 46182–46186. [DOI] [PubMed] [Google Scholar]
- Ho, C.K., Sriskanda, V., McCracken, S., Bentley, D., Schwer, B., and Shuman, S. 1998. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 273: 9577–9585. [DOI] [PubMed] [Google Scholar]
- Liang, X.H., Haritan, A., Uliel, S., and Michaeli, S. 2003. trans and cis splicing in trypanosomatids: Mechanism, factors, and regulation. Eukaryot. Cell 2: 830–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair, G., Ullu, E., and Tschudi, C. 2000. Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J. Biol. Chem. 275: 28994–28999. [DOI] [PubMed] [Google Scholar]
- Mao, X. and Shuman, S. 1996. Vaccinia virus mRNA (guanine-7-) methyltransferase: Mutational effects on cap methylation and AdoHcy-dependent photo-cross-linking of the cap to the methyl acceptor site. Biochemistry 35: 6900–6910. [DOI] [PubMed] [Google Scholar]
- Mao, X., Schwer, B., and Shuman, S. 1995. Yeast mRNA cap methyl-transferase is a 50-kilodalton protein encoded by an essential gene. Mol. Cell. Biol. 15: 4167–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1996. Mutational analysis of the Saccharomyces cerevisiae ABD1 gene: Cap methyltransferase activity is essential for cell growth. Mol. Cell. Biol. 16: 475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken, S., Fong, N., Rosonina, E., Yankulov, K., Brothers, G., Siderovski, D., Hessel, A., Foster, S., Shuman, S., and Bentley, D.L. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 11: 3306–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, K.P. and Agabian, N. 1992. Trypanosoma brucei spliced-leader RNA methylations are required for trans splicing in vivo. Mol. Cell. Biol. 12: 4844–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottram, J., Perry, K.L., Lizardi, P.M., Luhrmann, R., Agabian, N., and Nelson, R.G. 1989. Isolation and sequence of four small nuclear U RNA genes of Trypanosoma brucei subsp. brucei: Identification of the U2, U4, and U6 RNA analogs. Mol. Cell. Biol. 9: 1212–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaar, V., Dare, A.O., Hong, D., Ullu, E., and Tschudi, C. 1994. Upstream tRNA genes are essential for expression of small nuclear and cytoplasmic RNA genes in trypanosomes. Mol. Cell. Biol. 14: 6736–6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillutla, R.C., Yue, Z., Maldonado, E., and Shatkin, A.J. 1998. Recombinant human mRNA cap methyltransferase binds capping enzyme/RNA polymerase IIo complexes. J. Biol. Chem. 273: 21443–21446. [DOI] [PubMed] [Google Scholar]
- Saha, N., Schwer, B., and Shuman, S. 1999. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J. Biol. Chem. 274: 16553–16562. [DOI] [PubMed] [Google Scholar]
- Saha, N., Shuman, S., and Schwer, B. 2003. Yeast-based genetic system for functional analysis of poxvirus mRNA cap methyltransferase. J. Virol. 77: 7300–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer, B. and Shuman, S. 1994. Mutational analysis of yeast mRNA capping enzyme. Proc. Natl. Acad. Sci. 91: 4328–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer, B., Lehman, K., Saha, N., and Shuman, S. 2001. Characterization of the mRNA capping apparatus of Candida albicans. J. Biol. Chem. 276: 1857–1864. [DOI] [PubMed] [Google Scholar]
- Shuman, S. 2000. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66: 1–40. [DOI] [PubMed] [Google Scholar]
- Silva, E., Ullu, E., Kobayashi, R., and Tschudi, C. 1998. Trypanosome capping enzymes display a novel two-domain structure. Mol. Cell. Biol. 18: 4612–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.L., Levin, J.R., Ingles, C.J., and Agabian, N. 1989. In trypanosomes the homolog of the largest subunit of RNA polymerase II is encoded by two genes and has a highly unusual C-terminal domain structure. Cell 56: 815–827. [DOI] [PubMed] [Google Scholar]
- Tschudi, C. and Ullu, E. 2002. Unconventional rules of small nuclear RNA transcription and cap modification in trypanosomatids. Gene Expr. 10: 3–16. [PMC free article] [PubMed] [Google Scholar]
- Tschudi, C., Richards, F.F., and Ullu, E. 1986. The U2 RNA analogue of Trypanosoma brucei gambiense: Implications for a splicing mechanism in trypanosomes. Nucleic Acids Res. 14: 8893–8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu, E. and Tschudi, C. 1991. Trans splicing in trypanosomes requires methylation of the 5′ end of the spliced leader RNA. Proc. Natl. Acad. Sci. 88: 10074–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.P. and Shuman, S. 1997. Structure-function analysis of the mRNA cap methyltransferase of Saccharomyces cerevisiae. J. Biol. Chem. 272: 14683–14689. [DOI] [PubMed] [Google Scholar]
- Wang, S.P., Deng, L., Ho, C.K., and Shuman, S. 1997. Phylogeny of mRNA capping enzymes. Proc. Natl. Acad. Sci. 94: 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, Z., Maldonado, E., Pillutla, R., Cho, H., Reinberg, D., and Shatkin, A.J. 1997. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. 94: 12898–12903. [DOI] [PMC free article] [PubMed] [Google Scholar]