Abstract
Shoot branching is a major determinant of variation in plant stature. Branches, which form secondary growth axes, originate from stem cells activated in leaf axils. The initial steps by which axillary meristems (AMs) are specified and their stem cells organized are still poorly understood. We identified gain- and loss-of-function alleles at the Arabidopsis thaliana REGULATOR OF AXILLARY MERISTEMS1 (RAX1) locus. RAX1 is encoded by the Myb-like transcription factor MYB37 and is an Arabidopsis homolog of the tomato (Solanum lycopersicum) Blind gene. RAX1 is transiently expressed in a small central domain within the boundary zone separating shoot apical meristem and leaf primordia early in leaf primordium development. RAX1 genetically interacts with CUP-SHAPED COTYLEDON (CUC) genes and is required for the expression of CUC2 in the RAX1 expression domain, suggesting that RAX1 acts through CUC2. We propose that RAX1 functions to positionally specify a stem cell niche for AM formation. RAX1 also affects the timing of developmental phase transitions by negatively regulating gibberellic acid levels in the shoot apex. RAX1 thus defines a novel activity that links the specification of AM formation with the modulation of the rate of progression through developmental phases.
INTRODUCTION
Plant seedlings initially have a single growth axis for shoots and roots. Therefore, the diversity of plant stature observed in nature is largely due to two postembryonic processes: the formation of secondary growth axes and the timing of developmental phase transitions that govern meristem and, therefore, organ identity. Secondary growth axes require the establishment and activation of axillary meristems (AMs), which arise during modular plant growth in phytomers that comprise a leaf, an AM, and a stem segment. Once specified, rosette leaf AMs are activated in two patterns in Arabidopsis thaliana: an acropetal pattern of bud outgrowth, which is readily observed only in late flowering accessions, and a basipetal pattern of rosette paraclade formation upon the transition from vegetative growth to reproductive growth (Hempel and Feldman, 1994; Stirnberg et al., 1999; Grbic and Bleecker, 2000; Stirnberg et al., 2002). Once outgrowth has been activated, only a few cauline leaves are produced on the branch before its meristem forms an inflorescence.
The developmental origins of AMs have been controversial. On the basis of morphological and anatomical criteria, it has been proposed that the cells at the adaxial side of the boundary between incipient leaf primordium and shoot apical meristem (SAM) adopt AM fate de novo (Snow and Snow, 1942). Alternatively, it has been argued that these cells detach from the SAM early during leaf primordium development (Sussex, 1955). However, these arguments are based on morphology and do not take into account the complementary but distinct genetic functions required for meristem establishment and maintenance (Doerner, 2003). For example, studies with a molecular marker for indeterminate cell fate, such as SHOOT MERISTEMLESS (STM) (Long and Barton, 2000), have not clearly resolved the controversy because STM expression alone is insufficient to specify stem cell fate (Gallois et al., 2002; Lenhard et al., 2002), which requires coexpression of WUSCHEL (WUS) and possibly other genes of similar function (Haecker et al., 2004; Green et al., 2005).
Genetic approaches to identify the loci that control AM formation have proven powerful: Several genes have been identified in Arabidopsis and other model systems that control AM formation (for reviews, see McSteen and Leyser, 2005; Schmitz and Theres, 2005). Lateral suppressor (Ls) in tomato (Solanum lycopersicum) and its Arabidopsis and rice (Oryza sativa) homologs LAS and MONOCULM1, respectively, are required very early in AM development, as mutants lack any sign of AM development, including the stimulation of STM expression temporally associated with AM formation (Greb et al., 2003). LAS is expressed in the boundary zone separating the incipient leaf primordium from the SAM, similar to CUP-SHAPED COTYLEDON (CUC) and LATERAL ORGAN BOUNDARY (LOB) genes (Greb et al., 2003). This early onset of LAS expression suggests that the initial specification of a distinct AM identity occurs around the time of leaf primordium formation, possibly by maintaining cells in an indeterminate state as indicated by the STM marker. CUC genes may also play an important role in specifying AM identity as their overexpression stimulates adventitious shoot formation (Daimon et al., 2003), but the severe seedling phenotypes of hypomorphic cuc mutants has hindered detailed studies of their later functions in the plant life cycle. LAS encodes a putative transcription factor of the GRAS family. A further transcription factor required early in AM development was identified in tomato. The Blind (Bl) gene encodes a putative Myb-like transcription factor for which the Arabidopsis homolog has not yet been described. bl mutants lack AMs in many vegetative axils, and their response to SAM decapitation suggests that Bl is required early in AM development (Schmitz et al., 2002). Double mutants of ls and bl in tomato revealed additive phenotypes, suggesting the existence of at least two pathways involved in AM initiation (Schmitz et al., 2002).
Here, we report the identification of a gene we have designated REGULATOR OF AXILLARY MERISTEMS1 (RAX1), which is encoded by the Myb-like transcription factor MYB37, and is the putative Arabidopsis homolog of the tomato Bl gene. We show that RAX1 is expressed in a small central domain within the boundary zone separating SAM and leaf primordia during early leaf primordium development and is currently the earliest spatial marker for future AMs. RAX1, which is a transcriptional activator, genetically interacts with CUC genes and is required for the expression of CUC2 in the RAX1 expression domain. Our data suggest that RAX1 promotes early stages of AM formation and functions to establish or maintain an environment conducive for stem cell organization in the course of AM formation. Hence, RAX1 is involved in establishing the AM stem cell niche. RAX1 also affects the timing of developmental phase transitions by negatively regulating gibberellic acid levels in the shoot apex.
RESULTS
A Dominant Mutant That Affects Branching
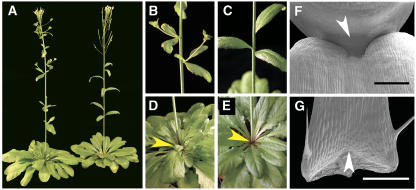
We generated a population of ∼5000 activation-tagged individuals in the FA4C reporter gene background established in the Columbia (Col-0) ecotype (Colón-Carmona et al., 1999). Using the pSKI015 vector, which carries four outward-facing 35S enhancers adjacent to the left border (Weigel et al., 2000), we screened for individuals with altered growth and vegetative development. A dominant mutant with reduced rosette branching was identified. This mutant defined a locus designated RAX1, and we refer to the dominant allele as rax1-1D. We observed two phenotypic classes in segregating populations, which corresponded to the hemizygous or homozygous state of the rax1-1D allele (Figure 1), suggesting that the effects of this allele were dosage dependent.
Figure 1.
rax1-1D/+ Plants Establish Supernumerary AM Organizing Centers.
(A) Wild-type (FA4C) plant midway through reproductive development, showing three rosette paraclades (arrowheads).
(B) rax1-1D/+ plant at a similar stage in development, completely lacking rosette paraclades.
(C) Close-up of previous individual, showing aerial rosettes and continued production of leaves on branches originating from cauline leaf axils.
(D) rax1-1D homozygous plant. After producing 6 to 10 true leaves, rax1-1D homozygous seedlings cease leaf production. Many plants die after resting at this stage for 2 to 3 weeks.
(E) A variable fraction of rax1-1D homozygous seedlings regenerates a SAM from the axils of the youngest initially produced leaves. This panel shows an early stage of such plant.
(F) Scanning electron micrograph of rosette leaf axil in FA4C. The inset shows the central domain at the basal end of the petiole in higher magnification, revealing the initiation of an AM.
(G) Scanning electron micrograph of a developing rosette AM in the FA4C wild-type background.
(H) Scanning electron micrograph of rosette leaf axil in rax1-1D/+ with two organizing centers of AM formation.
(I) A magnified view of the same specimen shows a much larger population of small proliferating cells organizing into two centers (arrowheads).
(J) Scanning electron micrograph of rosette leaf axil in rax1-1D/+ treated with GA3. GA3 was sprayed twice-weekly during vegetative growth as a solution of 100 μM GA3 and 0.02% Tween-20. AM development is further advanced, but branches did not develop.
(K) Scanning electron micrograph of a shoot apex of a plant similar to the one shown in (D), showing that the SAM has been completely consumed in the process of leaf organogenesis.
(L) Scanning electron micrograph of a plant similar to the one shown in (D), except that development of the final organ is incomplete.
(M) Scanning electron micrograph of plant similar to that shown in (E), revealing the initiation of new AMs.
Bars = 1 cm in (D), 2.5 mm in (E), 500 μm in (F) to (M), and 100 μm in (F) inset.
When compared with the parental FA4C line (Figure 1A), hemizygous individuals, which we refer to as rax1-1D/+, were slightly dwarfed and formed compact rosettes with smaller, rounder, and slightly wrinkly leaves (Figure 1B). After the transition to flowering, these plants formed significantly fewer rosette branches than the wild type (Table 1). However, slightly more cauline paraclades were formed on the elongating inflorescence stem when compared with FA4C (Table 1). Hemizygous individuals then produced flowers on long pedicels and fertile seed; however, rax1-1D/+ plants produced fewer flowers than wild-type siblings (see Supplemental Figure 1 online). This data suggested that RAX1 primarily regulates rosette branching.
Table 1.
Branching in Wild-Type and rax1-1D/+ Plants
| FA4C (n) | rax1-1D/+ (n) | |
|---|---|---|
| Rosette branches with IM | 5.41 ± 0.18 (29) | 1.11 ± 0.17* (28) |
| Cauline branches with VM | 0.03 ± 0.15 (29)a | 2.11 ± 0.48* (28) |
| Cauline branches with IM | 4.83 ± 0.15 (29) | 5.82 ± 0.31* (28) |
| Total branches | 10.24 ± 0.25 (29) | 9.04 ± 0.51** (28) |
| Rosette branches with IM after decapitation | 6.9 ± 0.37 (17) | 6.6 ± 1 (21) |
Branches with an inflorescence meristem at the apex (branches with IM) are distinguished from branches with vegetative identity that produce leaves only (branches with VM). Rosette branches originate from axils on the unexpanded stem, while cauline branches originated from the expanded segment of the stem. Errors are standard errors of the mean. One asterisk indicates significant differences (P < 0.01), while two asterisks indicates significant differences (P < 0.05) identified with Student's t test. Numbers in parentheses indicate the number of individuals tested.
One individual produced a single cauline branch with several leaves arranged as an aerial rosette.
rax1-1D homozygotes displayed a stronger phenotype that affected the shoot meristem: after germination, they were more dwarfed than rax1-1D/+ plants and never produced more than 6 to 10 leaves before arresting further development (Figure 1D). However, after a period of developmental stasis, some of these plants initiated new rosette branches of wild-type appearance (Figure 1E). These shoots eventually produced inflorescences with fertile flowers. However, this reversion was not permanent, as the progeny of these plants initially all displayed the severe phenotype.
To distinguish whether rax1-1D/+ individuals had defects in AM establishment or bud outgrowth, we examined axils in more detail by scanning electron microscopy. In wild-type FA4C plants, the first visible evidence of AM formation was the appearance of a cluster of small, proliferating cells at the basal end of the adaxial surface of the petiole (Figure 1F). Subsequently, these cells organized into an AM flanked by two leaf primordia (Figure 1G). In rax1-1D/+ plants, the field of small, proliferating cells was much enlarged laterally and distally into the petiole and frequently gave rise to more than one organizing center for an AM (Figures 1H and 1I). This suggested that RAX1 activity promoted early steps of AM formation.
In plants homozygous for rax1-1D, the scanning electron microscopy analysis revealed that the SAM was consumed in the formation of a terminal leaf primordium or led to the formation of small, radially symmetric structures with leaf epidermal cell types (Figures 1K and 1L). However, in individuals where the severe phenotype did not persist, new shoots were produced in existing leaf axils (Figure 1M).
However, stimulation of AM formation by RAX1 does not lead to increased rosette branching (Table 1). To test the hypothesis that the rax1-1D allele interfered with later stages of AM development or bud outgrowth, but not with the ability to specify and initiate AMs, we examined branching in plants that had been decapitated 2 weeks after the primary stem had begun to elongate. AMs are specified during vegetative development, but in Arabidopsis, buds are formed and branch outgrowth is activated only after the transition to flowering. The SAM suppresses branching, referred to as apical dominance (Cline, 1997). When rax1-1D/+ plants were decapitated, rosette branching increased strongly, while only a modest increase in branch number was seen in the wild type (Table 1). Overall branch number in these was very variable, and five of 22 decapitated rax1-1D individuals produced a much larger number of branches than the mean, suggesting that many more AMs were produced in this background. We concluded that RAX1 promotes early steps of AM formation and that reduced rosette branching observed in rax1-1D/+ plants was due to interference with later steps.
rax1-1D Is a Hypermorphic Allele of a R2R3-Type Myb Gene
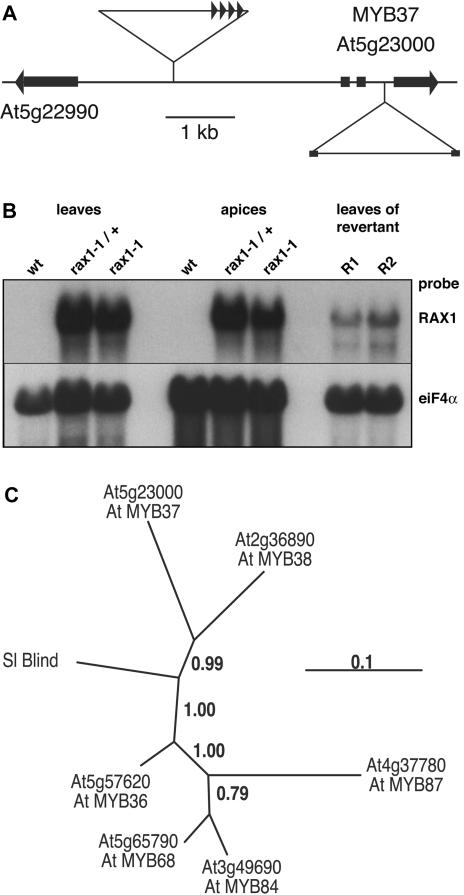
The rax1-1D phenotype cosegregated with BASTA resistance conferred by the pSKI015 activation tagging vector, indicating that the RAX1 locus was tagged. To identify the site of T-DNA insertion in the genome, we used thermal asymmetric interlaced PCR (Liu et al., 1995). Plants carrying the rax1-1D allele carried a simple T-DNA insertion on chromosome 5, 2381 bp upstream of the start codon of the gene At5g23000 (Figure 2A), which encodes MYB37, a Myb-like transcription factor of the R2R3 class (Stracke et al., 2001). To confirm that MYB37 is RAX1, we examined the expression of genes flanking the T-DNA insertion site and recapitulated the insertion of the activation-tagging vector. While the expression of the C2H2-type putative transcription factor gene described by the At5g22990 gene model was unaffected by the presence of the T-DNA, RNA levels of MYB37 were strongly elevated in individuals carrying the rax1-1D allele (Figure 2B): MYB37 transcripts were undetectable in RNA gel blots with wild-type samples but accumulated to high levels in leaf and shoot apices of rax1-1D plants. RT-PCR analysis revealed that MYB37 was not expressed in wild-type leaves (see Supplemental Figure 2 online), suggesting that this gene was ectopically expressed in leaves of plants carrying the rax1-1D allele. Interestingly, MYB37 transcript abundance was much reduced in the leaves of rax1-1D homozygous plants in which the severe phenotype did not persist, suggesting that very-high-level MYB37 expression was important for the rax1-1D phenotype. To test whether we could recreate rax1-1D phenotypes, we generated a construct encompassing four 35S enhancer elements, the putative MYB37 promoter, MYB37 gene, and 3′ terminator sequences. This was introduced into wild-type Arabidopsis, and 126 transformants were phenotypically analyzed. Approximately 20% of transformants recapitulated the rax1-1D hemizygous and homozygous phenotypes. The low frequency of phenotypic plants was not surprising, considering that MYB37 expression levels would have to be very high to manifest the hemizygous and homozygous phenotypes. We concluded that the rax1-1D/+ and rax1-1D phenotypes were caused by insertion of the activation tagging T-DNA upstream of MYB37 and, hence, that RAX1 was MYB37.
Figure 2.
RAX1 Is a Putative Myb-Like Transcription Factor Expressed at Low Levels.
(A) Schematic of genome organization around the At5g23000 gene model for MYB37. The orientation of the four 35S enhancer repeats in the activation-tagging T-DNA 2381 bp 5′ of the MYB37 start codon is indicated by triangles. The insertion site of the T-DNA inserted in the second intron of MYB37 is indicated below.
(B) RAX1 expression is very high in rax1-1D. RNA was extracted from leaves and microdissected shoot apices of rax1-1D hemizygous or homozygous individuals and from rax1-1D homozygous revertants. RNA gel blot analysis shows high levels of RAX1 RNA in rax1-1D individuals and much reduced levels in leaves of revertant homozygous rax1-1D plants. A 501-bp RAX1-specific probe corresponding to the third exon was used to detect RAX1 transcripts, and RNA loading was checked by hybridization to eukaryotic translation initiation factor 4A (eIF-4A) transcripts.
(C) Phylogenetic relationship between closely related Arabidopsis and tomato (Sl) Myb genes. The scale for branch lengths indicates the number of substitutions per amino acid residue. Genes are designated in accordance with the accompanying article.
MYB37 is closely related to several other R2R3 Myb genes in Arabidopsis as well as the recently identified Bl gene, which regulates sympodial branching in tomato (Stracke et al., 2001; Schmitz et al., 2002). MYB37 is most closely related to MYB38 (At2g36890) and tomato Bl (Figure 2C). The genetic function of most members of this clade is currently unknown, but MYB68 (At5g65790) is expressed in roots and is required for normal root growth (Feng et al., 2004). We therefore aimed to examine RAX1 genetic function by identifying loss-of-function alleles.
Loss of RAX1 Function Reduces AM Formation and Branching
To characterize RAX1 genetic function, we screened available T-DNA insertion collections. We identified the recessive rax1-2 allele in the Wassilewskija (Ws-2) background (Krysan et al., 1999). This allele carries a T-DNA insertion in the second MYB37 intron (Figure 2A). Using RNA gel blot analysis, we could not detect RAX1 transcripts in samples extracted from wild-type (Figure 2B) or rax1-2 shoot apices. We therefore used RT-PCR with two sets of primers, corresponding to targets 5′ or 3′ of the T-DNA insertion, to assess whether rax1-2 was a null allele. RAX1 expression could not be detected with either primer set in samples isolated from rax1-2, while it was readily detected in samples from Ws-2 (see Supplemental Figure 3 online). Therefore, rax1-2 is a null allele. We identified a further line (SALK_009859) in the SALK collection (Alonso et al., 2003) with a T-DNA insertion at the 3′ end of the gene, but homozygous individuals had only modestly reduced RAX1 RNA levels and no detectable phenotype. This line was not further characterized.
Individuals hemizygous for rax1-2 had no detectable phenotype, but homozygous rax1-2 plants had a branching phenotype: when grown in long- (LD) or short-day (SD) conditions, they produced fewer paraclades from rosette and cauline leaf axils when compared with the Ws-2 wild type (Table 2, Figures 3A to 3E). The phenotype was slightly more pronounced in SD (Table 2). The rosette paraclade phenotype of rax1-2 was less severe than that of rax1-1D/+ (Table 1). In contrast with rax1-1D/+ plants, the small clusters of proliferating cells characteristic for the early stages of AM formation were completely absent in most rosette or cauline leaf axils when analyzed by scanning electron microscopy (Figures 3F and 3G), confirming that RAX1 is required early in AM formation. Consistent with this interpretation, decapitated rax1-2 plants did not show increased branching (Table 2), as was seen for rax1-1D/+ (Table 1). Homozygous rax1-2 plants also produced fewer flowers than the matching wild type (see Supplemental Figure 1 online).
Table 2.
Branching in Wild-Type and rax1-2 Plants
| Ws-2 (n) | rax1-2 (n) | |
|---|---|---|
| Rosette branches with IM (LD) | 4.1 ± 0.15 (24) | 2.4 ± 0.17* (24) |
| Cauline branches with IM (LD) | 2.8 ± 0.10 (24) | 2.9 ± 0.10 (24) |
| Rosette branches with IM (SD) | 6.1 ± 0.20 (43) | 3.1 ± 0.27* (63) |
| Cauline branches with IM (SD) | 7.3 ± 0.13 (43) | 4.0 ± 0.18* (63) |
| Rosette branches with IM after decapitation (SD) | 6.7 ± 0.33 (15) | 3.1 ± 0.27* (18) |
Plants were grown in the photoperiodic conditions indicated. Rosette branches originate from axils on the unexpanded stem, while cauline branches originated from the expanded segment of the stem. Only branches topped by inflorescence meristems (IM) were observed. Errors are standard errors of the mean, and the asterisks indicate significant differences (P < 0.01) identified with Student's t test. Numbers in parentheses indicate the number of individuals tested.
Figure 3.
rax1-2 Plants Develop Less Rosette and Cauline Branches and Cannot Initiate or Maintain an AM.
(A) Wild-type (Ws-2) plant early during reproductive development (left) and rax1-2 plant at a similar developmental stage (right).
(B) to (E) Close-ups of the plants shown in (A).
(B) Ws-2 wild-type inflorescence stem showing developing cauline branches.
(C) Inflorescence stem of rax1-2 showing the absence of branches originating from cauline leaf axils.
(D) Ws-2 wild-type rosette with developing rosette paraclade (arrowhead).
(E) Rosette of rax1-2 plant lacking rosette paraclade (arrowhead).
(F) Scanning electron micrograph of a barren rax1-2 cauline leaf axil lacking an organizing center with proliferating cells (arrowhead).
(G) Scanning electron micrograph of barren rosette leaf axil in rax1-2 lacking an organizing center with proliferating cells (arrowhead). Bars = 500 μm in (F) and (G).
To unambiguously show that the rax1-2 allele was responsible for the phenotypes observed, we complemented the mutant by introduction of a wild-type copy of the RAX1 gene into rax1-2 homozygous plants. We obtained 52 transformed lines, of which 15 segregated a single T-DNA, and analyzed branching in two lines in more detail: In this experiment, the Ws-2 wild type produced 3.1 ± 0.27 rosette paraclades, while in rax1-2, 1.6 ± 0.24 rosette paraclades were made. The rax1-2 lines transformed with a wild-type copy of RAX1 formed 3.3 ± 0.28 and 3.2 ± 0.29 rosette paraclades, respectively, which was significantly different from the rax1-2 line (P < 0.001), but not different from Ws-2. We concluded that the phenotypes observed in rax1-2 were due to the insertional inactivation of RAX1.
Taken together, our data show that the rax1-1D and rax1-2 alleles of RAX1 have opposite effects on AM formation, and we conclude that RAX1 promotes early steps in AM formation, such as the specification of axillary identity or of meristematic competence in axillary cells.
RAX1 Is Expressed Transiently in the Axils of Leaf Primordia
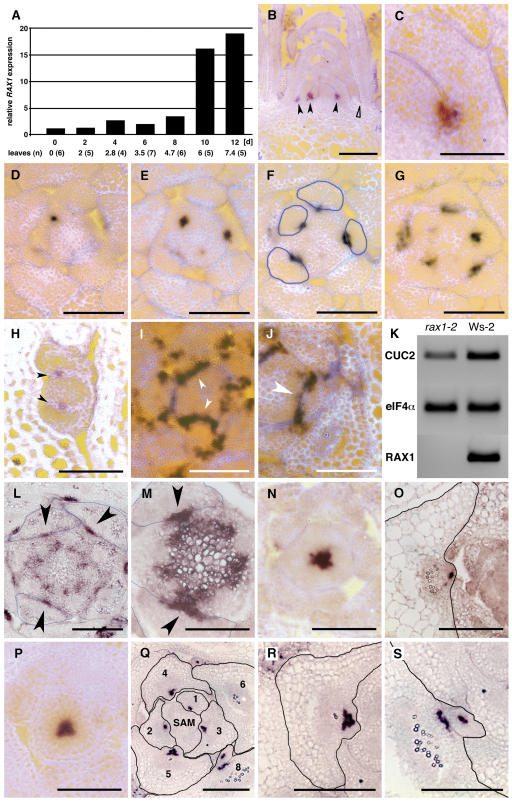
To establish how RAX1 promoted early steps in AM formation, we examined RAX1 expression and identified target genes. Using RT-PCR, we consistently detected RAX1 expression in shoots. In shoots, the RAX1 temporal expression pattern was biphasic. Low-level expression was observed from germination onward, but higher-level expression was restricted to the adult vegetative phase and became detectable upon initiation of the fifth leaf primordium, at the onset of the adult vegetative phase in Col-0 (Telfer et al., 1997) (Figure 4A). RAX1 is expressed transiently during leaf primordium development (Figures 4B to 4G) in a small domain at the center of the boundary between the SAM and developing leaf primordia (Figures 4D to 4G), marking the position of cells competent to initiate AMs in the axils of the primary shoot (Figure 3H). A similar pattern of expression is recapitulated in AMs (Figure 4H) after the formation of the lateral bud and stimulation of its outgrowth at the transition to flowering. Consistent with the results of decapitation experiments (Tables 1 and 2), this suggested that RAX1 functions early in AM development.
Figure 4.
RAX1 Is Transiently Expressed during Adult Vegetative Development in Young Leaf Primordia in a Pattern That Anticipates Future AM Position.
RAX1 mediates CUC2 expression in a central domain in leaf axils. Bars = 200 μm in (B), 25 μm in (C), 50 μm in (D) to (I) and (L) to (S), and 25 μm in (J).
(A) Seedlings were germinated in the dark for 5 d, transferred to light, and sampled every 2 d. Ten to twenty seedlings were fixed and dissected to count leaf primordia, and ∼30 seedlings were used to extract RNA for analysis of RAX1 RNA levels by RT-PCR. Mean number of vegetative leaves (not including cotyledons) that were counted at each time point is indicated below abscissa; the number of plants evaluated at each time point is given in parentheses.
(B) to (I) Vegetative wild-type (Col-0) shoot apices were processed for in situ hybridization with a 707-bp specific probe.
(B) Longitudinal section of a shoot apex shows accumulation of RAX1 RNA at the base of the adaxial side of young leaf primordia (closed arrowheads). RAX1 is not expressed at the equivalent position in older leaf primordia (open arrowhead).
(C) Close-up of RAX1 RNA accumulation pattern in young leaf primordium.
(D) to (G) Serial 8-μm cross sections of a wild-type SAM ∼8 μm below the top of the SAM (D), showing localized RAX1 RNA accumulation at the center of the adaxial face of the incipient leaf primordium, ∼16 μm below the top of the SAM (E), ∼24 μm below the top of the SAM (F) (outlines of leaf primordia are traced in blue), and ∼32 μm below the top of the SAM (G).
(H) RAX1 expression in an early AM, recapitulating the expression pattern at the shoot apex, with RAX1 RNA accumulation on the adaxial face of the incipient leaf primordia.
(I) Cross section of a wild-type (Col-0) shoot apex shows uniform accumulation of CUC2 RNA along the boundary between the SAM and incipient leaf primordia (arrowheads).
(J) Cross section of a rax1-2 shoot apex hybridized to CUC2 antisense probe shows that the accumulation of CUC2 RNA is interrupted at the center of boundary between the SAM and incipient leaf primordia. The gap in CUC2 RNA accumulation (arrowhead) is precisely where RAX1 RNA accumulates in the wild type (cf. with Figure 4F).
(K) RT-PCR analysis of CUC2 RNA levels in shoot apices of wild-type (Ws-2) and rax1-2 plants shows that CUC2 accumulates to ∼0.5× lower levels in the rax1-2 background.
(L) and (M) Cross sections hybridized to STM antisense probe.
(L) Ws-2 apex ∼80 μm below the top of the SAM, showing localized STM expression in axils at the onset of AM activation (arrowheads). Outlines of leaves are traced in blue.
(M) rax1-2 apex ∼72 μm below the top of the SAM with localized STM expression in axils at the onset of AM activation (arrowheads).
(N) to (S) Cross sections hybridized to WUS antisense probe.
(N) FA4C apex ∼24 μm below the top, showing WUS expression in a central domain of the SAM.
(O) FA4C leaf axil, with focused WUS expression in an incipient AM. Outline of the leaf is traced in black.
(P) rax1-1D/+ apex ∼24 μm below the top, showing WUS expression in a central domain of the SAM.
(Q) rax1-1D/+ apex ∼56 μm below the top, showing WUS expression at axillary positions in young leaves. Note how expression is in a larger domain when compared with the wild type in (O) in leaves numbered 3 to 5 and the presence of two discrete foci in leaf 8. Outlines of leaves traced in black.
(R) Close-up of leaf 5 shown in (Q) with a large domain of WUS expression.
(S) Close-up of the axil of leaf 8 in (Q), showing two separate zones of WUS expression.
Early AM development depends on the specification of axillary identity and the maintenance of meristematic competence in such cells (Schmitz and Theres, 2005). We therefore examined expression of LAS (Greb et al., 2003) and CUC genes (Aida et al., 1999), which are expressed at the boundary zone between the SAM and leaf primordia and are required for axillary or shoot meristem formation, respectively. The expression pattern of LAS was unaffected in rax1-2 plants when compared with the wild type (see Supplemental Figure 4 online).
By contrast, CUC2 expression, normally observed in a continuous band separating the SAM from incipient leaf primordia (Figure 4I), was absent at the position of future AM initials in the rax1-2 background (Figure 4J), precisely where RAX1 is expressed (Figure 4F). Moreover, CUC2 transcript levels were consistently reduced in rax1-2 (Figure 4K). Precise regulation of CUC gene expression patterns is required for maintenance of the boundary between meristems and leaf primordia (Laufs et al., 2004). Together, these data indicate that RAX1 is needed to promote CUC2 expression in a central domain of boundary cells in leaf axils. By contrast, expression of CUC3 was not affected (see Supplemental Figure 4 online).
CUC genes act through STM (Aida et al., 1999), which is also a potential target of LAS (Greb et al., 2003). Therefore, we examined the spatio-temporal pattern of STM transcript accumulation in the boundary zone between SAM and leaf primordia. STM expression in this zone is dynamic and complex (Long and Barton, 2000; Greb et al., 2003). Upon stimulation of AM activity, STM transcripts initially accumulated in a small group of cells at the base of leaf primordia in Ws-2 wild type (Figure 4L) and in rax1-2 (Figure 4M) apices. By contrast, STM does not accumulate at this position in a las background (Greb et al., 2003). However, while in the wild type, STM expression continued as AMs were activated, STM expression in rax1-2 did not persist, consistent with the notion that rax1-2 axils do not maintain meristematic cells. These observations support the notion that Ls (the LAS homolog in tomato) and Bl (a Myb gene closely related to RAX1; Figure 2C) are in two separate genetic pathways (Schmitz et al., 2002).
We also examined the expression of WUS, which mediates stem cell niche functions to control stem cell population size. The spatial pattern of WUS expression in FA4C (Figure 4N) and rax1-1D/+ (Figure 4P) SAMs was very similar. In contrast with the tightly focused WUS transcript accumulation in FA4C AMs (Figure 4O), the WUS expression domain in rax1-1D/+ AMs was larger (Figures 4Q to 4S) and frequently gave rise to two discrete foci (Figure 4S), suggesting the presence of supernumerary AM organizing centers, similar to those observed by scanning electron microscopy (Figures 1H and 1I).
To test whether CUC2 was an important target of the RAX1 pathway, we examined whether a reduction of CUC gene dosage would enhance the severity of the rax1-2 phenotype. Rosette paraclade formation in plants with reduced CUC gene dosage was indistinguishable from the wild type when in a RAX1 background (P > 0.1). However, in a rax1-2 background, rosette paraclade formation was dependent on CUC activity: progressive reduction of CUC gene dosage strongly enhanced the rax1-2 branching phenotype (Table 3). These data supported the notion that RAX1 acts through CUC genes. Based on the phenotype of rax1-2, its very early expression in leaf axils (Figures 4B to 4F), and the spatial pattern of expression (Figures 4E and 4J), we conclude that RAX1 is required to establish or maintain a stem cell niche for AM formation by spatial control of CUC2 expression.
Table 3.
Gene Dosage of CUC Genes Affects Branching in rax1-2
| Genotype | Ler (n) | rax1-2 (n) |
|---|---|---|
| CUC1/CUC1, CUC2/CUC2 | 4.54 ± 0.22 (54) | 2.0 ± 0.15* (72) |
| cuc1/cuc1, CUC2/CUC2 | 4.65 ± 0.27 (17) | 0.4 ± 0.11* (40) |
| cuc1/cuc1, cuc2/CUC2 | 4.81 ± 0.22 (42) | 0.08 ± 0.04* (60) |
Plants were grown in SD conditions. Errors are standard errors of the mean, and the asterisks indicate significant differences (P < 0.0001) identified with Student's t test. Numbers in parentheses indicate the number of individuals tested. Ler, Landsberg erecta.
RAX1 Is a Transcriptional Activator
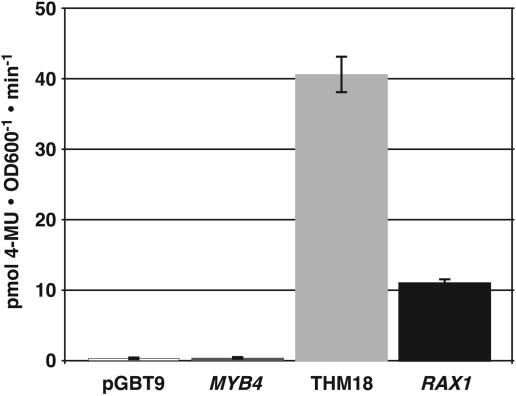
Reduced CUC2 expression in rax1-2 homozygous individuals suggested that RAX1 could be a transcriptional activator. We therefore examined the biochemical mechanism of RAX1 function in a heterologous yeast system. Myb-like genes comprise two domains: an N-terminal DNA binding domain (DBD) and a C-terminal activating or repression domain. We fused the entire coding sequence downstream of the Myb DBD to the yeast GAL4 DBD. As controls, similar constructs were made with the strong activating domain of the tomato THM18 Myb-like gene and the repression domain of the Arabidopsis MYB4 (At4g3620) gene (Schwechheimer et al., 1998; Jin et al., 2000). After these constructs were introduced into yeast cells, we analyzed their ability to transactivate expression of a β-galactosidase reporter gene. The RAX1 C-terminal domain transactivated reporter gene expression, albeit to only 25% of the levels observed with the strong THM18 activation domain (Figure 5). We concluded that RAX1 is a transcriptional activator.
Figure 5.
RAX1 Is a Transcriptional Activator.
The magnitude of transactivation conferred by the putative RAX1 transactivation domain C-terminal to the DBD was tested in yeast. The bars indicate the means of results obtained in quadruplicate measurements performed on each of four independently transformed yeast cultures; error bars indicate standard error of the mean. RAX1 transactivation strength is ∼25% of the strong activation conferred by THM18. 4-MU, 4-methyl umbelliferone.
RAX1 Modulates Progression through the Vegetative Phase and Affects GA Levels
In addition to rosette branching, hemizygous rax1-1D/+ and homozygous rax1-2 plants differed from wild-type plants in vegetative development: rax1-1D/+ hemizygous plants had an extended vegetative phase, while by contrast, vegetative development in SD conditions was shorter in homozygous rax1-2 plants (Table 4).
Table 4.
RAX1 Modulates Vegetative Phase and Plastochron Length
| Condition | Genotype (n) | Leaves/Day | Rosette Leaves Produced | Days to Flowering |
|---|---|---|---|---|
| LD | FA4C (42) | 0.38 | 9.02 ± 0.3 | 23.6 ± 1.1 |
| rax1-1D/+ (22) | 0.53* | 23.1 ± 1.1* | 44.2 ± 2.7* | |
| Ws-2 (24) | 0.31 | 7.4 ± 0.15 | 23.9 ± 0.15 | |
| rax1-2 (24) | 0.32 | 7.6 ± 0.13 | 23.6 ± 0.2 | |
| SD | FA4C (50) | 0.52 | 20.9 ± 1.4 | 41.8 ± 2.2 |
| rax1-1D/+ (18) | 0.52 | 43.9 ± 1.8* | 84.0 ± 4.6* | |
| Ws-2 (24) | 0.54 | 32.3 ± 0.62 | 60.2 ± 0.51 | |
| rax1-2 (22) | 0.47* | 25.9 ± 1.21* | 54.3 ± 0.87* |
Plants were grown in the photoperiod indicated. Errors are standard errors of the mean, and the asterisks indicate significant differences (P < 0.01) identified with Student's t test between mutants and the corresponding wild type. Numbers in parentheses indicate the number of individuals tested.
In rax1-1D/+ plants, time to flowering was almost twice as long compared with FA4C in LD or SD conditions (Table 4). These dwarfed plants produced more and darker green leaves than the wild type and did so at a higher rate (Table 4). Furthermore, the transition between vegetative and reproductive phases was protracted, exemplified by formation of aerial rosettes and cauline branches topped by meristems continuing to form leaves without formation of inflorescences (Figures 1B and 1C). Wild-type plants generally do not form such branches. These phenotypes suggested that rax1-1D/+ plants maintained vegetative identity and, hence, generated vegetative features after the onset of stem elongation. By contrast, in SD conditions, rax1-2 individuals flowered ∼10% earlier than Ws-2 after having produced ∼20% less leaves (Table 4). Hence, in SD, rax1-2 produces leaves at a lower rate than the wild type. Furthermore, these leaves were paler green than those in wild-type plants. Together, these data suggested that, in addition to regulating AM formation in rosettes, RAX1 promoted vegetative meristem identity during Arabidopsis development. RAX1 modulates the adult phase of vegetative development, as the duration of the juvenile phase was not significantly altered in rax1-1D/+ or rax1-2 plants (see Supplemental Figure 5 online).
In light of the conditional effect of rax1-2 on flowering time and the differences in leaf pigmentation, we examined whether RAX1 activity affects gibberellic acid (GA) accumulation or responses in the shoot apex. GA promotes vegetative phase transitions and in Arabidopsis steadily accumulates during the adult vegetative phase toward a critical threshold sufficient to activate the transition to flowering (Blazquez et al., 1998; Simpson and Dean, 2002) mediated by LEAFY (LFY) expression (Blazquez et al., 1998). Moreover, GA is necessary for flowering in SD conditions (Wilson et al., 1992).
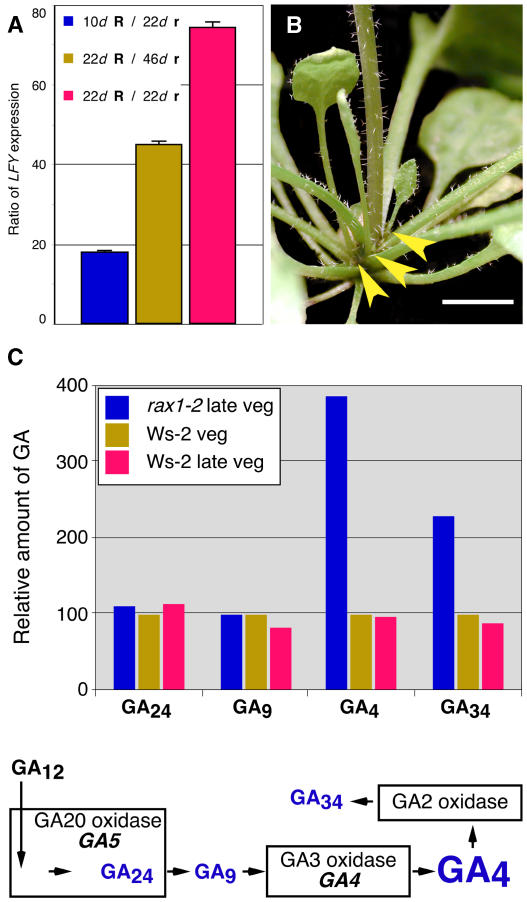
We first examined LFY transcript abundance, which responds to GA levels, in rax1-1D/+ individuals throughout vegetative development. LFY expression in rax1-1D/+ plants was strongly downregulated during vegetative development (Figure 6A). We then genetically tested the link between RAX1 and LFY activity using the lfy-9 mutant, an intermediate strength allele that sensitizes flowering to the endogenous rise of GA. In contrast with lfy-9 or rax1-1D/+ single mutants, rax1-1D/+ lfy-9 double mutants never produced flowers, consistent with the interpretation that endogenous levels of GA are lower in rax1-1D/+ than in the wild type. Furthermore, spraying of rax1-1D/+ plants with GA during vegetative growth suppressed the late-flowering phenotype (see Supplemental Figure 6 online), but these plants did not produce more branches than untreated controls. The formation of the AMs was further advanced in GA-treated rax1-1D/+ plants (Figure 1J) than in nonsprayed plants (cf. with Figures 1H and 1I) but not sufficient for the development of a functional lateral bud that could go on to form a branch (Figure 6B). GA-treated plants frequently formed a single leaf from axillary positions (Figure 6B), suggesting that while AMs could not be sustained in this background, cell expansion mediated by GA application sufficed to generate a macroscopically visible leaf organ. Together, these data suggested that GA levels might be reduced in the rax1-1D/+ plants but also indicated that alleviation of this defect was insufficient to restore branching.
Figure 6.
RAX1 Negatively Regulates GA Accumulation.
(A) During vegetative development, LFY is expressed at lower levels in rax1-1D/+ than in FA4C plants. FA4C (RAX1/RAX1, indicated by R) and rax1-1D/+ (indicated by r) plants were grown in LD conditions. RNA was extracted from plants in the middle (10 d, FA4C; 22 d, rax1-1D/+) and late in their vegetative development (22 d, FA4C; 46 d, rax1-1D/+), 5 to 6 d prior to the appearance of visible inflorescences. LFY RNA was analyzed by RT-PCR. Expression levels were normalized to eIF4α and then ratios of normalized LFY expression in wild-type and rax1-1D/+ samples calculated.
(B) LD-grown, GA-treated rax1-1D/+ plants frequently produce single leaves in leaf axils (arrowheads).
(C) Top: rax1-2 plants accumulate higher levels of GA4 in shoot apices. Leaves larger than ∼2 mm were removed, and GA levels were measured. The concentration of GA4, which stimulates flowering in Arabidopsis, was approximately fourfold elevated when compared with the wild type. Bottom: schematic of the GA biosynthetic pathways showing the pathway intermediates, products, and inactive catabolites measured in rax1-2 and wild-type plants.
We then examined whether earlier flowering in rax1-2, specifically in SD conditions, correlated with increased GA levels using gas chromatography–mass spectrometry selected reaction monitoring (Eriksson et al., 2000). We aimed to compare GA levels late in vegetative phase, just before the transition to flowering. We also sampled Ws-2 at the time rax1-2 flowered to compare GA levels in the early and normal flowering genotypes at this time point. Thus, we grew plants in SD conditions where inflorescences would become apparent in the rosette center of rax1-2 and Ws-2 individuals at 42 and 46 d, respectively, and harvested samples from both genotypes at 34 d and from Ws-2 only at 40 d. Levels of the active gibberellin GA4 were fourfold elevated in rax1-2 compared with Ws-2 at 34 d (Figure 6C), whereas levels of GA1, which does not induce the transition to flowering in Arabidopsis, were more modestly elevated (data not shown). We concluded that RAX1 antagonizes the accumulation of GA4, the biologically active GA required for flowering in Arabidopsis.
The inability to rescue branching in rax1-1D/+ plants sprayed with GA and the high level of GA in the rax1-2 plants indicated that the establishment of AMs and modulation of developmental phase transitions through control of GA levels were separate functions of RAX1. To examine which of these functions was mediated by CUC2 expression in the central domain of the boundary zone between the SAM and leaf primordia, CUC2 was expressed under control of the RAX1 promoter in a homozygous rax1-2 background. We examined flowering time and rosette branching in lines transformed with this ProRAX1:CUC2 construct (Table 5). Although we cannot exclude a general effect of CUC2 expression under control of the RAX1 promoter on flowering time or an altered expression pattern of RAX1 in a rax1-2 background, all eight homozygous lines examined differed significantly from the rax1-2 background and resembled Ws-2 with respect to flowering time. Only one line, A73-10, attained wild-type levels of branching as well. This indicated that expression of CUC2 in the RAX1 expression domain was necessary and sufficient to control wild-type timing of flowering but not sufficient to efficiently regulate the formation of AMs.
Table 5.
CUC2 Expression in the RAX1 Expression Domain in rax1-2 Plants Restores Wild-Type Flowering Time but Not Branching
| Line
|
n
|
Time to Flowering % of Wild Type
|
Rosette Paraclades % of Wild Type
|
|---|---|---|---|
| Ws-2 | 34 | 100%* | 100%* |
| rax1-2 | 37 | 92% | 45% |
| A22-7 | 35 | 100%* | 69% |
| A24-2 | 37 | 96%* | 32% |
| A29-8 | 16 | 98%* | 67% |
| A33-9 | 20 | 107%* | 75%* |
| A41-5 | 20 | 99%* | 44% |
| A42-17 | 21 | 108%* | 41% |
| A63-4 | 19 | 99%* | 71% |
| A73-10 | 19 | 101%* | 99%* |
Plants were grown in SD conditions. Days to flowering and rosette paraclade number were normalized to values observed for Ws-2. Data marked with asterisks are significantly different from the corresponding value of rax1-2 (P < 0.01; Student's t test).
DISCUSSION
RAX1 defines a novel genetic activity in Arabidopsis necessary for AM development, which also modulates the duration of vegetative development by controlling GA levels. RAX1 is part of a small gene family of Myb-like transcription factors in Arabidopsis with homology to the tomato Bl gene, which the accompanying article shows, has distinct but complementary functions in AM formation. RAX1 acts early in AM development and regulates CUC2 expression in a central axillary domain that anticipates the position of future AMs. We propose that RAX1 governs the spatial pattern of AM development by generating a tissue environment conducive for meristem establishment and therefore is involved in specifying the axillary stem cell niche.
Distinct Genetic Functions of LAS and RAX1 at Organ Boundaries
Cells recruited into organ primordia rapidly initiate expression of markers such as ASYMMETRIC LEAVES1 (Byrne et al., 2000) and AINTEGUMENTA (Elliott et al., 1996), which reflect the acquisition of determinate cell fate. By contrast, cells with indeterminate fate express markers such as STM (Long et al., 1996). A boundary zone that straddles the morphological demarcation between SAM and primordium is defined by the expression domains of LAS (Greb et al., 2003), CUC (Takada et al., 2001), and LOB (Shuai et al., 2002) genes. Cells can leave this dynamic zone; therefore, cell fate is not yet fixed (Laufs et al., 2004). AMs arise from the center of this boundary zone, consistent with the observation of low-level persistence of STM expression in the boundary domain early during primordium development, which suggests that boundary cells are not a priori determinate (Long and Barton, 2000). It is not clear how long this competent state is maintained. LAS is required for subsequent, focused, high-level STM expression upon onset of AM development (Greb et al., 2003), suggesting that LAS is required for reacquisition of indeterminate cell fate in axillary cells in the course of AM organization.
By contrast, RAX1 expression is spatially more restricted than LAS and is observed at a central position that anticipates the location of future AMs (Figure 4F). RAX1 is therefore the earliest known specific marker for AM position. RAX1 and LAS expression likely initiate at approximately the same stage of leaf primordium development (cf. Figures 4B and 4D to 4G with Figures 5E to 5H in Greb et al., 2003). High-level expression of RAX1, as observed in plants carrying the rax1-1D allele, which likely also somewhat expands its expression domain (Figure 2B; see Supplemental Figure 2 online), results in an enlarged zone of cells competent to initiate AMs, occasionally leading to two or more organizing centers for AM formation (Figures 1H and 1I). The expression domain of WUS in such incipient AMs is also expanded (Figures 4Q to 4S), suggesting higher stem cell–promoting activity. The analysis of rax1 mutant phenotypes indicates that RAX1 is necessary to specify AM position and sufficient to specify an AM stem cell niche within a boundary domain of cells competent to form AMs. Future experiments will determine whether RAX1 is sufficient to specify axillary stem cell identity. Our data indicate that RAX1 acts through CUC2. Interestingly, high-level expression of CUC1 is sufficient to stimulate formation of adventitious meristems on cotyledons (Takada et al., 2001).
It is tempting to speculate on the nature of the positional cue underpinning the RAX1 expression pattern. REVOLUTA, an HD-ZIP transcription factor, is required for formation of all lateral meristems (Talbert et al., 1995; Otsuga et al., 2001) and, in addition to other patterns of RNA accumulation, is expressed at adaxial positions of developing leaf primordia in a similar domain as RAX1 (Greb et al., 2003) and therefore could generate a positional signal for RAX1 expression. Perhaps differential accumulation of auxin directly provides a spatial cue: organogenesis requires auxin flux, which organizes a major conduit for auxin flow in the center of developing primordia (Reinhardt et al., 2003), and the RAX1 promoter has an auxin response element. Further experiments are necessary to understand the regulation of RAX1 expression.
Complex Phenotypes in rax1-1D/+ Plants
Two phenotypic aspects of the rax1-1D/+ plants appear paradoxical. First, while establishment of AM organizing centers is promoted (Figures 1H, 1I, and 4Q to 4S), rosette branching is highly reduced. We cannot exclude that this aspect of the rax1-1D/+ phenotype is neomorphic, caused by the expansion of the RAX1 expression domain (Figure 2B; see Supplemental Figure 2 online). Ectopic RAX1 expression might exacerbate local differences in GA levels between rax1-1D/+ mutants and wild-type plants. Furthermore, auxin is required for GA responses (Fu and Harberd, 2003); therefore, dynamic changes in basipetal auxin flux in the course of postembryonic development could further modify growth responses in rax1-1D/+ plants. Second, does our observation that the total number of branches (irrespective of their identity) in rax1-1D/+ compared with wild-type plants is only modestly reduced (Table 2) mean that branching is just shifted upwards in favor of cauline branches in the hemizygous mutants? Branch number is positively correlated with the length of vegetative development (Table 2, cf. Ws-2 in SD and LD). Furthermore, in rax1-1D/+, 0.25 ± 0.01 branches are made per leaf (rosette or cauline) on the primary stem, whereas in FA4C, 0.42 ± 0.07 branches are made. Thus, total branch number only appears similar because rax1-1D/+ plants have a longer vegetative phase while producing branches at a lower rate.
Is Control of Flowering Time a Direct Function of RAX1?
What is the significance of our observation that RAX1 represses GA accumulation in the shoot apex (Figure 6C)? GA is required for flowering in SD-grown Arabidopsis: GA-biosynthetic mutants are unable to flower under these conditions (Wilson et al., 1992), while treatments with exogenous GAs restore flowering. Modulation of GA levels by RAX1 has the effect of extending the vegetative phase, especially in SD conditions (Tables 2 and 4), which allows for the formation of more leaves and, hence, increased potential for rosette paraclade formation. Whether RAX1 is a selfish gene that sets up a positive feedback loop to promote vegetative identity remains to be determined. Modulation of GA levels by RAX1 could confer selective advantages if moderately late flowering in SD led to larger seed set or better dispersal by larger branch numbers.
However, GA does not only regulate flowering, but is also involved in enforcing determinate cell fate: KNOX transcription factors such as STM or KNAT1 suppress GA20-oxidase expression in indeterminate cells (Sakamoto et al., 2001); conversely, spindly, which shows constitutive GA responses, enhances the stm phenotype (Hay et al., 2002), suggesting that elevated GA levels or GA responses in the SAM impair indeterminacy. As GA can freely diffuse, these observations imply that low GA levels in the axillary boundary zone are critical to maintain indeterminacy of cells within it and the ability to organize AMs. Such control of GA levels in the axillary boundary could proceed by similar mechanisms as have been reported for the SAM by regulation of catabolic GA2-oxidase expression (Jasinski et al., 2005). Therefore, the observation of early flowering in rax1-2 could be simply a consequence of a diminished ability to block GA diffusion from developing leaf primordia to the SAM. Interestingly, our observation that restoration of CUC2 expression in the RAX1 expression domain in a rax1-2 background is sufficient to completely restore wild-type timing of flowering, but only partly suppresses the branching defect (Table 5), suggests that RAX1 has additional target genes necessary to promote AM formation and that AM formation and control of GA levels are mechanistically separable.
METHODS
Plant Materials, Mutants, and Growth Conditions
An activation-tagged collection was generated in the FA4C background (Colón-Carmona et al., 1999) using the pSKI015 vector (Weigel et al., 2000). Thermal asymmetric interlaced PCR was performed to identify sequences flanking T-DNA insertion sites, using four 35S enhancer-specific nested primers: 35S+21, 5′-ACGACACTCTCGTCTACTCCAAG-3′; 35S+53, 5′-GATACAGTCTCAGAAGACCAGAG-3′; 35S+93, 5′-AACAAAGGGTAATATCGGGAAAC-3′; and 35S+205, 5′-TAAAGGAAAGGCTATCGTTCAAG-3′ combined with the AD1, AD2, or AD3 degenerate primer (Liu et al., 1995). rax1-2 was isolated from the α T-DNA insert population (Krysan et al., 1999) by PCR-based screening (http://www.biotech.wisc.edu/arabidopsis/). The T-DNA insertion site in rax1-2 was identified by sequencing the PCR product amplified with the left border primer JL-202 and gene-specific primer myb37-6258: 5′-CCCATAAAACTGATCATAGTCGCTCTCTA-3′.
For genotyping, the wild-type RAX1 and rax1-1D alleles were detected by PCR using primer pairs: TF2001, 5′-GGTTTAACAGCCTGGCAAAAAACTTCAG-3′, and TF2561, 5′-CGATTGCATCAATCCCTTTCTCCTACG-3′, for the wild-type RAX1 allele and 35S+205 and TF2561 for the rax1-1D allele. The wild-type RAX1 and rax1-2 alleles were genotyped with primers myb37-4548, 5′-TCCTCCATAAACACAAAAAGTCCATCCTA-3′, and myb37-5684 for the wild-type allele and JL-202 and myb37-5684 for the rax1-2 allele. Genotypes for CUC1 and CUC2 loci were determined by PCR using primers as described (Takada et al., 2001).
Prior to genetic analysis, the rax1-1D and rax1-2 mutants, isolated in Col-0 or Ws-2 backgrounds, respectively, were backcrossed five times to Landsberg erecta or Col-0. rax1-2 was crossed into cuc1/cuc1 cuc2/+, all in the Landsberg erecta background.
Rosette branches were scored ∼3 weeks after onset of inflorescence stem elongation by visual inspection of individual plants. Only secondary growth axes elongated more than 5 mm were scored.
Plants were grown under LD (16 h light/8 h dark) or SD (9 h light/15 h dark) conditions in controlled environment rooms at 21°C on shelves with an average of 120 μE m−2 s−1 fluorescent light.
Plasmid Construction and Plant Transformation
All constructs were verified by DNA sequencing. To recapitulate the rax1-1D phenotype, genomic DNA was PCR amplified, comprising 4x35S enhancer elements, ∼2.3 kb 5′ of the RAX1 start codon, the entire coding region, including introns, and ∼0.6 kb 3′ untranslated region, and subsequently transformed into Col-0 plants. To complement rax1-2, the PCR-amplified transgene consisted of ∼1.9 kb 5′ of the RAX1 start codon, the entire coding region, including introns, and ∼0.6 kb of the 3′ untranslated region. To construct a ProRAX1:CUC2 fusion, an ∼2.1-kb genomic fragment encompassing the putative RAX1 promoter was amplified with primers myb37-2507 (5′-GTCGACAACATTACAACTCAAGGGCAGACG-3′) and myb37-4669 (5′-CACCCTAGGCTTCCCATTTCTCTCGTTAGTG-3′) and digested with SalI and AvrII. CUC2 was isolated as an ∼2.2-kb genomic NcoI-EcoRI fragment. These fragments were ligated together after addition of an AvrII-NcoI linker and introduced into the pSPTV50hyg plant transformation vector. The final construct was transformed into Col-0 plants via the Agrobacterium tumefaciens–mediated floral dip method (Clough and Bent, 1998).
Yeast Constructs and Methods
For RAX1, a cDNA fragment corresponding to amino acids 119 to 329 was cloned in frame downstream of the GAL4 DBD of the yeast expression vector pGBT9 (Clontech). The activation control construct GAL4DBD:THM18 with sequences encoding amino acids 163 to 256 of THM18 (Schwechheimer et al., 1998) and the repression control construct GAL4DBD:MYB4 with sequences encoding amino acids 163 to 282 of MYB4 (Jin et al., 2000) were made in a similar way. Yeast strain HF7c (Clontech) was transformed by the LiAc method (http://www.umanitoba.ca/faculties/medicine/biochem/gietz/).
The protocol for liquid culture β-galactosidase assays in yeast (Clontech; Yeast Protocol Handbook PT3024-1) was modified for use with 4-methyl umbelliferyl β-d-galactopyranoside as a substrate. After assay for 15 min at 30°C, the fluorescent product, 4-methyl umbelliferone, was detected in a spectrofluorometer (BMG Labtechnologies) using excitation and emission wavelengths of 360 and 460 nm, respectively. Enzyme activity was calculated as the amount of 4-methyl umbelliferone produced per OD600 per minute. This assay was repeated on four independently transformed lines.
GA Treatments and Quantification of Endogenous GA
Application of exogenous GA3 (G-7645; Sigma-Aldrich) during vegetative growth under LD and SD conditions was achieved by spraying soil-grown plants twice weekly with a solution of 100 μM GA3 and 0.02% Tween-20. Control plants were treated with the same concentration of surfactant in water. For GA measurements, shoot apices were microdissected by hand to remove all leaf primordia >2mm, stem segments, and roots. Endogenous GAs were extracted from apices and analyzed by gas chromatography–tandem mass spectrometry (JMS-MStation 700; JEOL) using 2H2-labeled GAs (L. Mander, Canberra, Australia) as internal standards as described (Eriksson et al., 2000). Averaged GA values were normalized to GA concentrations in vegetative apices of corresponding lines.
RNA Analysis, RT-PCR, and Quantitative PCR
RNA was isolated from apices and leaves using Trizol reagent (Invitrogen). For RNA gel blot analysis, 20 μg total RNA was separated by electrophoresis, transferred to Hybond N+ nylon membranes, hybridized, and washed following standard procedures (Ausubel et al., 1987). A RAX1-specific cDNA probe was amplified with primers TF258, 5′-AGGAAGCAGGTGGTCAATAATAGC-3′, and TF759, 5′-CCTTTTGTCCTCTGGTCAATGTGG-3′. RNA loading was checked by hybridization to eIF-4A transcripts.
Reverse transcription reactions were done with 1 μg of total RNA and 250 ng oligo(dT) or 10 pmol gene-specific primers in 10-μL reactions according to the manufacturer's protocol (ABgene). Diluted aliquots of the resulting reverse transcription reaction products were used for quantitative PCR (Q-PCR) analysis. Q-PCR was performed in an iCycler (Bio-Rad) using SYBR Green I (Molecular Probes) and ABsolute QPCR mix (ABgene) with 40 ng primers in a total of 20 μL per reaction. Quadruplicate Q-PCR reactions were averaged. The following primer pairs were used for RAX1 (At5g23000): myb37-4997, 5′-GCCATAGGAAGCAGGTGGTC-3′, and myb37-5593, 5′-GGGATTGTTGTTGGTGAGGT-3′, or myb37-5684, 5′-GTCACCAGCTTCGAAGCCATTG-3′. To confirm the absence of a truncated RAX1 transcript upstream of the T-DNA insertion in rax1-2, the primer pair myb37 5′ RT1, 5′-ACTTAGAGATTACATTGAAAAGTATGGT-3′, and myb37 5′ RT2, 5′-CAGCGAAGAGACTAAAAATGATC-3′, was used. Other specific primers were as follows: CUC2-243, 5′-GAAGTATCCGACGGGACTGA-3′, and CUC2-487, 5′-GATCACCCATTCATCCTTGGAG-3′, for CUC2 (At5g53950) and elFT22, 5′-TTCGCTCTTCTCTTTGCTCTC-3′, and elFB221, 5′-GAACTCATCTTGTCCCTCAAGTA-3′, for eIF-4A (At3g13920). Relative transcript levels in all samples were normalized using eIF-4A. A no-template control was included in each set of reactions to confirm the absence of contamination. The primers spanned at least one intron to distinguish cDNA amplification products from genomic DNA contamination.
In Situ Hybridization
Shoot apices were fixed in p-formaldehyde, paraffin embedded, sectioned to 8 μm, affixed to Probe Plus slides (Fisher Scientific) at 42°C overnight, and processed as described (http://www-ciwdpb.stanford.edu/research/barton/in_situ_protocol.html) with color substrate incubation overnight. For probes, a 707-bp RAX1 fragment was amplified with gene-specific primers myb37-5551, 5′-TTTCTCAGGATGTGAAAAGACCAACC-3′, and myb37-6258, 5′-CCCATAAAACTGATCATAGTCGCTCTCTA-3′; a 627-bp CUC2 template was synthesized with primers CUC2-1703, 5′-CCAGAAAACCACTTTAGCTAGC-3′, and CUC2-2330, 5′-TCAGTAGTTCCAAATACAGTCAA-3′; a 545-bp STM fragment was amplified from cDNA with primers stm-1, 5′-ATGGAGAGTGGTTCCAACAG-3′, and stm-1505, 5′-GATCAAGCCCTGGATCTTCA-3′; and a full-length cDNA generated from pMH16 was used as WUS probe (a gift from Martin Hobe). Fragments were cloned in sense and antisense orientation into pGEM-Teasy (Promega), and probes were synthesized by T7 RNA polymerase.
Phylogenetic Analysis
Conceptually translated cDNA sequences corresponding to the conserved DBD were aligned using ClustalW in the Lasergene suite (DNAStar) (see Supplemental Figure 7 online). The alignment file was used to generate a phylogenetic tree and to calculate the posterior probabilities of nodes with the Bayesian method implemented in MrBayes3.0 (Ronquist and Huelsenbeck, 2003; http://mrbayes.csit.fsu.edu/index.php). The program was run with the default settings for 100,000 generations.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AK175507 (MYB37), AJ012310 (WUS), NM_124774 (CUC2), AF543194 (CUC3), and NM_104434 (LAS).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Flower Number in Wild-Type, rax1-1D/+, and rax1-2 Plants.
Supplemental Figure 2. RAX1 Is Ectopically Expressed in rax1-1D/+ Leaves.
Supplemental Figure 3. RAX1 Transcripts Do Not Accumulate in rax1-2.
Supplemental Figure 4. Expression of LAS and CUC3 in rax1-1D/+ Shoot Apices
Supplemental Figure 5. The Juvenile Phase of Vegetative Development Is Not Altered in rax1 Mutants.
Supplemental Figure 6. GA Sprays during Vegetative Development Suppress Late Flowering in rax1-1D/+ Plants.
Supplemental Figure 7. ClustalW Alignment of Arabidopsis and Tomato Myb Genes.
Supplementary Material
Acknowledgments
We thank Shalu Mittal, Ratha You, Heng Phung, and Tal Heimovitch-Gal for help in generating the activation-tagged collection in the FA4C background. We also thank Marc Morgan, Stephanie Albouhair, Lydia Barth, Bavani Krishnan, and Angie Ng Ah Sock for help with characterizing the rax1 mutants. We thank Chris Jeffries for help with scanning electron microscopy analysis. We thank Ken-ichiro Hibara for seeds of the cuc1/cuc1 cuc2/+ mutant, Cathie Martin for the GAL4DBD:THM18 and GAL4DBD:At MYB4 clones and the Nottingham Arabidopsis Stock Centre for seed of various mutants. This work was supported by grants from Akkadix and the Biotechnology and Biological Science Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Peter Doerner (peter.doerner@ed.ac.uk).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.038588.
References
- Aida, M., Ishida, T., and Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126 1563–1570. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., More, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1987). Current Protocols in Molecular Biology. (New York: Greene Publishing Associates and Wiley-Interscience).
- Blazquez, M.A., Green, R., Nilsson, O., Sussman, M.R., and Weigel, D. (1998). Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. [DOI] [PubMed] [Google Scholar]
- Cline, M.G. (1997). Concepts and terminology of apical dominance. Am. J. Bot. 84 1064–1069. [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Colón-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20 503–508. [DOI] [PubMed] [Google Scholar]
- Daimon, Y., Takabe, K., and Tasaka, M. (2003). The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol. 44 113–121. [DOI] [PubMed] [Google Scholar]
- Doerner, P. (2003). Plant meristems: A merry-go-round of signals. Curr. Biol. 13 R368–R374. [DOI] [PubMed] [Google Scholar]
- Elliott, R.C., Betzner, A.S., Huttner, E., Oakes, M.P., Tucker, W.Q., Gerentes, D., Perez, P., and Smyth, D.R. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, M.E., Israelsson, M., Olsson, O., and Moritz, T. (2000). Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 18 784–788. [DOI] [PubMed] [Google Scholar]
- Feng, C., Andreasson, E., Maslak, A., Mock, H.P., Mattsson, O., and Mundy, J. (2004). Arabidopsis MYB68 in development and responses to environmental cues. Plant Sci. 167 1099–1107. [Google Scholar]
- Fu, X., and Harberd, N.P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421 740–743. [DOI] [PubMed] [Google Scholar]
- Gallois, J.L., Woodward, C., Reddy, G.V., and Sablowski, R. (2002). Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129 3207–3217. [DOI] [PubMed] [Google Scholar]
- Grbic, V., and Bleecker, A.B. (2000). Axillary meristem development in Arabidopsis thaliana. Plant J. 21 215–224. [DOI] [PubMed] [Google Scholar]
- Greb, T., Clarenz, O., Schafer, E., Muller, D., Herrero, R., Schmitz, G., and Theres, K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, K.A., Prigge, M.J., Katzman, R.B., and Clark, S.E. (2005). CORONA, a member of the class III homeodomain leucine zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell 17 691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker, A., Gross-Hardt, R., Geiges, B., Sarkar, A., Breuninger, H., Herrmann, M., and Laux, T. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131 657–668. [DOI] [PubMed] [Google Scholar]
- Hay, A., Kaur, H., Phillips, A., Hedden, P., Hake, S., and Tsiantis, M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hempel, F.D., and Feldman, L.J. (1994). Bi-directional inflorescence development in Arabidopsis thaliana: Acropetal initiation of flowers and basipetal initiation of paraclades. Planta 192 276–286. [Google Scholar]
- Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, I., Phillips, A., Hedden, P., and Tsiantis, M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15 1560–1565. [DOI] [PubMed] [Google Scholar]
- Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., Tonelli, C., Weisshaar, B., and Martin, C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs, P., Peaucelle, A., Morin, H., and Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131 4311–4322. [DOI] [PubMed] [Google Scholar]
- Lenhard, M., Jurgens, G., and Laux, T. (2002). The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 129 3195–3206. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8 457–463. [DOI] [PubMed] [Google Scholar]
- Long, J., and Barton, M.K. (2000). Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 218 341–353. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- McSteen, P., and Leyser, O. (2005). Shoot branching. Annu. Rev. Plant Biol. 56 353–374. [DOI] [PubMed] [Google Scholar]
- Otsuga, D., DeGuzman, B., Prigge, M.J., Drews, G.N., and Clark, S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25 223–236. [DOI] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Ronquist, F., and Huelsenbeck, J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 1572–1574. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., Kamiya, N., Ueguchi-Tanaka, M., Iwahori, S., and Matsuoka, M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, G., and Theres, K. (2005). Shoot and inflorescence branching. Curr. Opin. Plant Biol. 8 506–511. [DOI] [PubMed] [Google Scholar]
- Schmitz, G., Tillmann, E., Carriero, F., Fiore, C., Cellini, F., and Theres, K. (2002). The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc. Natl. Acad. Sci. USA 99 1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C., Smith, C., and Bevan, M.W. (1998). The activities of acidic and glutamine-rich transcriptional activation domains in plant cells: Design of modular transcription factors for high-level expression. Plant Mol. Biol. 36 195–204. [DOI] [PubMed] [Google Scholar]
- Shuai, B., Reynaga-Pena, C.G., and Springer, P.S. (2002). The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296 285–289. [DOI] [PubMed] [Google Scholar]
- Snow, M., and Snow, R. (1942). The determination of axillary buds. New Phytol. 41 13–22. [Google Scholar]
- Stirnberg, P., Chatfield, S.P., and Leyser, H.M. (1999). AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol. 121 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg, P., van de Sande, K., and Leyser, H.M.O. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129 1131–1141. [DOI] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 447–456. [DOI] [PubMed] [Google Scholar]
- Sussex, I.M. (1955). Morphogenesis in Solanum tuberosum L: Experimental investigation of leaf dorsoventrality and orientation of the juvenile shoot. Phytomorphology 5 286–300. [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128 1127–1135. [DOI] [PubMed] [Google Scholar]
- Talbert, P.B., Adler, H.T., Parks, D.W., and Comai, L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121 2723–2735. [DOI] [PubMed] [Google Scholar]
- Telfer, A., Bollman, K.M., and Poethig, R.S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124 645–654. [DOI] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.N., Heckman, J.W., and Somerville, C.R. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.