Abstract
Temperature compensation contributes to the accuracy of biological timing by preventing circadian rhythms from running more quickly at high than at low temperatures. We previously identified quantitative trait loci (QTL) with temperature-specific effects on the circadian rhythm of leaf movement, including a QTL linked to the transcription factor FLOWERING LOCUS C (FLC). We have now analyzed FLC alleles in near-isogenic lines and induced mutants to eliminate other candidate genes. We showed that FLC lengthened the circadian period specifically at 27°C, contributing to temperature compensation of the circadian clock. Known upstream regulators of FLC expression in flowering time pathways similarly controlled its circadian effect. We sought to identify downstream targets of FLC regulation in the molecular mechanism of the circadian clock using genome-wide analysis to identify FLC-responsive genes and 3503 transcripts controlled by the circadian clock. A Bayesian clustering method based on Fourier coefficients allowed us to discriminate putative regulatory genes. Among rhythmic FLC-responsive genes, transcripts of the transcription factor LUX ARRHYTHMO (LUX) correlated in peak abundance with the circadian period in flc mutants. Mathematical modeling indicated that the modest change in peak LUX RNA abundance was sufficient to cause the period change due to FLC, providing a molecular target for the crosstalk between flowering time pathways and circadian regulation.
INTRODUCTION
Circadian clocks provide organisms with a means of temporally organizing their daily metabolic and physiological activities relative to the day/night cycle. Such organization is believed to impart a selective advantage (Ouyang et al., 1998; Dodd et al., 2005). Nearly all eukaryotes and some prokaryotes possess circadian clocks, and comparison between diverse model species shows that although their components and construction are varied, they do share a unified set of defining properties: all circadian clocks are self-sustaining, entrainable, and temperature compensated (Pittendrigh, 1960). Transcription-translation feedback loops involving multiple positive and negative interacting components are important in the clock mechanisms of these model species (reviewed in Young and Kay, 2001). The Arabidopsis thaliana clock is putatively based on the feedback loop involving the genes TIMING OF CAB EXPRESSION1 (TOC1), CIRCADIAN CLOCK ASSOCIATED1 (CCA1), and LATE ELONGATED HYPOCOTYL (LHY), where TOC1 induces the transcription of LHY and CCA1, which are translated into proteins that feed back to repress the expression of TOC1 (Alabadi et al., 2001). Modeling of this clock suggested that it was insufficient to explain the experimental data (Locke et al., 2005a). As a result, Locke et al. (2005b) added two further components to the model: a gene X, which lies between TOC1 and LHY/CCA1, and a gene Y, which forms a coupled loop and is likely to correspond to the evening-expressed gene GIGANTEA (GI).
Temperature compensation, a defining feature of circadian rhythms, results in the period of the clock changing very little when measured over a broad range of constant temperatures (Pittendrigh, 1954; Rensing and Ruoff, 2002). Natural genetic variation in the Drosophila melanogaster central clock gene period was shown to affect the temperature compensation of the fly clock (Sawyer et al., 1997). Analysis of circadian period in Arabidopsis accessions revealed natural genetic variation in the temperature compensation of the plant clock (Edwards et al., 2005). This variation was used to map quantitative trait loci (QTL) for circadian period in recombinant inbred lines (RILs) derived from a cross between the accessions Landsberg erecta (Ler) and Cape Verde Islands (Cvi) (Edwards et al., 2005).
FLOWERING LOCUS C (FLC) was suggested as a candidate for the period QTL PerCv5b, mapped on the upper arm of chromosome 5 (Edwards et al., 2005). FLC is a MADS box transcription factor that inhibits the transition to flowering by repressing the expression of the floral integrators AGAMOUS LIKE20 (AGL20) and FLOWERING LOCUS T (Samach et al., 2000; Michaels et al., 2005). Prolonged cold treatment, such as a winter, represses the expression of FLC in a process called vernalization, allowing plants to flower in the subsequent warmer conditions (reviewed in Bastow et al., 2004; Sung and Amasino, 2004). FLC is also regulated by a suite of autonomous pathway genes, including FRIGIDA (FRI), LUMINIDEPENDENS (LD), and FLOWERING LOCUS D (FLD; Michaels and Amasino, 1999; He et al., 2003).The Ler allele of flc was shown to be weakly expressed as the result of a transposable element within the first intron of the gene (Gazzani et al., 2003), making it a strong candidate for a QTL in the Cvi crossed with Ler RILs. Indeed, Swarup et al. (1999) previously identified a circadian period QTL (named ANDANTE) linked to FLC and showed a slight (∼0.8 h) short circadian period phenotype in flc mutant seedlings, indicating that FLC could also affect the circadian clock. The molecular target of FLC function was unknown.
Microarray assays have enabled the large-scale identification of transcripts regulated by the circadian clock in Arabidopsis, Drosophila, and mouse (Harmer et al., 2000; Ceriani et al., 2002; Panda et al., 2002). Identifying rhythmic patterns in the short time courses and sparse samples typical of circadian array data has been challenging; a number of approaches have been adopted for different experimental designs (Akhtar et al., 2002; Langmead et al., 2002; Straume, 2004). Harmer et al. (2000) suggested that ∼6% of the Arabidopsis genome was regulated by the circadian clock. Clustering the rhythmic expression patterns using the time of the peak allowed the identification of an overrepresented regulatory sequence (the evening element) and of functional relationships among some coexpressed genes, though this study tested only ∼8000 genes (Harmer et al., 2000).
We now pursue the analysis of the PerCv5b QTL, using near-isogenic lines (NILs) and mutants to identify natural variation at FLC as the cause of the 27°C-specific QTL effect on circadian period. Using genome-wide transcriptomic analysis, we identify the likely mechanism by which FLC alters the period of the circadian clock at this higher temperature. Our results illustrate the benefits of functional genomics approaches, combined with dedicated data analysis methods and mathematical modeling, in understanding the quantitative molecular mechanisms downstream of a QTL of moderate effect.
RESULTS
FLC Causes the PerCv5b QTL
We previously mapped six temperature-dependent QTL for circadian period in the Cvi crossed with Ler RILs (Edwards et al., 2005). PerCv5b mapped to the top of Chromosome 5. The Cvi allele of this QTL was estimated to cause a 0.75-h period lengthening effect specifically at 27°C (Edwards et al., 2005). Figure 1 summarizes the mapping of the PerCv5b QTL to the MADS box transcription factor FLC.
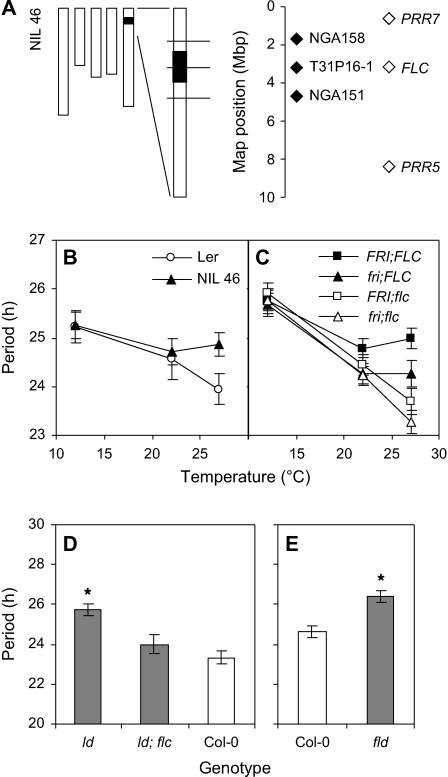
Figure 1.
FLC Alters the Period of the Clock in a Temperature-Dependent Manner.
(A) NIL46 summary map showing the five Arabidopsis chromosomes and an expanded view of the top of Chromosome 5. Genotype of chromosomal regions is displayed graphically for Ler (white) and Cvi (black), with recombination break points indicated at the midpoint between molecular markers. Position of molecular markers (closed diamonds) and clock-related genes (open diamonds) are shown at right. The leaf movement period of NIL46 and Ler (B) and the fri and flc single and double mutant combinations (C) were assayed at 12, 22, and 27°C. Symbols are indicated in inset legends. Bars represent se. Leaf movement period of ld, ld; flc (D), and fld (E) mutants along with wild-type Col-0 seedlings at 27°C. Bars represent variance-weighted se of period estimates. Asterisks (D) and (E) indicate t test P value < 0.05 compared with Col-0.
NILs, containing a small Cvi genomic region around the PerCv5b locus in an otherwise isogenic Ler background were constructed to isolate and confirm the effect of the QTL. NIL46 contained a Cvi introgression of <3 Mbp around PerCv5b (Figure 1A). Analysis of rhythmic leaf movement in this line showed a 27°C-specific period lengthening effect of 0.9 h relative to Ler, consistent with the QTL effect (Figure 1B, Table 1; Edwards et al., 2005). A second independently derived NIL with an equivalent Cvi introgression to NIL46 showed the same phenotype (data not shown). The clock-affecting genes PSEUDORESPONSE REGULATOR 7 (PRR7) and PRR5 (Nakamichi et al., 2005) were considered as candidates for the multiple Chromosome 5 QTL mapped in our work and a previous study (Michael et al., 2003). Neither of these genes lay within the Cvi introgression of NIL46, ruling them out as the cause of the QTL effect contained within this NIL (Figure 1A). FLC was an alternative candidate gene within the Cvi introgression of NIL46 (Figure 1A).
Table 1.
Mapping PerCv5b to FLC: NIL and Mutant Periods
| 12oC
|
22oC
|
27oC
|
||||
|---|---|---|---|---|---|---|
| Line | Period (h) | se | Period (h) | se | Period (h) | se |
| Ler | 25.22 | 0.32 | 24.56 | 0.41 | 23.94 | 0.32 |
| NIL46 | 25.27 | 0.29 | 24.72 | 0.27 | 24.86** | 0.25 |
| FRI; FLC | 25.76 | 0.21 | 24.78 | 0.20 | 24.99 | 0.22 |
| fri; FLC | 25.69 | 0.24 | 24.26* | 0.20 | 24.26* | 0.27 |
| FRI; flc | 25.91 | 0.23 | 24.43 | 0.21 | 23.69** | 0.26 |
| fri; flc | 25.76 | 0.25 | 24.22* | 0.20 | 23.27**a | 0.24 |
Leaf movement period estimates for NIL46 and fri and flc mutant combinations. Asterisks indicate P values of t test of period versus Ler for NIL46 and FRI; FLC for mutants at * <0.05 and ** <0.01.
P value <0.01 of Student's t test of fri; flc versus fri; FLC.
Period phenotypes in flc mutants (Swarup et al., 1999) and natural variation in the Ler allele of the gene (Gazzani et al., 2003) supported FLC as a candidate for PerCv5b. To test this possibility, we measured the period of flc mutant seedlings in combination with mutant or wild-type alleles of FRI, a positive regulator of FLC expression (Michaels and Amasino, 2001). No major period differences were shown between the lines at 12°C, but small differences were shown at 22°C, and by 27°C flc mutant seedlings had significantly shorter periods than plants with wild-type FLC alleles (Figure 1C, Table 1). This response was opposite to that shown by NIL46, and both data sets suggested that stronger expression of FLC resulted in longer circadian period at higher temperatures (Figures 1B and 1C).
Plants of FRI; FLC genotype showed a longer period than those of fri; FLC, suggesting that the upregulation of FLC by FRI might contribute toward the period lengthening (Figure 1C). To test this, we measured the circadian period of leaf movement in plants mutated in LD or FLD, genes that normally repress the expression of FLC (Michaels and Amasino, 1999; He et al., 2003). Figures 1D and 1E show that the period of ld and fld mutant seedlings at 27°C was significantly longer than wild-type seedlings. The period change was prevented when the ld mutant was combined with a mutant allele of flc, indicating that period lengthening in the ld mutant required FLC and was presumably mediated by increased FLC expression levels in the ld background (Figure 1D).
Mechanisms of FLC Effects on the Clock
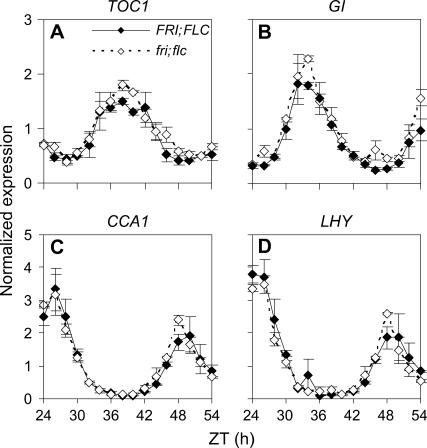
We tested whether FLC expression levels increased with temperature, thereby mediating the temperature-dependent period lengthening in FLC-expressing genotypes, but found little difference in the level of FLC transcript between 22 and 27°C (see Supplemental Figure 1 online). This suggested that altered FLC function rather than expression level caused the temperature dependence of the effect of FLC on the circadian clock. A simple possibility was that FLC might alter the period of the clock by regulating the transcription of a clock gene. To test this, the temporal expression patterns of TOC1, GI, CCA1, and LHY were compared between fri; flc and FRI; FLC seedlings at 27°C. The timing of expression of all rhythmic genes was expected to alter owing to the FLC-dependent change in period, but the target of FLC might additionally show altered expression levels. Figure 2 shows that neither the mean nor the peak level of transcript abundance at 27°C was clearly affected by FLC expression. It was therefore unlikely that one of these central clock genes was an early target of FLC regulation.
Figure 2.
Temporal Expression of Clock Genes in the FRI; FLC and fri; flc Genotypes at 27°C.
The temporal pattern in transcript abundance of the clock genes TOC1 (A), GI (B), CCA1 (C), and LHY (D) was analyzed by real-time PCR in FRI; FLC (closed diamonds) and fri; flc (open diamonds) genotype seedlings. Expression levels for each gene were normalized to the average for the fri; flc genotype with respect to ACTIN2 (ACT2). Data shown are the average of two biological replicates, with error bars representing the range.
Candidate Targets for FLC Regulation
To identify genes that might mediate the effect of FLC on the clock, global transcript profiles were compared in the fri; flc and FRI; FLC genotypes using the Affymetrix ATH1 microarray. RNA samples were taken from the two genotypes, under the conditions used for the leaf movement experiments at 27°C, and pooled from four time points spaced equidistantly across one circadian cycle (see Methods), in case FLC-dependent regulation was detectable only at specific circadian phases. Pooling may have reduced the effect shown by such genes, but it enabled a broader screen of genes peaking at different circadian phases. In the fri; flc genotype, FLC showed very low expression and AGL20 showed more than threefold increased expression compared with FRI; FLC (see Supplemental Table 1 online). As AGL20 is transcriptionally repressed by FLC in other conditions (Hepworth et al., 2002), this confirmed our ability to identify FLC-regulated genes.
Transcripts were ranked according to change in expression between the fri; flc and FRI; FLC genotypes, and the 1000 genes showing the greatest fold changes were termed FLC responsive (see Supplemental Table 1 online). This level was selected as an arbitrary cutoff to allow focus on potential candidate genes in the following microarray experiment. The FLC-responsive genes did not include any of the clock genes tested in Figure 2 nor any other genes thought to be important to circadian function at the time.
Global Analysis of Rhythmic Gene Expression
Regulators of the circadian clock are often themselves rhythmically regulated, so identifying rhythmic transcripts among the FLC-responsive genes could highlight possible targets of FLC regulation in the circadian clock mechanism. We therefore identified rhythmically regulated transcripts using the Affymetrix ATH1 array. Eight-day-old Columbia (Col-0) seedlings grown under 12-h-light/12-h-dark cycles (LD 12:12) were transferred to constant light at 22°C and harvested at 13 time points, covering two circadian cycles in 4-h intervals, starting 26 h after the last dark–light transition. This time is referred to as Zeitgeber time (ZT) 26, where ZT0 is the time of the last dark–light transition. Expression values were scored for circadian regulation using the modified cosinor analysis program COSOPT (Straume, 2004), which was previously used to score circadian expression of genes in Drosophila and mouse (Ceriani et al., 2002; Panda et al., 2002), and is similar to the algorithm used in the earlier array study in Arabidopsis (Harmer et al., 2000).
Previous studies using this method employed a threshold of 0.1 for the probability (pMMC-β) that the best-fit rhythm had a significant amplitude (Harmer et al., 2000; Panda et al., 2002). pMMC-β cutoff values of 0.10, 0.05, and 0.02 were considered to score rhythmic transcripts in our data (Table 2), and all three scored the putative central oscillator components TOC1, CCA1, and LHY as rhythmic. The remaining PRR genes were all scored rhythmic at pMMC-β <0.1, but PRR9 was not included at <0.05, and further circadian genes were also excluded at the cutoff of <0.02, suggesting this threshold was too stringent. Visual inspection of the expression patterns of genes around the thresholds suggested that 0.1 might be too liberal for our data; thus, the threshold of <0.05 was selected for scoring rhythmic transcripts.
Table 2.
Genes Scored Rhythmic by COSOPT
| pMMC-β | No. Rhythmic | Rhythmic (%) | Harmer et al. (2000) (%) | |
|---|---|---|---|---|
| <0.10 | 5127 | 22.54 | 88.94 | |
| <0.05 | 3504 | 15.40 | 82.80 | |
| <0.02 | 1729 | 7.60 | 61.43 | |
Number of rhythmic genes shown as total and percentage of genes on the array as well as the percentage of genes scored rhythmic by Harmer et al. (2000) at three pMMC-β thresholds.
At this threshold, 3503 genes were scored as rhythmic out of 22,746 probe sets represented on the array, suggesting that ∼16% of the Arabidopsis genome was regulated by the circadian clock. This rhythmic set included 83% of the rhythmic genes identified by Harmer et al. (2000) using an 8000-gene array (Table 2).
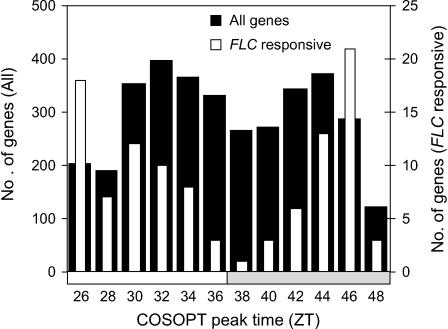
All circadian phases were well represented based on COSOPT estimates of peak phase times (Figure 3). As in Harmer et al. (2000), an increased number of genes peaked around the end of the subjective day (ZT30 to ZT36 h; subjective dawns are ZT24 and ZT48, subjective dusk is ZT36) and the second half of the subjective night (ZT42 to ZT46 h). However, fewer transcripts were shown to peak around the subjective dawns (ZT26 to ZT28 and ZT48), which was not observed in the previous study (Harmer et al., 2000). Of the FLC-responsive genes, 105 were included in the rhythmic set. The distribution of their peak phases followed the overall pattern but also showed an increased number peaking around ZT26 and ZT46 (Figure 3).
Figure 3.
Distribution of COSOPT Peak Phases.
Microarray time course expression profiles of Arabidopsis genes were scored for circadian rhythmicity with the program COSOPT. Phase estimates for all rhythmic genes (pMMC-β <0.05; closed bars) and rhythmic FLC-responsive genes (open bars) were binned into 2-h intervals. The number of genes was plotted for each bin, labeled with the lower period bound of the bin. Primary y axis (left) represents total number of genes and secondary axis (right) represents number of FLC-responsive genes. The bar on the x axis represents subjective day (white) and night (gray).
Promoter analysis of the COSOPT phase clusters supported the suggested role of the evening element regulatory sequence (AAAATATCT; Harmer et al., 2000; Harmer and Kay, 2005), with this sequence, and a one base variant (AAATATCT), being overrepresented in genes peaking late in the subjective day (ZT34 to ZT38; P < 2 × 10−6). Also, the G-box core sequence (CACGTG) was overrepresented in genes peaking at the end of the subjective night (ZT44 to ZT46; P < 0.004), suggesting that G-box binding factors may play a role in determining this phase of expression.
COSOPT expression phase clusters were also tested for overrepresentation of groups of functionally related genes using Gene Ontology (GO) annotations. Among the most significant patterns, genes involved in photosynthesis were overrepresented around the middle of the subjective day (ZT30; P < 0.001), while genes involved in phenylpropanoid metabolism were overrepresented just before subjective dawn (ZT46; P < 0.001), consistent with previous findings (Harmer et al., 2000). Conversely, genes involved in glucose and alcohol catabolism (P = 0.001 and 0.002, respectively) and carbon utilization (P = 0.011) showed overrepresentation in the middle of the subjective night (ZT42), ∼6 h later than previously suggested for genes involved in carbon metabolism (Harmer et al., 2000).
The COSOPT analysis provided a robust method of detecting rhythmic genes and their associated properties but has limited capacity to discriminate among rhythmic waveforms. To assist the identification of candidate circadian regulators, rather than downstream clock-regulated targets, we applied a complementary clustering method.
Clustering of Expression Patterns Using Fourier Series
Fourier analysis provides a well-understood, rapid, and flexible means of characterizing rhythmic waveforms in terms of a combination of sine and cosine waves. The Fourier coefficients measure the contribution of sine and cosine waves with differing periods (for our time series, the six harmonics used are 48, 24, 16, 12, 9.6, and 8 h) to the rhythmic patterns in the data. This efficiently captures the rhythmic properties of interest to us, so we developed an agglomerative, hierarchical method of clustering our gene expression patterns based on the Fourier coefficients using a Bayesian statistical approach (see Methods). Bayesian Fourier clustering (BFC) can discriminate among circadian-regulated patterns based on the amplitude and waveform of the rhythm, in addition to the phase. Circadian-regulated expression profiles were identified by the dominant contribution of the sine and cosine waves with a 24-h period. For our data, this was reflected in the Fourier coefficients for the second harmonic, which we measured using the circadian score (see Methods).
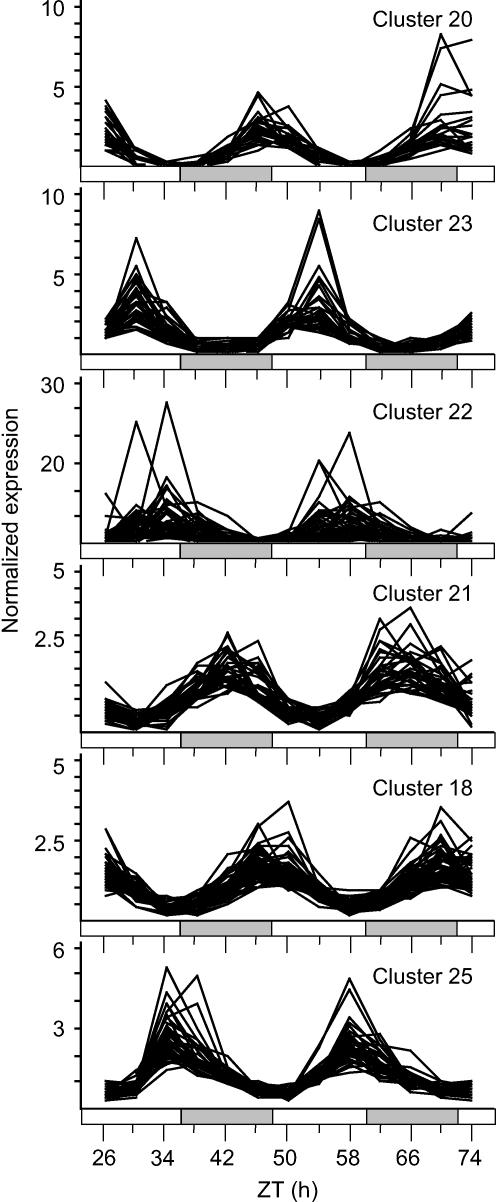
We identified 27 circadian-regulated clusters comprising 3063 genes (13% of transcripts represented on the array). These accounted for ∼70% of the genes scored rhythmic by Harmer et al. (2000). Of the genes in circadian clusters, 65% were also scored as rhythmic by COSOPT. This rose to 86% in the six clusters with highest amplitude and fell to 50% in the six lowest amplitude clusters. Figure 4 shows the expression patterns of all genes in the six clusters of highest amplitude. All of the clusters are summarized in Table 3 and are shown in Supplemental Figure 2 online; the distribution of cluster phases, amplitudes, and gene numbers around the circadian cycle are shown in Figure 5. The clusters varied significantly in the similarity of the individual expression profiles and in the robustness of their circadian rhythmicity but readily discriminated between genes with different amplitudes at the same peak phase (Figure 5; see Supplemental Figure 2 online).
Figure 4.
Bayesian Clustering of Rhythmic Genes.
BFC was applied to microarray time course data to identify rhythmic genes. Graphs in vertical order showing expression profiles of genes in the six clusters scored with the highest amplitude. Cluster numbers are shown in the top right of each graph. Bars on x axes represent subjective day (white) and night (gray). The full analysis results are available from www.amillar.org.
Table 3.
BFC Identified Clusters
| No. | Circadian Score | Amplitude | Phase (ZT) | No. Genes | Rhythmic Circadian Score (%) | Rhythmic COSOPT (%) | Clock-Related Genes |
|---|---|---|---|---|---|---|---|
| 1 | 0.54 | 0.11 | 35.3 | 181 | 56 | 51 | |
| 2 | 0.45 | 0.11 | 42.8 | 311 | 45 | 32 | |
| 3 | 0.71 | 0.24 | 43.3 | 208 | 81 | 72 | PHYE |
| 4 | 0.76 | 0.18 | 43.7 | 72 | 97 | 94 | PHYB |
| 5 | 0.61 | 0.20 | 47.7 | 280 | 78 | 68 | SPA1 |
| 6 | 0.60 | 0.25 | 50.0 | 54 | 81 | 74 | |
| 7 | 0.64 | 0.19 | 29.5 | 306 | 74 | 59 | |
| 8 | 0.62 | 0.24 | 30.3 | 53 | 94 | 75 | CRY1 |
| 9 | 0.78 | 0.29 | 32.0 | 292 | 87 | 73 | PHYA, WNK1 |
| 10 | 0.56 | 0.24 | 42.7 | 98 | 77 | 79 | |
| 11 | 0.67 | 0.34 | 41.3 | 46 | 100 | 100 | |
| 12 | 0.46 | 0.24 | 34.2 | 47 | 49 | 38 | |
| 13 | 0.71 | 0.35 | 35.4 | 60 | 100 | 100 | |
| 14 | 0.50 | 0.23 | 34.5 | 325 | 41 | 34 | ELF4, ELF3 |
| 15 | 0.69 | 0.44 | 48.9 | 26 | 100 | 100 | |
| 16 | 0.70 | 0.38 | 45.7 | 63 | 100 | 97 | |
| 17 | 0.73 | 0.53 | 45.0 | 126 | 97 | 85 | |
| 18* | 0.76 | 0.73 | 46.8 | 59 | 100 | 95 | COL1 |
| 19 | 0.67 | 0.54 | 50.0 | 20 | 95 | 100 | |
| 20* | 0.80 | 1.60 | 46.9 | 24 | 100 | 100 | LHY, CCA1 |
| 21* | 0.70 | 0.84 | 42.4 | 34 | 100 | 94 | |
| 22* | 0.76 | 0.96 | 34.1 | 58 | 74 | 57 | GI, PRR5 |
| 23* | 0.78 | 1.06 | 30.2 | 30 | 100 | 87 | PRR7, PRR9, PIF4, PIL6, EPR1 |
| 24 | 0.39 | 0.37 | 26.0 | 21 | 0 | 5 | |
| 25* | 0.64 | 0.70 | 35.1 | 38 | 100 | 97 | TOC1, PRR3, FKF1, LUX, CCR2 |
| 26 | 0.72 | 0.62 | 33.2 | 131 | 89 | 77 | |
| 27 | 0.70 | 0.52 | 30.2 | 100 | 97 | 87 | NPH1 |
BFC clusters, showing the circadian score, amplitude, and phase estimates for the average trace for each cluster, the number of genes as a total and percentage scored rhythmic by circadian score (>0.4) and COSOPT (pMMC-β <0.05), and a list of clock-associated genes in each cluster. The asterisks indicate the six highest-amplitude clusters.
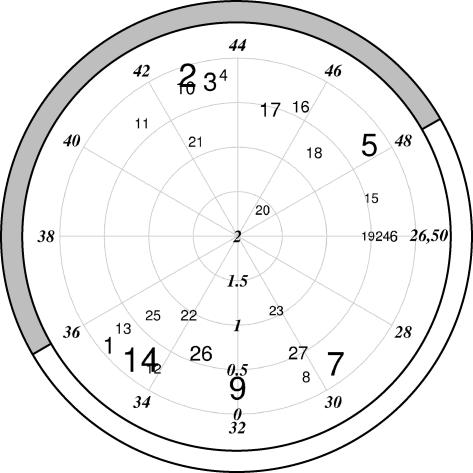
Figure 5.
Phase and Amplitude of BFC-Identified Clusters.
Polar plot of phase versus amplitude for each of the rhythmic clusters identified. Clusters are represented by their identity numbers (as in Table 3). Phase estimates for clusters are shown clockwise in italics around the circumference from ZT26 to ZT50, and amplitudes are shown in italics on the radial axis. Size of cluster identity numbers represents number of genes in each cluster. Subjective night (gray) and day (white) represented by the band around the outside of the plot.
As each cluster includes a range of expression patterns around the average, we performed a Fourier analysis of individual gene expression patterns. A total of 783 genes had a circadian score below our threshold (0.4; see Methods) despite being placed in the circadian clusters, indicating a potentially high number of false positives. The vast majority of these fell in the clusters with low amplitude: 75% of these were in the six lowest-amplitude clusters (40% of all the genes in these clusters), whereas only 2% were in the six clusters of highest amplitude (6% of all genes in these clusters).
None of the BFC clusters were scored as peaking between ZT36 and ZT40 (Figure 5). This gap in phase expression was not indicated by the phase estimates from COSOPT (Figure 3). Comparison of the expression profiles of genes in BFC clusters either side of this window suggests that genes peaking between ZT36 and ZT40 may have been pulled into the surrounding clusters. The spread of phases estimated by COSOPT in the clusters supports this (see Supplemental Figure 3 online), suggesting that the lack of genes peaking between ZT36 and ZT40 may be an artifact of the clustering.
BFC clusters were also tested for functionally related groups of genes based on GO terms and showed similar patterns for genes involved in photosynthesis and phenylpropanoid biosynthesis as COSOPT. No overrepresentation of carbon metabolism genes was seen in the middle of the subjective night for BFC, but sulfate assimilation was overrepresented in clusters peaking around the end of the subjective night (clusters 5, ZT48, P < 0.02 and 17, ZT45, P < 0.001) as suggested previously by Harmer et al. (2000). Water channel activity genes were overrepresented in the middle of the subjective day (cluster 7, ZT30, P < 0.001) followed by genes responsive to water and water deprivation ∼3 h later (cluster 26, ZT33, P < 0.05). This pattern in water responses suggests a possible selective advantage of circadian clocks in plants.
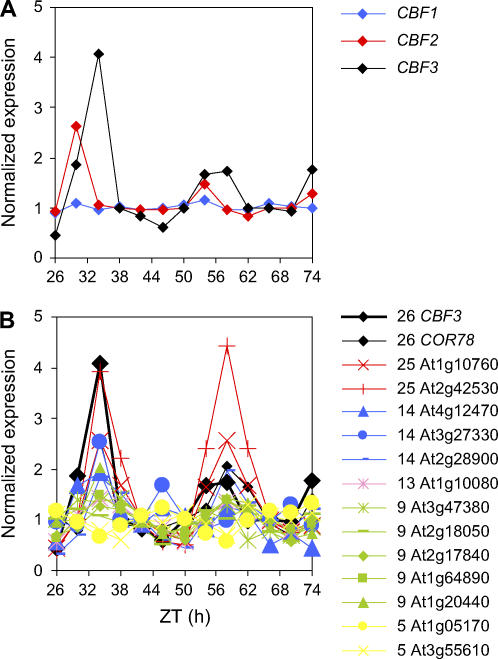
Another interesting result was the overrepresentation of transcription factors in clusters 20 (seven transcription factors out of 24 genes), 23 (10 out of 30 genes), and 27 (16 out of 100 genes), suggesting that these genes may play a regulatory role in the clock or in output from it. Clusters 20 and 23 were scored with the two highest amplitudes of all the clusters. The high amplitude clusters tended to include fewer genes, whereas several of the low amplitude clusters were much larger (Figure 5, Table 3). This is consistent with the notion that genes in the smaller, higher-amplitude clusters might regulate the expression of genes in lower-amplitude clusters, particularly those at a similar phase. This is supported by the location of genes clustered by BFC and identified as regulated by the cold response transcription factor C REPEAT BINDING FACTOR3 (CBF3) primarily in clusters of lower amplitude and/or later phase than this gene.
CBF3 (also called DREB1a) is part of the three-gene CBF family (CBF1-3), which regulates a large number of cold-responsive genes (Fowler and Thomashow, 2002; Vogel et al., 2005). Harmer et al. (2000) showed this gene to be rhythmically expressed previously and suggested that this regulation might explain the circadian rhythm in cold resistance of cold-sensitive plants. Cold induction of all three CBF genes, along with two of their known targets, was recently shown to be gated by the circadian clock (Fowler et al., 2005), suggesting that clock control of the CBF genes may indeed be important to induction of cold resistance. CBF3 was placed in BFC cluster 26, along with the known CBF target gene COLD REGULATED78 (COR78; Jaglo-Ottosen et al., 1998). However, neither CBF1 nor CBF2 was clustered by BFC or scored rhythmic by COSOPT, although CBF2 did appear potentially rhythmic by eye (Figure 6A). Fourteen genes, including COR78, from a list of 41 previously identified as CBF responsive (Fowler and Thomashow, 2002), were located in BFC clusters. Of these, only two genes were in a cluster with a markedly different peak phase to CBF3. Of the remaining 12, four were in clusters 13 and 14 (later phased and lower amplitude than CBF3's cluster), two were in cluster 25 (later phased but higher amplitude), and five were in the lower amplitude but slightly earlier phased cluster 9. Visual inspection of the genes' profiles supported the suggestion that CBF3 may be regulating the output of the other genes (Figure 6B). The earlier phasing of the five genes in cluster 9 could be explained by coregulation by the other CBF genes, particularly as the expression of CBF2 showed an earlier peak in expression than CBF3 (Figure 6A).
Figure 6.
Circadian Expression of CBF Genes and Their Targets.
Microarray expression profiles of CBF1 (blue), CBF2 (red), and CBF3 (black) genes (A) plus CBF3 and CBF target genes (identified in Fowler and Thomashow, 2002) in BFC clusters (B). See keys to right of graphs for gene identification. Cluster numbers shown as prefix to Arabidopsis Genome Initiative numbers in keys (B) and gene profiles colored by cluster.
Many known clock genes (including LHY, CCA1, GI, and TOC1) were placed within the six clusters of highest amplitude (Table 3). Two notable absentees from this list included EARLY FLOWERING3 (ELF3) and ELF4, in which mutations can cause arrhythmic circadian phenotypes (Hicks et al., 1996; Doyle et al., 2002). Similarly, other genes, such as ZEITLUPE, are known to affect the function of the clock but are not clock regulated at the transcript level (Somers et al., 2000). Thus, the high-amplitude clusters did not provide a comprehensive list of important clock genes, but they did provide a means of identifying a subset of potentially important clock genes from our list of FLC-responsive genes. The results of our COSOPT and BFC analyses are available from www.amillar.org.
Testing FLC-Responsive Genes
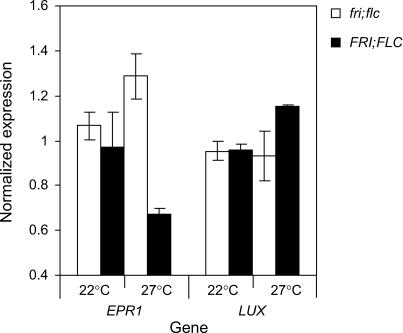
Of the FLC-responsive genes, 132 were in BFC clusters, with 32 falling in the six highest amplitude clusters (see Supplemental Table 2 online). Four of the 32 genes were transcription factors, highlighting them as candidates to potentially affect clock function. Three of the transcription factors were SHAQKYF-type MYB genes, like the core clock components LHY and CCA1 (Schaffer et al., 1998; Wang and Tobin, 1998), and the other was a member of the WRKY family. One of the MYB genes, EARLY PHYTOCHROME RESPONSIVE1 (EPR1), was previously shown to be rhythmically regulated and suggested as a component of a slave oscillator regulating some output pathways from the circadian clock (Kuno et al., 2003). EPR1 transcript abundance in fri; flc and FRI; FLC seedlings was tested by quantitative PCR of the pooled samples used in the microarray experiments at 27°C, along with a comparative set grown at 22°C. Figure 7 shows that, as suggested by the array data, FLC downregulated the expression of EPR1 at 27°C. Overexpression of EPR1, however, reportedly did not show any period effects (Kuno et al., 2003), and similarly little or no period phenotype was shown by leaf movement analysis of the epr1 T-DNA insertion mutant SALK_047716 at either 22 or 27°C (see Supplemental Figure 4 online), suggesting that EPR1 did not mediate FLC's effect on the clock.
Figure 7.
EPR1 and LUX Expression in the FRI; FLC Genotypes.
The average expression of EPR1 and LUX was analyzed by real-time PCR in pooled samples for FRI; FLC (closed bars) and fri; flc (open bars) genotype seedlings at 22 and 27°C. Expression levels for each gene are based on the average of two independent biological replicates and normalized to the average for all samples with respect to ACT2. Error bars show range of the replicates.
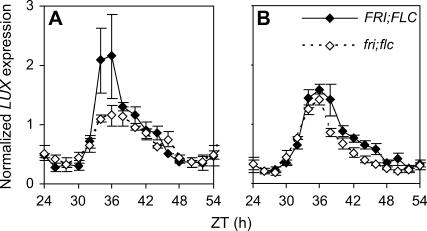
A more likely candidate was offered by another of the MYB transcription factors, At3g46640. This gene was recently identified as LUX ARRHYTHMO (LUX; also called PHYTOCLOCK1). Arrhythmic circadian phenotypes were shown by lux mutants, suggesting that the gene is important to the function of the clock (Hazen et al., 2005; Onai and Ishiura, 2005). As for EPR1, the expression of LUX in fri; flc and FRI; FLC seedlings was tested by quantitative PCR. LUX showed increased expression in the FRI; FLC genotype at 27°C, but no difference was apparent in its expression levels between the lines at 22°C (Figure 7). Figure 8 shows the temporal expression of LUX in the fri; flc and FRI; FLC genotypes at 27 and 22°C. LUX was clustered along with TOC1 by BFC (cluster 25; Table 3), and this was supported by the temporal expression pattern shown for both genes by real-time PCR (Figures 2A and 8). Trough levels of LUX expression were comparable between the two genotypes at both temperatures. The peak in LUX expression was approximately twofold higher in the FRI; FLC seedlings at 27°C but only slightly higher at 22°C (Figure 8). This small increase in LUX RNA at 22°C was nonetheless consistently observed over multiple time points in replicated experiments. Thus, FLC-dependent LUX expression correlated with period change at both 22 and 27°C, with only small differences in peak LUX expression level (Figure 8) and period (Figure 1C, Table 1) between the FRI; FLC genotypes at 22°C compared with the larger changes at 27°C. We could not test the requirement for LUX to mediate the period change caused by FLC by constructing the flc lux double mutant because the lux single mutant was already arrhythmic (Hazen et al., 2005). We therefore sought an alternative quantitative test.
Figure 8.
FLC Alters the Peak Level of LUX in a Temperature-Specific Manner.
The temporal pattern in LUX transcript abundance was analyzed by real-time PCR in FRI; FLC (closed diamonds) and fri; flc (open diamonds) genotype seedlings at 27°C (A) and at 22°C (B). LUX expression levels were normalized with respect to ACT2, and the average for the fri; flc genotype at each temperature across ZT32 to ZT40 was set to 1. Data shown are the average of two to three biological replicates, with technical triplicates (see Methods). Error bars represent se.
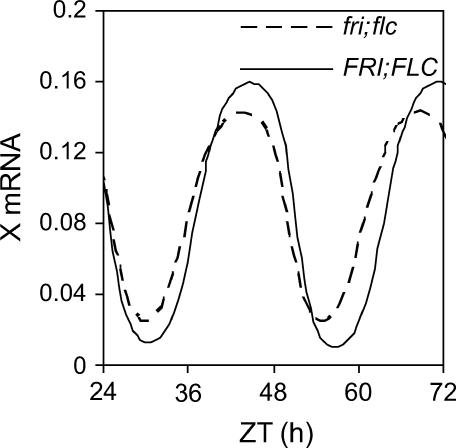
The evening phase of LUX expression and its role as a transcription factor suggests it may help to close the loop between TOC1 and LHY/CCA1 in the clock mechanism (Hazen et al., 2005). Locke et al. (2005b) proposed a TOC1-activated factor, modeled as a gene X, to explain the delay between peak expression of TOC1 and the increase in expression of LHY/CCA1. LUX alone is unlikely to be X, as the expression pattern of LUX RNA is similar to TOC1 RNA; however, LUX may contribute to the synthesis or assembly of active X. We therefore modeled the increase in peak expression of LUX by increasing the parameter that specifies the maximum transcription rate of gene X by 10% in the model by Locke et al. (2005b). This elevated the peak levels of X RNA by 12% and caused a 1.6-h period lengthening in the model under constant light (Figure 9), similar to the period lengthening in FRI; FLC plants relative to fri; flc at 27°C.
Figure 9.
Modeling the Effect of LUX on the Clock.
Graph showing temporal expression of X (mRNA abundance) for fri; flc (dashed line) and FRI; FLC (solid line) genotypes as predicted by the model by Locke et al. (2005b). Initial parameters were taken from this model, and maximum transcription rate of X mRNA was increased by 10% under constant light conditions to simulate the increased peak expression of LUX mRNA in the FRI; FLC genotype compared with fri; flc. This resulted in a 1.6-h increase to free running period of the model.
DISCUSSION
FLC's involvement in the circadian clock was first suggested by QTL mapping (Swarup et al., 1999), and further analysis revealed the PerCv5b QTL overlapping FLC's map location, with a high-temperature-specific effect on circadian period (Edwards et al., 2005). The similarity of temperature specificity, direction, and extent of the period phenotype in the QTL, in NILs carrying the weak FLC-Ler allele or functional FLC-Cvi allele, as well as in single or double mutant combinations of induced flc and fri alleles strongly support FLC as the cause of the PerCv5b QTL. Temperature compensation keeps the circadian period relatively constant over a wide temperature range and is a ubiquitous property of circadian rhythms. Shorter periods are commonly observed at the upper end of the physiological temperature range due to increased biochemical reaction rates (Rensing and Ruoff, 2002). FLC expression contributes to normal temperature compensation of the Arabidopsis circadian clock by counteracting the period shortening observed in flc mutants at 27°C.
As aberrant circadian timing impairs plant growth (Dodd et al., 2005), this suggests that the circadian function of FLC is likely to be relevant to fitness in some habitats. The period change caused by FLC at 27°C is at least as great as the effects of natural allelic variants in Drosophila that are distributed in a latitudinal cline (Sawyer et al., 1997). FLC expression could be lost either by epigenetic repression of FLC following vernalization (Bastow et al., 2004; Sung and Amasino, 2004) or in the early flowering accessions that carry mutations of FLC or its activator FRI (Michaels and Amasino, 1999; Johanson et al., 2000; Gazzani et al., 2003). Selective pressure for such downregulation of FLC function in flowering time appears to have overridden secondary effects of altered circadian timing, at least in these cases. The multiple QTL that affect circadian period at 27°C (Edwards et al., 2005) might in part reflect compensating mechanisms to balance selection on flowering time and circadian timing.
Transcriptomic analysis comparing the fri; flc and FRI; FLC genotypes at 27°C was used to identify FLC-responsive genes. A list of 1000 candidate genes showing greatest change in expression between the two genotypes included FLC and AGL20, but few other genes known to be involved in the regulation of flowering time or, for that matter, of the circadian clock. Further microarray assays were used to identify circadian-regulated transcripts and highlight a subset of candidate genes to mediate FLC's effect on the clock. Some differences were apparent between the two methods of scoring rhythmic transcripts, COSOPT and BFC. Overall, COSOPT gave a more reliable indication of rhythmicity, which suggested that ∼16% of the Arabidopsis genome was clock regulated. Clustering by BFC enabled identification of genes with higher amplitude profiles, similar to those of known important clock genes, as a means of targeting potentially important rhythmic regulators. Even the high-amplitude BFC clusters included several genes that were not scored as rhythmic by COSOPT (Table 3). A consensus of the two methods indicated that a minimum of 8.7% of the genes on the array showed a circadian expression pattern under our experimental conditions.
GO and promoter analysis of the genes scored rhythmic by COSOPT and clustered by BFC revealed that the data contained considerable information on functional clustering and potential regulatory sequences. Both methods used in concert enabled wider sampling of this information. Clearly, not all the important clock-affecting genes are rhythmically regulated at the transcript level, but BFC provides a means of targeting a few genes from a large number, as is often required from array data. This identified LUX as a strong candidate to mediate FLC's effect on the clock. Mathematical modeling suggested that a modest increase in peak LUX expression, as observed in FRI; FLC, would be sufficient to explain FLC's effect on the clock at 27°C. One caveat to this is that plants constitutively overexpressing LUX did not show a long period phenotype but rather wild-type period oscillations that damped into arrhythmia (Onai and Ishiura, 2005). However, rhythmic overexpression of LUX, as in FRI; FLC, may alter the clock in a different way to constitutive overexpression of the gene. Indeed, increasing rhythmically expressed LUX gene dosage does appear to increase circadian period, consistent with our prediction, because plants heterozygous for a lux mutation have a shorter period than wild-type LUX homozygotes (Onai et al., 2004).
Natural genetic variation is a valuable resource, and its importance for understanding plant biology is increasingly being recognized (reviewed in Koornneef et al., 2004). Pinpointing the mechanisms of small effect QTL, starting with the identification of the underlying genes, is not a trivial matter (Weigel and Nordborg, 2005). As demonstrated for FLC, mathematical modeling provides a useful complement to experiments in understanding quantitative changes in plant response networks.
METHODS
Plant Materials and Growth Conditions
NIL46 was produced by genotypic selection from a backcross of NIL187 (donated by M. Koornneef) to Ler. The fri; flc, FRI; FLC, fri; FLC, and FRI; flc genotypic combinations used for leaf movement and RNA time courses are in the Col background with combinations of either wild-type Col or flc-3 and FRI-SF2 alleles (Michaels and Amasino, 1999, 2001). The ld-1 and ld-1; flc-3 mutants have been described (Michaels and Amasino, 2001), as have the fld-3 mutants (He et al., 2003). Wild-type Col-0 seedlings were used for the microarray circadian time-course experiment. Unless otherwise stated, seedlings were sterilized and grown as described (Edwards et al., 2005).
Measuring Circadian Rhythms
Circadian rhythms were measured by video imaging of leaf movement under constant light and analyzed in the BRASS interface (Edwards et al., 2005; http://www.amillar.org/Downloads.html). Mean period estimates for each genotype in Figures 1B and 1C are based on 10 to 50 leaf traces from two to four independent experiments at each temperature, analyzed using REML (Patterson and Thompson, 1971) in the statistical package GENSTAT 5 (Payne et al., 1993). Mean period estimates in Figures 1D and 1E are from representative experiments analyzed in BRASS.
Quantitative RT-PCR
Approximately 100 seedlings were ground under liquid nitrogen per time point, and total RNA was extracted using a Plant RNeasy kit and RNase-free DNase (Qiagen) according to the manufacturer's instructions. cDNA samples for real-time PCR applications were reverse transcribed from 1 μg of RNA using the RevertAid first-strand cDNA synthesis kit (Fermentas, Helena Biosciences) according to the manufacturer's instructions, and the cDNA product was diluted 1:5 in RNase-free water. Transcript abundance of TOC1, CCA1, LHY, GI, EPR1, and LUX were assessed by quantitative real-time PCR in either an ABI PRISM 7700 (Applied Biosystems) or Bio-Rad iCycler IQ using ABI SYBR Green PCR Master Mix (Applied Biosystems) in 15-μL reactions. Transcript levels were normalized to ACT2 using a cDNA dilution series for each primer set in each experiment. Each RNA sample was assayed in triplicate. Primers for GI and ACT2 have been described previously (Locke et al., 2005b). Primer sequences to assess other transcripts are shown below: TOC1 forward, 5′-ATCTTCGCAGAATCCCTGTGATA-3′; TOC1 reverse, 5′-GCACCTAGCTTCAAGCACTTTACA-3′; CCA1 forward, 5′-CTGTGTCTGACGAGGGTCGAA-3′; CCA1 reverse, 5′-ATATGTAAAACTTTGCGGCAATACCT-3′; LHY forward, 5′-CAACAGCAACAACAATGCAACTAC-3′ LHY reverse, 5′-AGAGAGCCTGAAACGCTATACGA-3′; EPR1 forward, 5′-CCAAGATGGCTCAGGAAGCT-3′; EPR1 reverse, 5′-AAGGATGTGCCGGTTTTCTCT-3′; LUX forward, 5′-GACGATGATTCTGATGATAAGG-3′; LUX reverse, 5′-CAGTTTATGCACATCATATGGG-3′.
Data presented in Figure 8 are based on the average of three independent biological replicates for all time points aside from ZT24 to ZT30 and ZT42 to ZT54 in Figure 8A, which are based on independent biological duplicates.
Microarrays
Total RNA was extracted as described above. Seedlings for the fri; flc versus FRI; FLC microarray were grown as described for leaf movement and placed into constant conditions at 27°C. Four samples were taken at 6-h intervals starting from 24 h into constant conditions (ZT24, ZT30, ZT36, and ZT42), and equal amounts of RNA were pooled from each time point for each genotype. An independent biological repeat was performed for both genotypes. Seedlings for the Col-0 time course were sterilized and grown as described above, with the exception that they were placed immediately into LD 12:12 and grown for 7 d at 22°C. At dawn on the 8th day, they were placed into constant 60 to 65 μmol m−2 s−1 cool white fluorescent light. Samples were taken over two circadian cycles at 4-h intervals starting from ZT26. Samples were assayed on the Affymetrix GeneChip oligonucleotide ATH1 array (Affymetrix) according to the manufacturer's instructions. Background correction and normalization and gene expression analysis of the array data were performed using the GC-RMA routine (Wu et al., 2004) in GeneSpring version 7.2 (Silicon Genetics). This results in the normalization of expression values to the average expression of all time points for that probe set. The normalized values are then used to detect rhythms using COSOPT or BFC. Promoter analysis of the immediate upstream regions (1000 bp from start codon) of the transcripts was performed in GeneSpring, and GO terms were analyzed using FuncAssociate (Berriz et al., 2003).
Scoring Circadian Transcripts Using COSOPT
COSOPT was used as previously described (Straume, 2004), with the exception of the removal of the initial sampling-density-weighted linear regression detrending. The sparse time points and short time course, covering only two cycles of circadian period, enabled the linear regression to skew the data. An increased number of genes were scored rhythmic at the same pMMC-β threshold when no detrending of the data occurred, but ∼80% of the rhythmic genes for a given pMMC-β were common to both methods. Phase estimates presented in Figure 3 and Supplemental Figure 3 online have been translated into ZT within the first circadian cycle of the real array data.
Scoring Circadian Transcripts Using BFC
BFC employs Bayesian techniques to cluster time series data according to a standard linear model (Heard et al., 2006). Curves were clustered together by BFC if they appear to have been drawn from a joint distribution with parameters β and σ2, where Y = Bβ + ɛ and Y holds the expression levels. ɛ is a noise term, which is normally distributed with mean zero and variance σ2. Design matrix B was chosen to contain Fourier basis functions for identification of rhythmic genes. β holds the Fourier coefficients for the average profile of each cluster (these values produce the average profile, seen as the blue line in Supplemental Figure 2 online, and the circadian score, amplitude, and phase values in Table 3). Thus, each cluster of genes is characterized by a different β and σ2. The clustering is exceptionally fast because σ2 was chosen to be inverse γ distributed and, given σ2, β is multivariate normal. The algorithm was used to perform an agglomerative hierarchical clustering; each gene expression profile was initially put in a separate cluster and then the two clusters most similar in covariance structure were merged repeatedly until all profiles formed one cluster. At each merger, the clustering was scored; the highest score was obtained for 27 clusters for 3063 genes. To search the massive space of potential clusters effectively (Anderson et al., 2005), the 22,810 gene profiles were arbitrarily split into four groups. Each of the groups was clustered as described, then clusters from each group that contained rhythmic genes were clustered with the rhythmic clusters from another group, until all remaining gene profiles could be clustered in a single group.
Circadian clusters were identified by dominance of the second harmonic (24-h period). This dominance was measured by the circadian score, which might more accurately be termed the second harmonic ratio: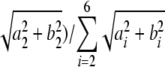 , where ai is the coefficient of the ith cosine term and bi the coefficient of the ith sine term. Thus, a cluster with a high circadian score indicated that the 24-h period characterized the average expression pattern more than the shorter periods. A circadian score of >0.4 was used as a guide to determine which clusters were retained at each stage of the hierarchical clustering. On this basis, BFC identified 26 clusters of circadian genes together with a 27th cluster with a circadian score of 0.38 that appeared circadian regulated by eye. Amplitude of rhythms was provided by the second harmonic (
, where ai is the coefficient of the ith cosine term and bi the coefficient of the ith sine term. Thus, a cluster with a high circadian score indicated that the 24-h period characterized the average expression pattern more than the shorter periods. A circadian score of >0.4 was used as a guide to determine which clusters were retained at each stage of the hierarchical clustering. On this basis, BFC identified 26 clusters of circadian genes together with a 27th cluster with a circadian score of 0.38 that appeared circadian regulated by eye. Amplitude of rhythms was provided by the second harmonic (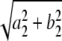 ), and phase was determined as the maximum point of the average expression profile within the first cycle of data.
), and phase was determined as the maximum point of the average expression profile within the first cycle of data.
Accession Numbers
Data for the microarray experiments described in Methods are available from the NASCArrays database (http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl) under the accession numbers NASCARRAYS-334 (FRI; FLC versus fri; flc) and NASCARRAYS-108 (circadian time course).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. FLC mRNA Expression at 22 and 27°C.
Supplemental Figure 2. BFC Clusters.
Supplemental Figure 3. COSOPT and BFC Peak Time Comparisons.
Supplemental Figure 4. EPR1 Leaf Movement.
Supplemental Table 1. FLC-Responsive Genes.
Supplemental Table 2. FLC Clock Candidate Genes.
Supplementary Material
Acknowledgments
N.S.S. performed leaf movement analysis of ld and ld; flc mutant seedlings. REML analysis of all leaf movement period estimates was performed by J.R.L. M.S. modified the linear regression function of COSOPT for this analysis. Samples for the fri; flc versus FRI; FLC array experiment were produced by A.H., and the arrays were conducted by the Molecular Biology Service (University of Warwick). The Col-0 circadian time course array experiment was conducted by the Nottingham Arabidopsis Stock Centre Affymetrix facility (University of Nottingham, UK). P.E.A. and J.Q.S. developed and applied the Bayesian clustering method BFC. Modeling was performed by J.C.W.L. All other work was performed by K.D.E. The authors thank Rick Amasino, Maarten Koornneef, and Susan Gibson for provision of seed stocks and Nazir Shariff and Victoria Hibberd for expert technical assistance. K.D.E., N.S.S., and J.C.W.L. were supported by PhD studentships from the Biotechnology and Biological Science Research Council (BBSRC), by a CASE award from Horticulture Research International, and by the Gatsby Charitable Foundation, respectively. Research funds were provided by BBSRC awards G13967 and G19886 to A.J.M. and by the Engineering and Physical Sciences Research Council/BBSRC award to the Interdisciplinary Programme for Cellular Regulation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Andrew J. Millar (andrew.millar@ed.ac.uk).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.038315.
References
- Akhtar, R.A., Reddy, A.B., Maywood, E.S., Clayton, J.D., King, V.M., Smith, A.G., Gant, T.W., Hastings, M.H., and Kyriacou, C.P. (2002). Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 12 540–550. [DOI] [PubMed] [Google Scholar]
- Alabadi, D., Oyama, T., Yanovsky, M.J., Harmon, F.G., Mas, P., and Kay, S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1. Science 293 880–883. [DOI] [PubMed] [Google Scholar]
- Anderson, P.E., Smith, J.Q., Edwards, K.D., and Millar, A.J. (2005). Bayesian Clustering and Model Exploration. (Coventry, UK: University of Warwick).
- Bastow, R., Mylne, J.S., Lister, C., Lippman, Z., Martienssen, R.A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164–167. [DOI] [PubMed] [Google Scholar]
- Berriz, G.F., King, O.D., Bryant, B., Sander, C., and Roth, F.P. (2003). Characterizing gene sets with FuncAssociate. Bioinformatics 19 2502–2504. [DOI] [PubMed] [Google Scholar]
- Ceriani, M.F., Hogenesch, J.B., Yanovsky, M., Panda, S., Straume, M., and Kay, S.A. (2002). Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 22 9305–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd, A.N., Salathia, N., Hall, A., Kevei, E., Toth, R., Nagy, F., Hibberd, J., Millar, A.J., and Webb, A.A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309 630–633. [DOI] [PubMed] [Google Scholar]
- Doyle, M.R., Davis, S.J., Bastow, R.M., McWatters, H.G., Kozma-Bognar, L., Nagy, F., Millar, A.J., and Amasino, R.M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419 74–77. [DOI] [PubMed] [Google Scholar]
- Edwards, K.D., Lynn, J.R., Gyula, P., Nagy, F., and Millar, A.J. (2005). Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics 170 387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, S., and Thomashow, M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, S.G., Cook, D., and Thomashow, M.F. (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 137 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani, S., Gendall, A.R., Lister, C., and Dean, C. (2003). Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113. [DOI] [PubMed] [Google Scholar]
- Harmer, S.L., and Kay, S.A. (2005). Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17 1926–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen, S.P., Schultz, T.F., Pruneda-Paz, J.L., Borevitz, J.O., Ecker, J.R., and Kay, S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., Michaels, S.D., and Amasino, R.M. (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302 1751–1754. [DOI] [PubMed] [Google Scholar]
- Heard, N.A., Holmes, C.C., and Stephens, D.A. (2006). A quantitative study of gene regulation involved in the immune response of Anopheline mosquitoes: An application of Bayesian hierarchical clustering of curves. J. Am. Stat. Assoc., in press.
- Hepworth, S.R., Valverde, F., Ravenscroft, D., Mouradov, A., and Coupland, G. (2002). Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21 4327–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, K.A., Millar, A.J., Carré, I.A., Somers, D.E., Straume, M., Meeks-Wagner, D.R., and Kay, S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274 790–792. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280 104–106. [DOI] [PubMed] [Google Scholar]
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290 344–347. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., and Vreugdenhil, D. (2004). Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55 141–172. [DOI] [PubMed] [Google Scholar]
- Kuno, N., Moller, S.G., Shinomura, T., Xu, X., Chua, N.H., and Furuya, M. (2003). The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell 15 2476–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, C.J., McClung, C.R., and Donald, B.R. (2002). A maximum entropy algorithm for rhythmic analysis of genome-wide expression patterns. Proc. IEEE Comput. Soc. Bioinform. Conf. 1 237–245. [PubMed] [Google Scholar]
- Locke, J.C.W., Millar, A.J., and Turner, M.S. (2005. a). Modelling genetic networks with noisy and varied experimental data: The circadian clock in Arabidopsis thaliana. J. Theor. Biol. 234 383–393. [DOI] [PubMed] [Google Scholar]
- Locke, J.C.W., Southern, M.M., Kozma-Bognar, L., Hibberd, V., Brown, P.E., Turner, M.S., and Millar, A.J. (June 28, 2005. b). Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. 1 http://dx.doi.org/10.1038/msb4100018. [DOI] [PMC free article] [PubMed]
- Michael, T.P., Salome, P.A., Yu, H.J., Spencer, T.R., Sharp, E.L., McPeek, M.A., Alonso, J.M., Ecker, J.R., and McClung, C.R. (2003). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302 1049–1053. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., Himelblau, E., Kim, S.Y., Schomburg, F.M., and Amasino, R.M. (2005). Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol. 137 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi, N., Kita, M., Ito, S., Sato, E., Yamashino, T., and Mizuno, T. (2005). The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol. 46 609–619. [DOI] [PubMed] [Google Scholar]
- Onai, K., and Ishiura, M. (2005). PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10 963–972. [DOI] [PubMed] [Google Scholar]
- Onai, K., Okamoto, K., Nishimoto, H., Morioka, C., Hirano, M., Kami-ike, N., and Ishiura, M. (2004). Large-scale screening of Arabidopsis circadian clock mutants by a high-throughput real-time bioluminescence monitoring system. Plant J. 40 1–11. [DOI] [PubMed] [Google Scholar]
- Ouyang, Y., Andersson, C.R., Kondo, T., Golden, S.S., and Johnson, C.H. (1998). Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. USA 95 8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda, S., Antoch, M.P., Miller, B.H., Su, A.I., Schook, A.B., Straume, M., Schultz, P.G., Kay, S.A., Takahashi, J.S., and Hogenesch, J.B. (2002). Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109 307–320. [DOI] [PubMed] [Google Scholar]
- Patterson, H.D., and Thompson, R. (1971). Recovery of inter-block information when block sizes are unequal. Biometrika 58 545–554. [Google Scholar]
- Payne, R.W., Lane, P., Digby, P., Harding, S., Leech, P., Morgan, G., Todd, A., Thompson, R., Tunnicliffe, W., Welham, S., and White, R. (1993). Genstat 5 Release 3 Reference Manual. (Oxford: Oxford University Press).
- Pittendrigh, C.S. (1954). On temperature independance in the clock system controlling emergence time in Drosophilla. Proc. Natl. Acad. Sci. USA 40 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh, C.S. (1960). Circadian rhythms and the circadian organisation of living systems. Cold Spring Harb. Symp. Quant. Biol. 25 159–184. [DOI] [PubMed] [Google Scholar]
- Rensing, L., and Ruoff, P. (2002). Temperature effect on entrainment, phase shifting, and amplitude of circadian clocks and its molecular bases. Chronobiol. Int. 19 807–864. [DOI] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sawyer, L.A., Hennessy, J.M., Peixoto, A.A., Rosato, E., Parkinson, H., Costa, R., and Kyriacou, C.P. (1997). Natural variation in a Drosophila clock gene and temperature compensation. Science 278 2117–2120. [DOI] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101 319–329. [DOI] [PubMed] [Google Scholar]
- Straume, M. (2004). DNA microarray time series analysis: Automated statistical assessment of circadian rhythms in gene expression patterning. Methods Enzymol. 383 149–166. [DOI] [PubMed] [Google Scholar]
- Sung, S., and Amasino, R.M. (2004). Vernalization and epigenetics: How plants remember winter. Curr. Opin. Plant Biol. 7 4–10. [DOI] [PubMed] [Google Scholar]
- Swarup, K., Alonso-Blanco, C., Lynn, J.R., Michaels, S.D., Amasino, R.M., Koornneef, M., and Millar, A.J. (1999). Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J. 20 67–77. [DOI] [PubMed] [Google Scholar]
- Vogel, J.T., Zarka, D.G., Van Buskirk, H.A., Fowler, S.G., and Thomashow, M.F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41 195–211. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Nordborg, M. (2005). Natural variation in Arabidopsis. How do we find the causal genes? Plant Physiol. 138 567–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z., Irizarry, R.A., Gentleman, R., Murillo, F.M., and Spencer, F. (2004). A Model Based Background Adjustment for Oligonucleotide Expression Arrays. (Baltimore, MD: Johns Hopkins University), http://www.bepress.com/jhubiostat/paper1.
- Young, M.W., and Kay, S.A. (2001). Time zones: A comparative genetics of circadian clocks. Nat. Rev. Genet. 2 702–715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.