Abstract
Arabidopsis thaliana is a host to the powdery mildew Erysiphe cichoracearum and nonhost to Blumeria graminis f. sp hordei, the powdery mildew pathogenic on barley (Hordeum vulgare). Screening for Arabidopsis mutants deficient in resistance to barley powdery mildew identified PENETRATION3 (PEN3). pen3 plants permitted both increased invasion into epidermal cells and initiation of hyphae by B. g. hordei, suggesting that PEN3 contributes to defenses at the cell wall and intracellularly. pen3 mutants were compromised in resistance to the necrotroph Plectosphaerella cucumerina and to two additional inappropriate biotrophs, pea powdery mildew (Erysiphe pisi) and potato late blight (Phytophthora infestans). Unexpectedly, pen3 mutants were resistant to E. cichoracearum. This resistance was salicylic acid–dependent and correlated with chlorotic patches. Consistent with this observation, salicylic acid pathway genes were hyperinduced in pen3 relative to the wild type. The phenotypes conferred by pen3 result from the loss of function of PLEIOTROPIC DRUG RESISTANCE8 (PDR8), a highly expressed putative ATP binding cassette transporter. PEN3/PDR8 tagged with green fluorescent protein localized to the plasma membrane in uninfected cells. In infected leaves, the protein concentrated at infection sites. PEN3/PDR8 may be involved in exporting toxic materials to attempted invasion sites, and intracellular accumulation of these toxins in pen3 may secondarily activate the salicylic acid pathway.
INTRODUCTION
Nonhost resistance is the type of nonspecific resistance that an entire plant species exhibits against all genotypes within a pathogen species (Thordal-Christensen, 2003). Although until recently little was known about the biochemical defenses that contribute to nonhost resistance, cytological descriptions suggested that a majority of inappropriate fungal pathogens were unable to breach the plant cell wall and infiltrate host cells (Yun et al., 2003; Zimmerli et al., 2004). However, some variation in penetration efficiency exists among different host and fungal pathogen combinations (Mellersh and Heath, 2003). These studies also showed that plants respond actively to attack by inappropriate pathogens and do not rely solely on preformed and constitutive barriers for protection against inappropriate pathogens (Meyer and Heath, 1988; Zimmerli et al., 2004). Furthermore, it appears that operationally, nonhost resistance can be divided into penetration resistance, barriers limiting entry of the pathogen into cells, and postpenetration resistance, mechanisms that act intracellularly if penetration resistance is overcome (Fernandez and Heath, 1991; Huitema et al., 2003; Mellersh and Heath, 2003; Yun et al., 2003; Zimmerli et al., 2004; Lipka et al., 2005).
Both surveys of mutants with defects in various defense functions and screens for mutants specifically compromised in nonhost resistance have identified a diverse group of genes that contribute to nonhost resistance. Among the mutants with defects in the salicylic acid (SA) signal transduction pathway, nonhost resistance was diminished in eds1 mutants and transgenic plants with the bacterial NahG gene relative to wild-type plants (Parker et al., 1996; Mellersh and Heath, 2003; Yun et al., 2003; Zimmerli et al., 2004). Using virus-induced gene silencing, the chaperone and chaperone-like proteins HSP70, HSP90, and SGT1, which are thought to stabilize resistance (R) proteins and act upstream of EDS1, were shown to contribute to nonhost resistance in Nicotiana benthamiana (Peart et al., 2002; Kanzaki et al., 2003). Mutants with defects in the ethylene/jasmonate (ET/JA) signal transduction pathways exhibited wild-type levels of nonhost resistance in Arabidopsis thaliana, even though there is evidence from microarray studies that this pathway is preferentially induced after inoculation of plants with some inappropriate pathogens (Huitema et al., 2003; Zimmerli et al., 2004). These data suggest that the ET/JA pathway and components of the SA pathway acting downstream of eds1 play no role or only a limited role in nonhost resistance. Alternatively, it is possible that these pathways contribute to resistance to inappropriate pathogens only after the other defenses have been defeated (Jones and Takemoto, 2004). Because EDS1 contributes to basal resistance (Parker et al., 1996), defenses that act to limit the growth of appropriate pathogens even in compatible interactions, it seems likely that there is some overlap between basal resistance and nonhost resistance.
Additional host components, for which no clear role in basal resistance has been demonstrated to date, contribute to nonhost resistance. NHO1, a glycerol kinase, is required for nonhost resistance in Arabidopsis to a bacterial pathogen of bean (Phaseolus vulgaris), Pseudomonas syringae pv phaseolicola (Kang et al., 2003). In physiological studies using inhibitors, the actin cytoskeleton was shown to be required for nonhost resistance against barley (Hordeum vulgare) and wheat (Triticum aestivum) powdery mildews in pea (Pisum sativum) and Arabidopsis, respectively (Kobayashi et al., 1997; Yun et al., 2003). These genes do not fit into a single pathway or process, but their identification does suggest that diverse processes contribute to nonhost resistance. This is consistent with an early model of nonhost resistance in which it was proposed that this form of resistance consists of several barriers operating in parallel to limit pathogen colonization (Heath, 2000).
To identify the components required for nonhost resistance, we screened for Arabidopsis mutants allowing increased penetration by the barley powdery mildew Blumeria graminis f. sp hordei, assuming that such mutants would carry defects in those components of nonhost resistance that limit pathogen entry into host cells. The syntaxin PEN1 (=SYP121) and the glycosyl hydrolase PEN2 were recovered from this and related screens (Collins et al., 2003; Lipka et al., 2005). Here, we describe a third mutant isolated from this screen and show that PEN3 encodes the putative ATP binding cassette (ABC) transporter PDR8. Subcellular localization of PEN3 in the plasma membrane and extensive gene interaction studies lead us to speculate that PEN3 mediates the targeted export of toxins to penetration sites.
RESULTS
pen3 Lacks Penetration Resistance to Three Inappropriate Pathogens
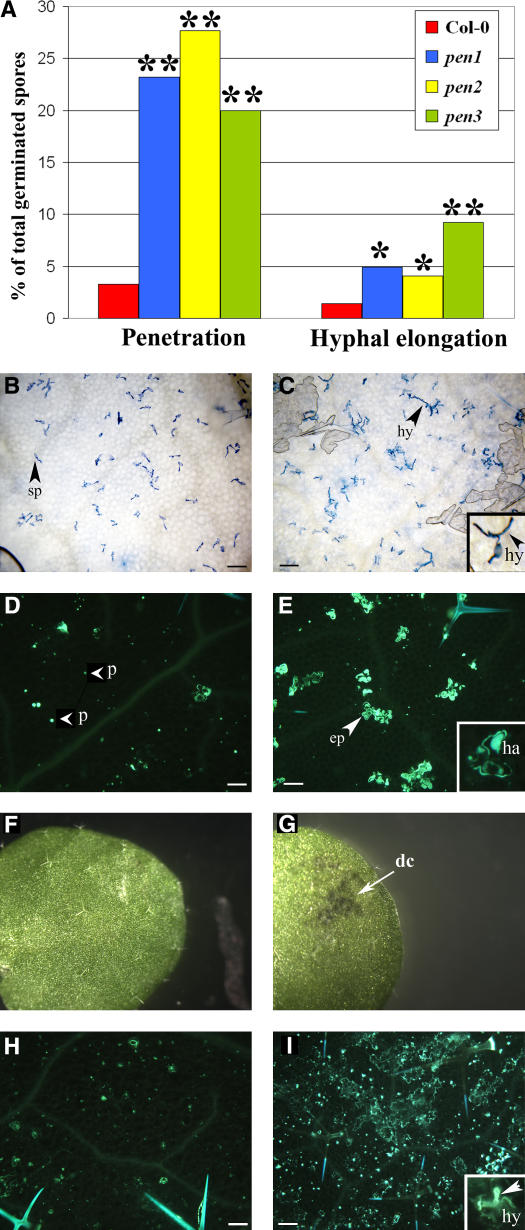
We conducted a screen for the loss of penetration resistance by screening for Arabidopsis mutants that allowed B. g. hordei to form haustoria within epidermal cells at a higher frequency than on wild-type plants. Haustoria, which are fungal feeding structures encased in a specialized host membrane within epidermal cells, are produced once the fungal germlings have successfully breached the host cell wall. The few B. g. hordei haustoria that formed in Arabidopsis were rapidly encased in callose and could be recognized by their distinctive oval shape in inoculated leaves stained with the callose stain aniline blue (Figure 1E, inset). Nine pen mutants were recovered from ∼12,000 M2 plants from ethyl methanesulfonate–mutagenized seeds. Mapping experiments and crosses among the mutants were used to place the pen mutants in complementation groups. When the identities of the PEN genes became known, allelism among the various mutants was confirmed by sequencing the appropriate gene (i.e., PEN1, PEN2, or PEN3). pen1-4 (Collins et al., 2003), pen2-3 (Lipka et al., 2005), pen3-1, and pen3-2 were recovered from this screen. pen3-1, like the pen1 and pen2 mutants, supported a higher frequency of fungal penetration (Figure 1A). Another feature of the phenotype conferred by pen3 was that callose deposition lining the entire periphery of invaded epidermal cells was more common than in wild-type plants (Figures 1D and 1E). Compared with wild-type plants, pen3-1 plants also allowed increased frequency of formation of elongating secondary hyphae, an indication that the underlying haustoria were functional (Figures 1A to 1C). Secondary hyphae are the hyphae that emerge from the conidium after the first haustorium is established. Germ tube growth is supported by reserves in the conidium, whereas the emergence and growth of secondary hyphae are thought to be dependent on nutrients acquired from the host via the haustorium (Masri and Ellingboe, 1966; Ellingboe, 1972). Therefore, pen3-1 mutants were partially compromised not only in penetration resistance against B. g. hordei but also in a mechanism restricting haustorium function. Loss of PEN3 function does not allow B. g. hordei to complete its life cycle and form asexual conidia on Arabidopsis as it does on barley, suggesting that additional factors contribute to nonhost resistance to B. g. hordei in Arabidopsis.
Figure 1.
pen3 Nonhost Resistance Phenotypes.
(A) Mean of the frequency of B. g. hordei penetration (left) and hyphal elongation (right) on Arabidopsis at 2 DAI, expressed as a percentage of total germinated spores. Wild-type controls are represented in red, pen1-4 plants in blue, pen2-3 plants in yellow, and pen3-1 plants in green. Asterisks denote statistically significant differences between mutants and the wild type by Student's t test (** P < 0.0001, * P< 0.001).
(B) and (D) B. g. hordei inoculation on Columbia-0 (Col-0). Conidia (sp; arrowhead) germinate and produce appressoria but seldom are able to penetrate and establish functional haustoria (B). Papillae (p; arrowhead), callose-rich cell wall appositions, form at penetration sites whether or not the powdery mildew pathogen is able to successfully breach the cell wall and form a haustorium (D). Papillae were used as markers for attempted penetration sites.
(C) and (E) B. g. hordei inoculation on pen3-1. A higher proportion of conidia are able to penetrate, establish haustoria, and form elongating secondary hyphae (hy) on pen3 plants relative to Col-0 ([C] and inset). Haustorial formation (ha) is associated with callose deposition, encompassing the entire invaded host epidermal cell (ep; arrowhead) ([E] and inset).
(F) to (I) Response to P. infestans inoculation. Cell death (dc; arrow) is evident macroscopically after inoculation of pen3 (G) but not in Col-0 (F). A small amount of callose is deposited in Col-0 (H), whereas widespread callose deposition and occasional intracellular hyphae (hy; arrowhead) are observed in pen3 ([I] and inset).
In (B) and (C), samples were stained with trypan blue at 2 DAI and visualized by bright-field microscopy. In (D), (E), (H), and (I), samples were stained with aniline blue at 2 DAI to detect callose and visualized by fluorescence microscopy. Bars = 30 μm.
To determine whether increased pathogen entry was limited to interactions with B. g. hordei, pen3-1 plants were inoculated with two other inappropriate pathogens. Erysiphe pisi (tribe Erysipheae), the powdery mildew pathogenic on pea, belongs to a tribe distinct from Erysiphe cichoracearum (tribe Golovinomyceteae) and B. g. hordei (tribe Blumerieae) (Saenz and Taylor, 1999; Braun et al., 2002). Similar to B. g. hordei, E. pisi was able to initiate the formation of elongating secondary hyphae at a higher frequency on pen3-1 than on wild-type plants (Figure 5D). Likewise, Phytophthora infestans, which is a hemibiotrophic oomycete pathogen and the causal agent of late blight in potato (Solanum tuberosum), formed invasive hyphae, which were encased in callose (Figure 1I, inset), more often in pen3-1 mutants than in wild-type plants (Figure 1H). Inoculation with P. infestans elicited little callose deposition in wild-type plants, whereas dramatic callose deposition was observed in pen3-1 epidermal cells (Figures 1H and 1I). When it occurred, penetration by P. infestans into Arabidopsis epidermal cells was often accompanied by local cell death at sites of infection (Huitema et al., 2003). Although the wild type appeared healthy where droplets of P. infestans zoospores had been applied (Figure 1F), pen3-1 exhibited macroscopic cell death (Figure 1G), consistent with the idea that a greater proportion of oomycete individuals had penetrated into host cells and elicited cell death. Thus, pen3-1 had a diminished capacity to restrict pathogen entry by three different inappropriate pathogens.
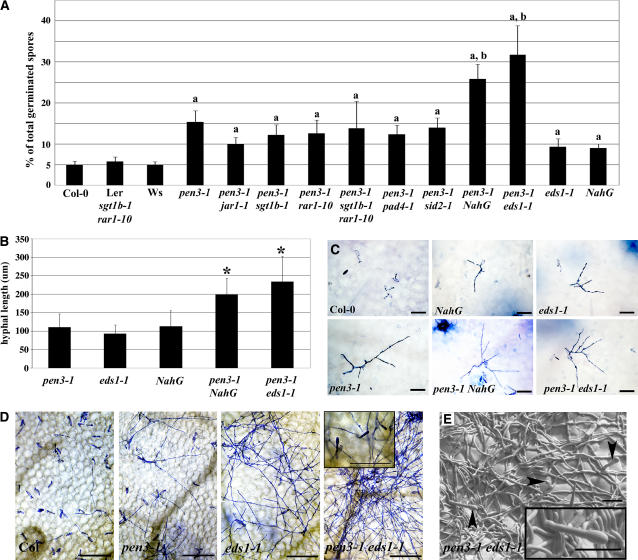
Figure 5.
Combined Mutations in PEN3 and EDS1or Introduction of the NahG Transgene Compromise Nonhost Resistance to B. g. hordei and E. pisi.
(A) Mean of the frequency of B. g. hordei penetration on Arabidopsis, expressed as a percentage of total germinated spores. Wassilewskija (Ws) is the wild-type control for eds1-1; Ler sgt1b-1 rar1-10 is the control for double mutants with sgt1b-1 and rar1-10; and Col-0 is the wild-type control for npr1-1, sid2-1, pad4-1, jar1-1, and NahG. Eight leaves per genotype were stained with aniline blue at 2 DAI, and the occurrence of callose-encased haustoria was monitored as a measure of penetration efficiency. a Significantly different from the wild type; b significantly different from pen3 using Student's t test (P < 0.001).
(B) Average hyphal length of B. g. hordei colonies growing on different mutants. Leaves were stained with trypan blue at 10 DAI, and hyphal lengths per colony were measured from photographs using ImageJ software. A minimum of 10 colonies were measured for hyphal length. This experiment was repeated twice. * Significantly different from the corresponding value for pen3 using Student's t test (P < 0.001).
(C) Examples of B. g. hordei colonies growing on different lines. Infected leaves were stained with trypan blue at 10 DAI. Bars = 30 μm.
(D) E. pisi growth on Col-0 leaves and the indicated mutant genotypes at 7 DAI. Light microscopic images were taken after visualization of fungal structures using Coomassie Brilliant Blue. Bars = 200 μm. The inset shows a close-up view of E. pisi conidiophores on the pen3-1 eds1-1double mutant. Bar = 50 μm.
(E) Cryogenic scanning electron micrograph of E. pisi growth on pen3-1 eds1-1 at 7 DAI. The inset shows an E. pisi conidiophore. Bars = 50 μm.
PEN3 Encodes a PDR-Like ABC Transporter
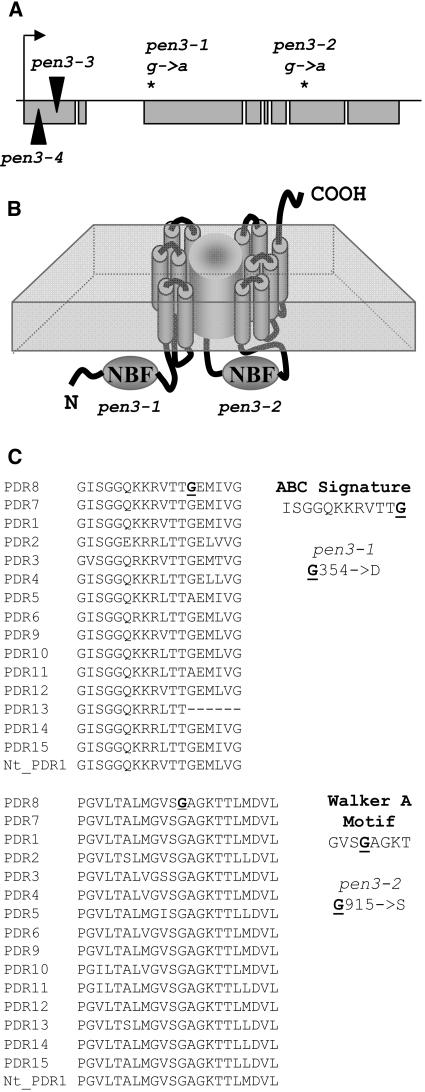
Mapping populations were generated by crossing pen3-1 and pen3-2 to Landsberg erecta (Ler). Mutant plants identified in the F2 generation were used for bulk segregant mapping (Lukowitz et al., 2000). In both crosses, PEN3 mapped to the bottom of chromosome 1 between markers nga280 and ciw1. These two markers were used to identify additional recombinants in this genetic interval from ∼3600 F2 plants. In this manner, the interval containing PEN3 was delimited to a region encompassed by BAC F23H11. Nineteen T-DNA mutant populations with insertions in 1 of 15 genes in this interval were screened for the phenotype conferred by pen after inoculation with B. g. hordei (Ecker, 2002). Two of these T-DNA populations (SALK__110926 and SALK_000578) segregated plants with a penetration-deficient phenotype. These mutants had predicted insertions in a PDR-like ABC transporter (At1g59870). Both pen3-1 and pen3-2 carried single nucleotide substitutions at nucleotides 1419 and 3335, respectively, from the A of the ATG initiation codon of the genomic sequence for this gene (Figure 2A). Two T-DNA alleles, pen3-3 (SALK_110926) and pen3-4 (SALK_000578), had insertions near the 5′ end of the coding region (Figure 2A). The PEN3 gene had previously been annotated as PDR8 based on its homology with yeast PDR transporters (van den Brule and Smart, 2002). It encodes a 4.3-kb cDNA and is predicted to encode a 1469–amino acid protein with 13 transmembrane domains (Figure 2B). Arabidopsis has 15 PDR family members, which are highly similar at the amino acid level (van den Brule and Smart, 2002). An alignment of these members showed that the point mutations in pen3-1 and pen3-2 occurred in highly conserved regions of the nucleotide binding folds (Figure 2C). The nucleotide binding folds are thought to mediate ATP binding and consist of a Walker A motif followed by an ABC signature motif and a Walker B motif (van den Brule and Smart, 2002). The mutation in pen3-1 converted a Gly to an Asp in the ABC signature motif of the first nucleotide binding fold, whereas the mutation in pen3-2 converted a Gly to a Ser in the Walker A motif of the second nucleotide binding fold. PEN3 transcript was detected in pen3-1 and pen3-2 but not in the T-DNA mutants pen3-3 and pen3-4 (data not shown). Given that all four pen3 alleles have identical phenotypes, we assume that proteins encoded by pen3-1 and pen3-2 are nonfunctional, at least in nonhost resistance.
Figure 2.
Molecular Identity of PEN3 and of the pen3 Alleles.
(A) PEN3 gene structure indicating exons (gray boxes) and introns (black lines connecting exons). The genomic sequence of PEN3 is 6.1 kb. Sequencing of the pen3-1 and pen3-2 alleles revealed single nucleotide changes (asterisks) that result in amino acid substitutions. Two T-DNA alleles (black arrowheads) carry insertions early in the gene.
(B) Predicted architecture of the PEN3 protein. PEN3 is predicted to have 1469 amino acids. The amino acid changes in pen3-1 and pen3-2 occur in the ABC signature motif and the Walker A motif of the first and second nucleotide binding folds (NBF), respectively.
(C) Alignment of amino acids of the two nucleotide binding folds of the 15 members of the Arabidopsis PDR family and of tobacco PDR1. Mutations in pen3-1 and pen3-2 result in changing highly conserved Gly (boldface, underlined G) to larger amino acids, Asp and Ser, respectively. Both changes occur in pockets thought to mediate ATP binding.
The Arabidopsis PDR family of ABC transporters has been studied via RNA gel blotting and RT-PCR to determine in which plant organs these transporters are expressed (van den Brule and Smart, 2002). In addition, publicly available databases provide microarray expression profiles of different Arabidopsis organs, tissues, and environmental treatments (Rhee et al., 2003; Zimmermann et al., 2004). These resources showed that PEN3 was expressed in all tissues, but most highly in leaves. Most other PDR family members were expressed at low levels and in specific plant organs (see Supplemental Table 1 online) (Rhee et al., 2003). In leaves, PEN3 transcript accumulated to levels comparable to those of metabolic housekeeping genes, such as cytosolic glyceraldehyde-3-phosphate, and 1 to 2 orders of magnitude higher than the other PDR family members (see Supplemental Table 1 online) (Shih et al., 1991). The gene encoding PDR7, the ABC transporter most similar to PEN3 in sequence (90% amino acid similarity), was expressed primarily in roots (see Supplemental Table 1 online), and T-DNA insertion mutations in PDR7 (SALK_134725 and SALK_008761) did not exhibit the penetration-deficient phenotype (data not shown). These microarray data, together with RNA gel blot analyses, indicate that PEN3 is highly and ubiquitously expressed in plant tissues. These data further suggest that there is limited functional redundancy among the PDR genes and PEN3.
Publicly available expression data were mined to determine whether PEN3 expression was modified after pathogen attack (see Supplemental Tables 2 and 3 online) (Zimmermann et al., 2004). Induction was modest after inoculation with virulent pathogens (e.g., E. cichoracearum, Agrobacterium tumefaciens, and the necrotroph Plectosphaerella cucumerina), inappropriate pathogens (e.g., B. g. hordei, P. infestans), and nonspecific elicitors (e.g., chitin fragments, flg22 peptide) but dramatic after inoculation with avirulent bacteria and the phloem-feeding aphid Myzus persicae (see Supplemental Table 2 online) (Glombitza et al., 2004; Zimmermann et al., 2004; De Vos et al., 2005; Ramonell et al., 2005). Induction by the flg22 peptide, a synthetic version of a conserved region of flagellin, was abolished in plants lacking FLS2, a receptor-like kinase required for flagellin perception (see Supplemental Table 2 online) (Gomez-Gomez and Boller, 2000). Exposure to methyl jasmonate, ozone, and the herbicides primisulfuron and prosulfuron also induced PEN3 expression (see Supplemental Table 2 online) (Glombitza et al., 2004; Zimmermann et al., 2004). Although these two herbicides interfere with the synthesis of branched-chain amino acids, an herbicide that targets photosystem II, bromoxynil, did not induce PEN3 expression (Glombitza et al., 2004). Abscisic acid and heat shock treatments downregulated the expression of PEN3 (see Supplemental Table 2 online) (Busch et al., 2005). Thus, PEN3 expression is altered by pathogens and a subset of abiotic stresses.
Resistance to E. cichoracearum and Cell Death in pen3 Are SA-Dependent
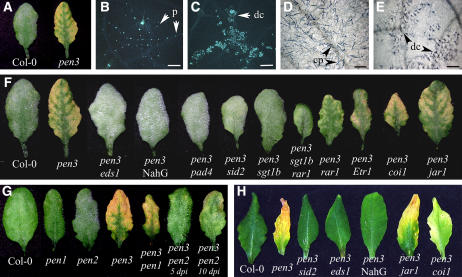
To examine the possibility that pen3-1 plants were also defective in basal resistance to virulent pathogens, they were infected with the Arabidopsis powdery mildew, E. cichoracearum. Surprisingly, pen3-1 plants underwent chlorosis and cell death after E. cichoracearum infection (Figure 3A). Closer inspection indicated that cell death was fungus-associated and did not spread extensively beyond areas of fungal colonization (Figures 3C and 3E). By 5 d after inoculation (DAI), E. cichoracearum hyphae had elongated on wild-type leaves, leaving a trail of callose-containing papillae at each penetration site (Figure 3B). By contrast, hyphal elongation on pen3-1 plants was accompanied by extensive callose deposition, preceding the death of penetrated cells (Figures 3C and 3E). By 7 DAI, extensive conidiation was observed on wild-type plants, whereas colonies that formed on pen3-1 were stunted, lacked conidiophores, and were overlaid with patches of dead cells (Figure 3E).
Figure 3.
pen3-1 Mutants Are Resistant to E. cichoracearum and Become Chlorotic after E. cichoracearum Infection.
(A) Single leaves at 7 DAI with the Arabidopsis powdery mildew, E. cichoracearum. Col-0 (left) supports good fungal conidiation, which is evident by eye as the whitish powdery appearance of inoculated leaves. Col-0 does not develop lesions, whereas pen3-1(right) undergoes chlorosis and cell death.
(B) and (C) On Col-0, E. cichoracearum infection elicits discrete callose deposits at papillae (p) (B), whereas penetration attempts on pen3-1 plants elicit widespread callose deposition and cell death (dc) (C). Leaves were stained with aniline blue at 5 DAI. Bars = 100 μm.
(D) and (E) Lesions (dc) can be observed in pen3-1 (E) but not in Col-0 (D) under conidiophores (cp) and hyphae. Leaves were stained with trypan blue at 7 DAI. Bars = 100 μm.
(F) Double mutant analysis to evaluate the role of the SA and ET/JA signal transduction pathways on resistance to E. cichoracearum in pen3-1 plants. On double mutants with pen3-1 and mutants or transgenic plants with blocks in the SA pathway (e.g., eds1-1, NahG, pad4-1, sid2-1), powdery mildew growth and conidiation exceed those observed in wild-type plants. The double mutant pen3-1 sgt1b-1 and the triple mutant pen3-1 sgt1b-1 rar1-10 support an intermediate level of powdery mildew conidiation that is slightly less than that observed on the wild type. Mutations in the ET pathway genes (e.g., Etr1-1), the JA pathway genes (e.g., coi1-1, jar1-1), and in rar1-10 were not able to suppress the chlorotic and necrotic phenotype that develops on pen3-1 mutants after infection with the Arabidopsis powdery mildew. Plants were photographed at 7 DAI.
(G) Double mutant analysis of the effect of pen1-1 and pen2-3 on pen3-1–associated resistance to E. cichoracearum. Plants were photographed at 6 DAI unless indicated otherwise.
(H) Double mutant analysis of the effect of the SA signal transduction pathway (eds1-1, NahG, sid2-1) and of the JA signal transduction pathway (coi1-1, jar1-1) on light-induced chlorosis of pen3-1 plants. Plants were 4 weeks old when the photograph was taken.
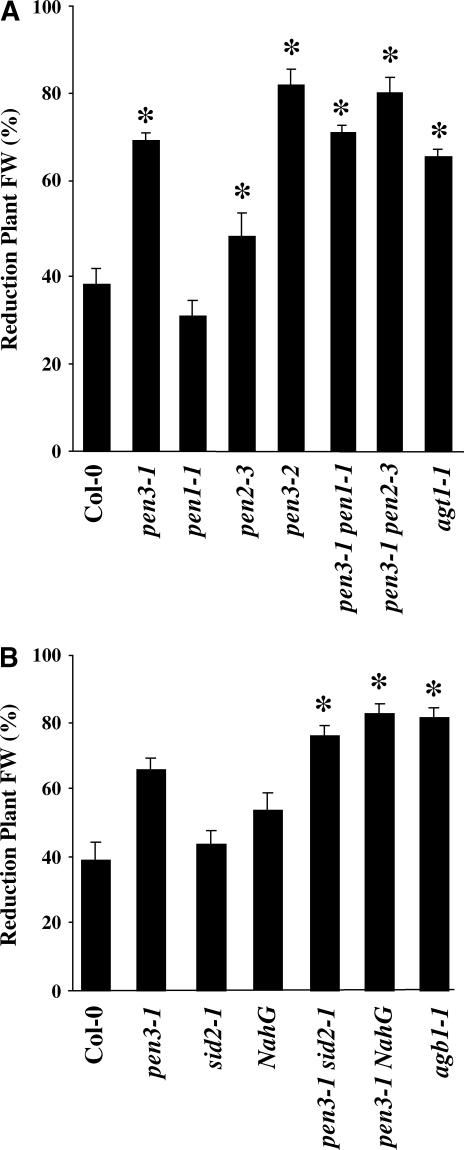
Additionally, pen3 plants were inoculated with the broad host range necrotrophs Botrytis cinerea and P. cucumerina (Berrocal-Lobo et al., 2002). pen3-1 and pen3-2 mutants were more susceptible to P. cucumerina than wild-type plants (Figure 4). Leaf tissue damage and fresh weight reduction caused by this fungus were greater in pen3 mutants than in wild-type plants and similar to the damage observed in the highly susceptible agb1-1 mutant, which is impaired in the β-subunit of Arabidopsis heterotrimeric G protein (Figure 4A) (Llorente et al., 2005). Resistance of pen3-1 plants to B. cinerea did not differ from that of wild-type plants (data not shown).
Figure 4.
pen3 Mutants Are More Susceptible to the Broad Host Range, Necrotrophic Pathogen P. cucumerina.
(A) pen3 and pen2 are highly susceptible to P. cucumerina. Plants were inoculated with a suspension of 2 × 106 spores/mL and scored at 7 DAI. * Significantly different from Col-0 using Student's t test (P < 0.05).
(B) The susceptibility to P. cucumerina of the pen3-1 mutant does not depend on the SA signal transduction pathway. Plants were inoculated with a suspension of 1 × 106 spores/mL and scored at 10 DAI. * Significantly different from pen3-1 using Student's t test (P < 0.05).
The susceptibility of plants was scored by fresh weight reduction (mean ± sd). At least 30 plants per genotype were inoculated with each pathogen, and the experiment was repeated four times. The highly susceptible agb1-1 mutant was used as a positive control for disease development. FW, fresh weight.
Cell death in response to pathogen attack is often associated with the activation of the SA defense pathway (Greenberg and Yao, 2004). Similarly, chlorosis is associated with high levels of ET production (Schaller and Keiber, 2001). Double mutants were generated between pen3-1 and mutants affecting the SA and ET/JA pathways to determine whether these defense signaling pathways were necessary for the pathogen-induced chlorosis and necrosis phenotype. Transgenic NahG plants, as well as mutants lacking a functional SA pathway, such as eds1, pad4, and sid2, suppressed the cell death and resistance phenotypes of pen3-1, allowing E. cichoracearum to conidiate (Figure 3F). The effect of two other SA-related mutants, rar1 and sgt1b, on resistance to E. cichoracearum in pen3-1 was also investigated. Mutations in RAR1 did not affect the occurrence of cell death in inoculated pen3-1 (Figure 3F). However, in pen3-1 sgt1b double mutants, conidiation was observed initially but chlorosis and cell death were delayed to ∼10 DAI (data not shown). Triple pen3-1 sgt1b rar1 mutants resembled pen3-1 sgt1b double mutants. Mutations that affect the ET/JA pathway, such as jar1, coi1, and Etr1, did not alter the pen3-1 cell death phenotype. Thus, pen3-1 resistance to the Arabidopsis powdery mildew is SA-dependent and ET/JA-independent.
Disease development after P. cucumerina infection was modestly enhanced in the SA-deficient lines sid2-1 and NahG, as reported previously (Berrocal-Lobo et al., 2002), although to a lesser extent than in pen3-1 (Figure 4B). The enhanced susceptibility to P. cucumerina observed in pen3 mutants was further enhanced in pen3-1 sid2-1 or pen3-1 NahG double mutant lines, suggesting that PEN3 acts additively with the SA pathway to limit P. cucumerina infections.
We considered the possibility that a propensity to become chlorotic and necrotic could indirectly result in increased pathogen entry. To address this question, a set of 34 Arabidopsis mutants that are resistant to the host powdery mildew pathogen were examined for pen3-like phenotypes (see Supplemental Table 4 online) (Greenberg and Ausubel, 1993; Frye and Innes, 1998; Rate et al., 1999; Clough et al., 2000; Petersen et al., 2000; Vogel and Somerville, 2000; Clarke et al., 2001; Mach et al., 2001; Rate and Greenberg, 2001; Maleck et al., 2002; Vogel et al., 2002, 2004; Liang et al., 2003). This collection included published cpr, acd, agd, and pmr mutants as well as a set of mil (for mildew-induced lesion) mutants (M. Nishimura, J. Vogel, and S. Somerville, unpublished data). Only one of these mutants, mil9, exhibited both increased penetration by B. g. hordei and resistance to E. cichoracearum (see Supplemental Table 4 online). The PEN3 gene from mil9 contained a point mutation at nucleotide 3985, which is predicted to change a Trp codon (TGG) to a stop codon (TGA). Thus, mutants prone to chlorosis and cell death do not commonly allow increased invasion by inappropriate pathogens.
Unlike pen3, pen1 and pen2 were not resistant to E. cichoracearum and did not become chlorotic upon infection (Collins et al., 2003; Lipka et al., 2005). As shown by the more rapid appearance of visible symptoms, pen2 was more susceptible to E. cichoracearum than the wild type, indicating that PEN2 is important for both basal and nonhost resistance (Figure 3G). The pen1 mutation was not able to suppress the E. cichoracearum–induced cell death of pen3-1 (Figure 3G). In a pen2-3 pen3-1 double mutant, however, cell death was delayed and resistance was compromised relative to the pen3-1 mutant (Figure 3G). Although some conidiation occurred, pen2-3 pen3-1 leaves underwent cell death and chlorosis at ∼10 DAI (Figure 3G, data not shown). Thus, pen3 resistance to E. cichoracearum plants is partially dependent on PEN2. In addition, pen2, but not pen1, was more susceptible to P. cucumerina than the wild type, but to a lesser degree than pen3-1 and pen3-2 (Figure 4A). The double mutants pen3-1 pen1-1 and pen3-1 pen2-3 resembled the pen3 single mutants in their susceptibility to P. cucumerina (Figure 4A) (Lipka et al., 2005).
E. cichoracearum infection was not the only stress capable of inducing chlorosis and cell death in pen3-1 plants. Although pen3-1 plants were indistinguishable from wild-type plants up to 4 weeks after germination, eventually they became chlorotic and senesced earlier than wild-type plants (data not shown). This phenotype was exacerbated by growth in continuous high light (24 h at 900 μE·m−2·s−1) (Figure 3H). High light–induced chlorosis and senescence were suppressed by mutations in the SA pathway but not by those in the ET/JA pathway (Figure 3H). Thus, the SA pathway appears to be necessary for heightened sensitivity to abiotic stress in pen3-1 plants as well.
Effects of SA and ET/JA Pathway Mutations on Residual Nonhost Resistance in pen3
None of the mutations in the SA and ET/JA pathways tested reversed the loss of penetration resistance to B. g. hordei (Figure 5). However, plants carrying the NahG transgene and plants carrying mutations in EDS1 were slightly compromised in their ability to restrict B. g. hordei entry into epidermal cells relative to wild-type plants (Figure 5) (Zimmerli et al., 2004). Microscopic inspection of the invasion and growth of B. g. hordei on double and single mutants revealed that the frequencies of fungal penetration on pen3-1 NahG and pen3-1 eds1-1 were higher than those on the single mutants (Figure 5A). Colonies growing on these double mutant combinations were typically twice the average size than those growing on the single mutants (Figure 5B). This increase in growth was accompanied by an increase in hyphal branching (Figure 5C). Conidiation by B. g. hordei was not observed.
E. pisi epiphytic growth was increased on eds1 and pen3-1 mutants compared with wild-type plants (Figure 5D). A cumulative effect was seen on pen3-1 eds1-1 double mutants, on which E. pisi colonies consisted of dense leaf-spanning mycelia, accompanied by occasional conidiophore formation (Figures 5D and 5E, insets). E. pisi conidia recovered from Arabidopsis double mutants were able to successfully infect pea and cause disease (data not shown).
Expression Profiling of pen3-1 Plants
We compared the transcript profiles of wild-type and pen3-1 plants at 1 DAI with E. cichoracearum or B. g. hordei using full-genome Affymetrix microarrays. Of the 22,810 probe sets (genes) on the microarray, 4240 exhibited some change in expression in this experiment. When considering the trends observed in the expression patterns of these genes, two main clusters were observed: genes induced by and genes repressed by pathogen attack (see Supplemental Figure 1 online). As described previously (Zimmerli et al., 2004), B. g. hordei elicited a more dramatic response than E. cichoracearum. Transcripts with increased levels after inoculation with the fungal pathogens included defense-related transcripts primarily, whereas transcripts with reduced levels consisted in large part of transcripts encoding photosynthetic and metabolic components (Zimmerli et al., 2004). It is possible that plants respond more dramatically to B. g. hordei because it cannot evade or suppress basal defenses as efficiently as the host powdery mildew, E. cichoracearum. In addition, responses to inoculation were more dramatic in pen3-1 than in the wild type. Included among these genes were several SA-associated genes (Table 1). These included genes of the SA pathway, such as PAD4, SID2, EDS1, and EDS5, as well as downstream SA pathway markers, such as PR-4, PR-5, chitinases, and glucanases (Shah, 2003). This hyperactivation of SA defenses, together with the double mutant analysis, suggests that the basis for the resistance to E. cichoracearum observed in pen3-1 plants is an enhanced activation of the SA pathway.
Table 1.
Examples of SA-Induced or SA Pathway–Associated Genes That Are Hyperinduced in pen3-1 Mutants after Inoculation with Either the Arabidopsis or the Barley Powdery Mildew
| Col-0 | Col-0 | pen3 | pen3 | pen3 | ||
|---|---|---|---|---|---|---|
| Locus | Gene Description | Ec | Bgh | Unin | Ec | Bgh |
| At1g65690 | Harpin-induced protein–related | 3.04 | 6.67 | 1.35 | 7.42 | 17.87 |
| At1g74710 | Isochorismate synthase1 (ICS1 = SID2) | 2.23 | 5.67 | 1.15 | 4.13 | 10.14 |
| At1g75040 | Pathogenesis-related protein5 (PR-5) | 4.89 | 9.18 | 2.55 | 12.81 | 26.13 |
| At2g43570 | Chitinase, putative similar to chitinase class IV | 2.88 | 7.50 | 1.59 | 8.24 | 24.39 |
| At2g43590 | Chitinase, putative similar to basic endochitinase CHB4 | 1.11 | 1.48 | 1.12 | 2.59 | 5.95 |
| At2g46400 | WRKY family transcription factor | 2.16 | 4.80 | 1.52 | 3.63 | 8.42 |
| At3g04720 | Pathogenesis-related protein4 (PR4) | 1.36 | 1.58 | 1.00 | 2.24 | 4.46 |
| At3g12500 | Basic endochitinase | 1.05 | 1.28 | 1.14 | 2.37 | 8.39 |
| At3g48090 | Disease resistance protein, lipase-like (EDS1) | 2.33 | 3.49 | 1.34 | 3.50 | 4.11 |
| At3g50480 | Broad-spectrum mildew resistance RPW8 | 2.61 | 4.97 | 1.52 | 5.35 | 11.19 |
| At3g52430 | Phytoalexin-deficient4 (PAD4) | 3.20 | 5.48 | 1.42 | 5.02 | 9.75 |
| At3g57240 | β-1,3-Glucanase (BG3) | 5.43 | 6.07 | 1.78 | 7.11 | 6.67 |
| At4g01700 | Chitinase, putative similar to peanut type II chitinase | 3.79 | 6.91 | 1.36 | 6.28 | 8.26 |
| At4g39030 | Enhanced disease susceptibility5 (EDS5 = SID1) | 2.44 | 5.43 | 1.13 | 4.84 | 10.33 |
| At4g39830 | l-Ascorbate oxidase | 5.29 | 11.70 | 1.78 | 9.58 | 14.99 |
| At5g24210 | Lipase class 3 family protein | 2.80 | 4.62 | 1.17 | 3.92 | 8.86 |
| At5g47120 | Bax inhibitor1 putative | 1.57 | 2.23 | 1.15 | 2.13 | 3.19 |
Ec, 1 DAI with Erysiphe cichoracearum; Bgh, 1 DAI with Blumeria graminis f. sp hordei; Unin, uninoculated. Based on the analysis of variance with a Benjamini and Hochberg multiple text correction, the P values for these genes were <0.0008 for the factor genotype and for the factor infection. Values shown indicate average fold induction relative to uninoculated Col-0 (n = 4).
A single transcript (At3g30720) was highly upregulated in pen3-1–independent infection status (P = 1.7 × 10−8). This observation was confirmed by semiquantitative RT-PCR using independent samples (data not shown). In addition, the expression of this gene did not change very much upon inoculation with pathogens in either pen3 or the wild type (i.e., Col-0 uninoculated, 78 intensity units [average of four replicates]; Col-0 + E. cichoracearum, 99; Col-0 + B. g. hordei, 74; pen3-1 uninoculated, 1082; pen3-1 + E. cichoracearum, 916; pen3-1 + B. g. hordei, 978). No other gene exhibited this expression pattern. This gene is predicted to encode a 59–amino acid polypeptide with no significant homology with any known protein. ESTs have been found for this transcript, and array data show that this transcript responds to high CO2 and is constitutively upregulated in the ET-insensitive Etr1 mutant (Zimmermann et al., 2004). Unlike pen3-1, Etr1 mutants resemble wild-type plants in their ability to restrict the entry of B. g. hordei into epidermal cells, and Etr1 plants are susceptible to E. cichoracearum (data not shown). Thus, high levels of expression of this uncharacterized gene are not sufficient to account for the mutant phenotypes of pen3-1.
Of the PDR genes, only three, including PEN3, changed expression after inoculation with pathogens (see Supplemental Table 3 online) (Nishimura et al., 2003). Although PDR4 was repressed after infection, PDR12 was induced, and its induction was dramatically enhanced in B. g. hordei–inoculated pen3-1 plants.
Localization of PEN3 to Penetration Sites after Pathogen Attack
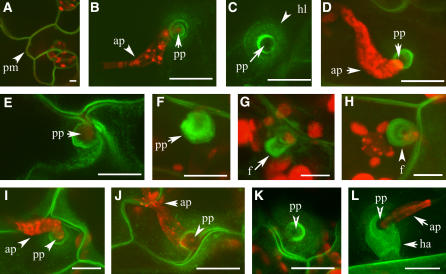
In plants, ABC transporters have been localized to the tonoplast, mitochondria, chloroplasts, and plasma membrane (Kushnir et al., 2001; Moller et al., 2001; Goodman et al., 2004; Pighin et al., 2004). Recently, proteomic studies of different organelles have placed PEN3 in the plasma membrane, mitochondria, and chloroplasts (Brugiere et al., 2004; Kleffmann et al., 2004; Nühse et al., 2004). Given its high abundance, PEN3 is likely a common contaminant in organelle preparations and unlikely to be localized in all three subcellular structures. To localize PEN3, a translational fusion to green fluorescent protein (GFP) was made, transformed into pen3-1 plants, and shown to complement the phenotypes conferred by the pen3-1 mutant (data not shown). PEN3-GFP was localized to the plasma membrane; no GFP signal was associated with chloroplasts (Figure 6A). After inoculation with B. g. hordei, PEN3-GFP accumulated to high levels in regions of the plasma membrane just under fungal appressoria, in the shape of compact disks surrounded by diffuse halos approximately the size of papillae (Figures 6B to 6D). Occasionally, this disk of PEN3-GFP signal was surrounded by a concentric circle of intense PEN3-GFP label within the diffuse halo (Figure 6B). Closer inspection of PEN3-GFP fluorescence at attempted entry sites revealed a three-dimensional bubble-like shape, presumably reflecting the invagination of the plant plasma membrane by the growing fungal penetration peg and newly developing haustorium (Figures 6E and 6F). A funnel-like structure, which resembled a hollow tube and extended into the plant cell, was sometimes observed at penetration sites at later stages (Figures 6G and 6H). Localization of PEN3-GFP after E. cichoracearum infection was similar to that observed after inoculation with B. g. hordei. PEN3-GFP accumulated in compact disks beneath appressoria, as well as in diffuse halos around this domain, and in bubble-like structures (Figures 6I to 6K). When E. cichoracearum haustoria were formed, the PEN3-GFP marker was also observed partially surrounding the haustorium (Figure 6L).
Figure 6.
Localization of PEN3-GFP.
(A) PEN3-GFP localizes to plant plasma membranes (pm) in uninfected leaves.
(B) to (H) Localization of PEN3-GFP in leaves inoculated with B. g. hordei.
(B) Conidia germinate and produce appressoria (ap), which attempt penetration via penetration pegs (pp).
(C) Green channel image of an attempted penetration site, accompanied by the accumulation of PEN3-GFP directly around the peg and by diffuse accumulation in a halo-like structure (hl).
(D) PEN3-GFP accumulates preferentially beneath penetration pegs.
(E) and (F) Accumulation of PEN3-GFP around the penetration peg persists in a bubble-like structure as penetration continues.
(G) and (H) At later stages, PEN3-GFP is seen in a funnel-like structure (f) around haustorial initials. This structure extends deep into the cell (side view [G]) and does not fully encase the haustorial initial (bottom view [H]).
(I) to (L) Localization of PEN3-GFP in leaves inoculated with E. cichoracearum.
(I) to (K) Attempted penetrations by E. cichoracearum elicit PEN3-GFP aggregation around penetration pegs and in diffuse halos.
(L) At later stages, PEN3-GFP aggregates around the upper part of the haustoria (ha).
PEN3-GFP appears green; chloroplast autofluorescence and propidium iodide–stained fungal structures appear red. Bars = 5 μm.
DISCUSSION
Like pen1 and pen2, the pen3 mutant allowed increased entry into epidermal cells and haustorium formation by the inappropriate pathogen B. g. hordei (Collins et al., 2003; Lipka et al., 2005). In addition, pen3 supported increased hyphal elongation by the inappropriate fungus, indicating that a higher proportion of haustoria remained functional long enough to support the establishment and growth of secondary hyphae (Figure 1).
Nonhost resistance to B. g. hordei was also slightly compromised in the eds1 mutant and in NahG transgenic plants, as shown for B. g. tritici, the powdery mildew pathogenic on wheat (Figure 5) (Yun et al., 2003; Zimmerli et al., 2004). Double mutants of pen3-1 and eds1 allowed additional penetration and growth by two separate inappropriate biotrophic pathogens, B. g. hordei and E. pisi, and the necrotroph P. cucumerina, than either mutant alone (Figures 4 and 5). Furthermore, E. pisi was able to reproduce asexually on plants with both pen3-1 and eds1-1 mutations (Figures 5D and 5E). Although EDS1 and NahG affect the functioning of the SA pathway, they both have additional poorly characterized but SA-independent effects on resistance (Feys et al., 2001; Heck et al., 2003; van Wees and Glazebrook, 2003). Because sid2 and its double mutant with pen3 did not implicate the SA pathway in nonhost resistance, it seems likely that the SA-independent responses of EDS1 and NahG contribute to nonhost resistance. Furthermore, double mutant combinations of pen3 and eds1 or NahG exhibited additive rather than epistatic interactions, suggesting that PEN3 operates independently of EDS1 and NahG in nonhost resistance. Therefore, it appears that only two barriers, penetration resistance (e.g., PEN2 or PEN3) and postpenetration or basal resistance (e.g., EDS1 or PAD4), are sufficient to limit E. pisi invasion, growth, and asexual reproduction on Arabidopsis (Lipka et al., 2005). However, an additional barrier(s) appears to limit B. g. hordei infections of Arabidopsis.
Unexpectedly, pen3 mutants were more resistant to the Arabidopsis powdery mildew pathogen. This resistance was associated with the development of chlorosis and necrosis late in the infection sequence (Figure 3). A survey of 34 Arabidopsis mutants that exhibited chlorosis and necrosis upon pathogen infection showed that these phenotypes are not commonly associated with a lack of nonhost resistance (see Supplemental Table 4 online). Both double mutant analysis and microarray expression profiling suggested that the SA pathway was hyperactivated in pen3 plants relative to wild-type plants and that this was responsible for the chlorotic and E. cichoracearum–resistant phenotypes of pen3 (Table 1, Figure 3). SGT1b has a role in cell death signaling in addition to its role in modulating R protein levels, which is consistent with the attenuation of chlorosis and cell death observed in pen3 sgt1b double mutants (Figure 3) (Holt et al., 2005). Because powdery mildews are obligate biotrophs that grow only on living host tissue, the necrosis and chlorosis are likely responsible for the resistance to E. cichoracearum in pen3 plants.
PEN3 encodes a putative PDR-like ABC transporter previously designated PDR8. The Arabidopsis PDR gene family comprises 15 members, of which PEN3 is the most widely and highly expressed. Although other ABC transporter families in plants are well characterized and their members have been implicated in the transport of auxin, lipids, pigments, and chlorophyll precursors, the PDR family is less well studied (Martinoia et al., 2002). Among the plant PDR subgroup of ABC transporters, studies of two Nicotiana species transporters, N. plumbaginifolia PDR1/ABC1 and tobacco (Nicotiana tabacum) PDR1, which are most similar in sequence to the Arabidopsis PDR12, suggest that they may function in the export of toxic secondary plant metabolites and/or in the detoxification of pathogen toxins. Similar to PEN3, the expression of N. tabacum PDR1and N. plumbaginifolia PDR1 is induced by defense-related signals such as pathogen elicitors and methyl jasmonate, and N. plumbaginifolia PDR1 is also induced by the phytoalexin sclareol (Jasinski et al., 2001; Sasabe et al., 2002). Arabidopsis does not produce sclareol; however, PDR12 is induced by fungal pathogens, and plants carrying a T-DNA insertion in PDR12 are reportedly more sensitive to exogenous sclareol and to lead (see Supplemental Table 3 online) (Campbell et al., 2003; Lee et al., 2005). Given the broad range of compounds and small peptides that can be transported by ABC transporters, these studies of tobacco PDR transporters cannot be used to suggest a candidate substrate for PEN3. A single gene, encoding a small unknown protein, was strongly upregulated in the pen3 mutant independent of infection status. In yeast, an ABC transporter is responsible for exporting the 38–amino acid mating factor A (Kuchler et al., 1989). Thus, it is possible that the small unknown protein is a substrate for PEN3. Alternatively, this gene could be a novel marker for the stress experienced by pen3 plants. In spite of the expression responses, loss of PDR12 function in Arabidopsis or PDR1 function in N. plumbaginifolia did not lead to increased susceptibility to several fungal and bacterial pathogens (Campbell et al., 2003; Stukkens et al., 2005). However, N. plumbaginifolia plants lacking PDR1 were highly susceptible to the necrotroph B. cinerea (Stukkens et al., 2005). With this result and the cloning of PEN3, PDR-type ABC transporters have been implicated in defense, adding a new role to the diverse repertoire of the PDR-like ABC transporter family.
ABC transporters have been shown to require phosphorylation for activity or regulation (Kolling, 2002). A recent study of phosphorylated plasma membrane proteins identified PEN3 as a target of flagellin-induced phosphorylation (Nühse et al., 2004), supporting the idea that PEN3 functions in nonhost resistance and defenses elicited by nonspecific elicitors such as flagellin. Consistent with this idea, PEN3 expression was 2.5-fold higher in flg22 peptide–treated seedlings compared with control seedlings, and this induction was dependent on the FLS2 receptor-like kinase (see Supplemental Table 2 online) (Gomez-Gomez and Boller, 2000).
Localization of PEN3 to the plasma membrane and its accumulation at penetration sites support the idea that PEN3 exports defense compounds in a focal manner at attempted invasion sites (Figure 6). PEN1 (=SYP121 syntaxin), the barley PEN1 homolog ROR2, and the barley MLO protein also localize in a similar pattern to the plasma membrane after powdery mildew infection (Assaad et al., 2004; Bhat et al., 2005). In addition, PEN2-GFP localizes to peroxisomes that accumulate near sites of invasion (Lipka et al., 2005). This marshalling of defense components at sites of attempted pathogen invasion illustrates the importance of subcellular dynamics in plant responses to pathogens.
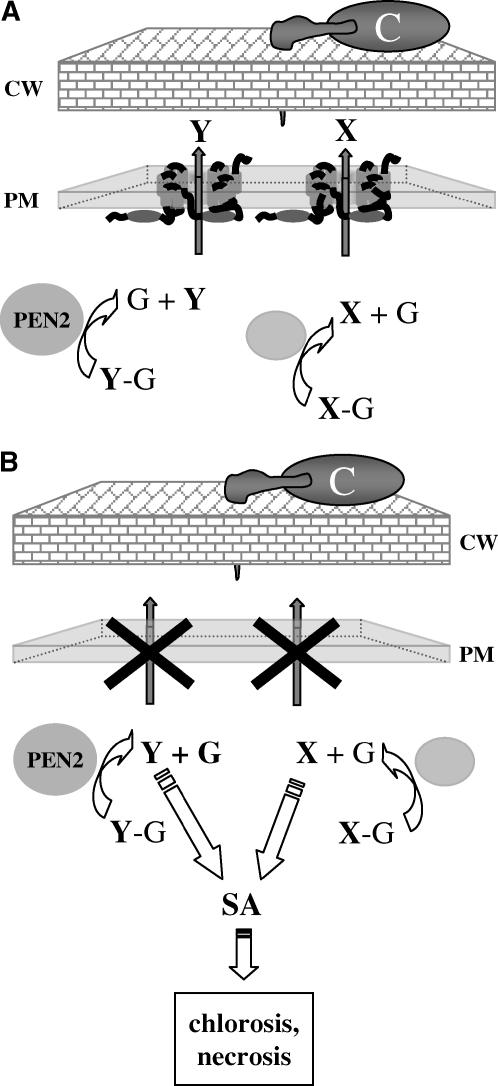
pen3 and pen2 mutants have similar phenotypes that are not observed in pen1 mutants. For example, only pen2 and pen3 plants were more susceptible to the inappropriate pathogen P. infestans (Figures 1G and 1I) and the necrotroph P. cucumerina (Figure 4) (Lipka et al., 2005). However, pen3 mutants differed from pen2 mutants by their resistance to the Arabidopsis powdery mildew (Figure 3A). Mutations in PEN2 partially suppressed cell death and chlorosis in E. cichoracearum–infected pen3 mutants, suggesting that PEN2 and PEN3 functions might be interdependent. We propose a biochemical model for the role of PEN3 in penetration resistance (Figure 7A) in which PEN2 converts a nontoxic substrate to a toxic product, which is then exported either directly or after further modification to the apoplast by PEN3, poisoning the fungal penetration peg as it attempts to cross the cell wall. This model predicts that the substrate(s) normally exported by PEN3 accumulates to higher levels intracellularly in E. cichoracearum–infected pen3 plants than in pen3 plants inoculated with B. g. hordei, because E. cichoracearum continues to grow and repeatedly penetrate the epidermis, providing a greater stimulus for the production of PEN3 substrate(s) (Figure 7B). Based on the double mutant analysis (Figure 3) and microarray data (Table 1), it seems likely that the PEN3 substrate(s) spuriously activates the SA pathway. Because pen3 but not pen2 mutants develop chlorotic and necrotic patches with E. cichoracearum infections, we further propose that another enzyme(s), acting in parallel with PEN2, generates a related toxin(s) that is also exported by PEN3. To date, one phytoalexin, camalexin, has been identified in Arabidopsis (Tsuji et al., 1992). However, it is unlikely that this compound is solely responsible for the phenotypes conferred by pen3, because phytoalexin-deficient3 (pad3) and pad4 mutants lack the phenotype conferred by pen (data not shown). A postpenetration role for PEN3 can be explained by suggesting that PEN3 exports toxin(s) to the extrahaustorial matrix, poisoning the haustorium, thereby limiting the initiation and growth of secondary hyphae. This suggestion is supported by the localization of PEN3 protein to haustorial complexes (Figure 6). High-light stress presumably leads to the accumulation of toxic materials that would normally be exported by PEN3. The data supporting this model are incomplete as yet, and it is also possible that the loss of PEN2 attenuates the activation of the SA pathway in pen3 mutants or that PEN3 exports a fungal toxin or a fungal suppressor of defenses.
Figure 7.
Model for a Role for PEN3 in Penetration Resistance.
(A) We hypothesize that the PEN3 ABC transporter exports toxic secondary metabolites or other toxic materials (X and Y) to the apoplast at sites of attempted invasion. We speculate that the barley and pea powdery mildews are sensitive to these toxins, whereas the Arabidopsis powdery mildew is either less sensitive to this toxin or fails to fully activate this defense mechanism. Because some ABC transporters can transport a variety of related compounds, we assume that PEN3 exports a family of chemically related compounds, which may have varying toxicity. The glycosyl hydrolase PEN2 may activate one of these toxins (Y) by hydrolysis of a nontoxic precursor metabolite (Y-G). We assume that other enzymes activate related toxins (X).
(B) In the pen3 mutant, the sustained intracellular accumulation of PEN2-activated and related toxin(s) in interactions with the Arabidopsis powdery mildew leads to the activation of the SA pathway and the development of chlorosis and necrosis.
C, conidium; CW, cell wall; PM, plasma membrane.
Plants are resistant to most pathogens in their environment and succumb to only a small number of highly adapted pathogen species. Collectively, the isolation of the pen1, pen2, and pen3 mutants highlights the importance of defenses operating at the cell periphery. Only a subset of pathogens are able to overcome these defenses and attempt to colonize plants when additional defenses, operating intracellularly, come into play. For some inappropriate pathogens, only a limited number of barriers limit growth and asexual reproduction, suggesting that nonhost resistance may not be as complex as originally thought and that these defenses from nonhost plants can be introduced into host plants to provide stable, broad host range resistance to difficult pathogens.
METHODS
Growth Conditions and Inoculations
Arabidopsis thaliana and squash (Cucurbita maxima, Hybrid Kuta; Park Seed, Greenwood, SC) were grown in growth chambers at 22°C with a 14-h photoperiod of ∼125 μE·m−2·s−1 in the 400- to 700-nm range. Host powdery mildew (Erysiphe cichoracearum UCSC1) was cultured on squash for 10 to 12 d and then applied to Arabidopsis using settling towers (Vogel and Somerville, 2000). Barley powdery mildew (Blumeria graminis f. sp hordei CR3) was grown on barley (Hordeum vulgare) line Algerian-S (CI-16138) and inoculated onto Arabidopsis using the methods described by Zimmerli et al. (2004). Phytophthora infestans isolate 1306 (A1 mating type; provided by H. Judelson, University of California, Riverside) was maintained and prepared for inoculations as described by Zimmerli et al. (2004). Plants to be infected with Erysiphe pisi were grown in growth chambers at 20 to 23°C with a 12-h photoperiod and a light intensity of ∼150 μE·m−2·s−1 on a turf substrate mix (Stender Substrate; Wesel-Schermbeck) containing 0.001% Confidor WG70 (Bayer). Three-week-old plants were inoculated with E. pisi (Birmingham Isolate) using a settling tower (Lipka et al., 2005). For the growth of Plectosphaerella cucumerina and Botrytis cinerea, seeds were surface-sterilized, sown on square, 12.5 × 12.5-cm Petri dishes containing Murashige and Skoog basal salt mixture medium with 0.8% phytagel (Sigma-Aldrich), transferred to a phytochamber, and grown as described previously (Berrocal-Lobo et al., 2002; Berrocal-Lobo and Molina, 2004; Llorente et al., 2005). For high-light treatments, plants were germinated and grown at 22°C under a continuous light regime of ∼900 μE·m−2·s−1 in the 400- to 700-nm range.
M2 seeds were derived from an ethyl methanesulfonate–mutagenized population of Col-0 and screened for loss of penetration resistance to B. g. hordei by identifying individuals supporting an increased frequency of haustorium formation. The B. g. hordei haustoria became encased in callose and were visualized by staining for callose as described below
Cytology and Quantification of Fungal Growth
For penetration assays, 3-week-old Arabidopsis plants were inoculated with B. g. hordei and leaf samples were taken at 2 DAI. For P. infestans assays, 2-week-old Arabidopsis plants were sprayed with P. infestans zoospore suspensions and leaves were sampled at 2 DAI (Zimmerli et al., 2000). The deposition of callose was visualized by staining with aniline blue (Vogel and Somerville, 2000). For E. cichoracearum infections, 3-week-old Arabidopsis plants were inoculated as described above, and phenotypes were monitored visually at 4, 5, 6, 7, and 10 DAI.
To quantify fungal growth, eight Arabidopsis leaves per genotype were stained with aniline blue as described by Adam and Somerville (1996), with 250 mg of trypan blue added per milliliter. Haustoria, which become encased in callose in incompatible interactions, were visualized with aniline blue staining. Secondary hyphal elongation was quantified in nonoverlapping fields of view at ×200 magnification, avoiding the edge and midvein regions of the leaf. Numbers were expressed as percentages of total germinated conidia and were assessed for differences between mutant and the wild type with Student's t test. Fungal growth, the occurrence of lesions, and the accumulation of autofluorescent compounds were assessed as described by Vogel and Somerville (2000). To quantify hyphal length per colony, B. g. hordei–inoculated leaves were harvested at 10 DAI, cleared, and stained with trypan blue (Vogel and Somerville, 2000). At least 10 individual colonies were photographed under bright-field illumination at ×200 magnification with a Leica D500 digital camera attached to a Leica Leitz DMRB compound microscope. Hyphal lengths were measured using ImageJ software (version 1.30) (Abramoff et al., 2004; http://rsb.info.nih.gov/ij/).
Leaves infected with E. pisi were fixed and cleared in ethanol:acetic acid (3:1). Epiphytic fungal growth was visualized by staining of fungal structures with an ethanolic solution containing 0.6% Coomassie Brilliant Blue R 250 (Lipka et al., 2005). Cryogenic scanning electron microscopy of E. pisi–infected plants was performed as described by Sturaro et al. (2005).
Assays of P. cucumerina disease development were done by spraying 10-d-old plants with a spore suspension (at either 1 × 106 or 2 × 106 spores/mL, 1.5 mL/plate). Mock inoculations were done by spraying the plants with sterile water. After inoculation, plants were kept under the growth conditions described above; 10 d later, the percentage reduction of plant fresh weight caused by the fungal infection was calculated as described (Berrocal-Lobo and Molina, 2004). Inoculations of wild-type plants and pen3-1 mutants with B. cinerea were performed as described previously (Llorente et al., 2005), and the percentage of decayed plants was determined at different days after inoculation. The highly susceptible agb1-1 mutant was included in these experiments as a control for disease development (Llorente et al., 2005). At least 30 plants per genotype were inoculated with each pathogen, and the experiment was repeated four times. Differences between genotypes were identified using Student's t test.
Mapping and Cloning
Rough mapping markers, PCR conditions, and DNA preparation were as described previously (Lukowitz et al., 2000). Briefly, F2 populations of a mutant crossed to Arabidopsis Ler were sown in 96-well flats and then moved to a greenhouse. Plants were inoculated with B. g. hordei, as described above, at ∼3 weeks after germination when they had at least four true leaves. At 2 DAI, the third or fourth true leaf from each plant was harvested. Leaves were stained individually, mounted, and examined at ×50 magnification as described below. Tissue was harvested from plants with mutant phenotypes for DNA preparation and bulk segregant PCR (Lukowitz et al., 2000). Key individuals were retested for their pen3 phenotype in the F3 generation. New markers within the mapping interval were generated using the CEREON database of Col-0 and Ler polymorphisms (Jander et al., 2002) and the Whitehead Institute online primer design software, Primer3 (Rozen and Skaletsky, 2000; http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). All available T-DNA insertion lines for genes in a small interval on chromosome 1 were identified from the Salk Institute Genomic Analysis Laboratory's T-DNA project (http://signal.salk.edu/cgi-bin/tdnaexpress) (Ecker, 2002). Two SALK lines, SALK_110926 (pen3-3) and SALK_000578 (pen3-4), with insertions in the At1g59870 gene segregated pen3 phenotypes. The At1g59870 gene was sequenced from the pen3-1, pen3-2, and mil9 mutant lines on an ABI 310 Sequenator (Applied Biosystems).
Alignments of PDR family members were made using DNAStar MEGALIGN software with the ClustalW method and default parameters. Microarray data mining was done using the web tool GENEVESTIGATOR (Zimmermann et al., 2004).
Double Mutant Construction
The mutant alleles used for the construction of double mutants with pen3-1 were coi1-1 (Col-6 background [Feys et al., 1994]), npr1-1 (Cao et al., 1994), sid2-1 (Nawrath and Métraux, 1999), pad4-1 (Glazebrook and Ausubel, 1994), eds1-1 (Ws-0 background [Parker et al., 1996]), Etr1-1 (Bleecker et al., 1988), jar1-1 (Staswick et al., 1992), edr1 (Frye and Innes, 1998), sgt1b-1 (Ler background [Peart et al., 2002]), rar1-10 (Ler background [Muskett et al., 2002]), NahG (Lawton et al., 1995), pen1-1 (Collins et al., 2003), and pen2-3 (Lipka et al., 2005). All of the mutant lines, unless noted otherwise, were in the Col-0 background. We generated the double mutants by standard genetic crosses, following the mutations with cleaved-amplified polymorphic sequence (CAPS) markers. When the mutation did not lead to a change in a restriction site, we generated derived CAPS markers using the dCAPS web tool (Neff et al., 1998). The Etr1-1 (Hua and Meyerowitz, 1998), NahG (Morris et al., 2000), rar1-10 (Muskett et al., 2002), sgt1b (Austin et al., 2002), coi1-1 (Xie et al., 1998), pad4-1 (Nishimura et al., 2003), and npr1-1 (Nishimura et al., 2003) primers have been described previously. See Supplemental Table 5 online for a listing of primers used in genotyping mutations.
Expression Profiling
Samples of Col-0 and pen3-1 rosettes were collected from uninfected plants, plants infected with E. cichoracearum, or plants inoculated with B. g. hordei at 1 DAI. Each sample represented a pool of the rosettes from 16 plants grown in one pot under the growth conditions outlined above and inoculated as described above. Four pots (each pot a biological replicate) were grown for each treatment × genotype combination for a total of 24 pots. Total RNA was extracted from the plants using the Trizol method (Chomczynski and Sacchi, 1987) (Gibco BRL) and purified (Qiagen; RNeasy) essentially as described (Ramonell et al., 2002). Biotinylated complementary RNA (20 μg) was prepared as described (Hernan et al., 2003). The resulting complementary RNA was used to hybridize ATH1 Arabidopsis GeneChips (Affymetrix) using the manufacturer's protocols. The array images were analyzed with Affymetrix Microarray Suite 5.0 with the target intensity set to 500. The expression levels of the genes were analyzed with GeneSpring 4.2 software (Silicon Genetics), and the chip-to-chip signal variation was minimized by normalizing signal intensities to the averaged intensity values of uninoculated Col-0 using the expression levels of the top 50th percentile of probe sets. Differentially expressed genes were identified using two-way analysis of variance and a Benjamini and Hochberg multiple testing correction (GeneSpring 4.2). Genes were considered differentially expressed at P ≤ 0.001.
PEN3-GFP Localization
To create a PEN3 fusion to GFP, a 3′ fragment encoding the C terminus (nucleotides 4380 to 5681 of the unspliced transcript) of PEN3 was amplified by PCR using primers containing a C-terminal adaptor to keep the protein sequence in-frame with GFP in the vector pEGAD (Cutler et al., 2000). The primer sequences were 5′-CGGGGTACCGGACACTGGAAGAACCGTGGTC-3′ and 5′-ACGCGTCGACCGTCCCAACATGGAAACTCTTGTATC-3′. The 35S promoter was removed from pEGAD using SacI, before ligating the PCR-amplified and KpnI-BamHI–digested C-terminal PEN3 fragment in-frame with eGFP. BAC F23H11 was ordered from the ABRC and digested with KpnI, yielding a 10-kb genomic fragment containing the PEN3 gene as well as 2132 nucleotides upstream of the start codon and 2542 nucleotides downstream of the stop codon. This fragment was gel-purified and redigested using DraIII. The resulting digest was cloned into the pEGAD vector containing the PEN3 sequences encoding the C terminus fused with eGFP and digested with KpnI and DraIII, to give the full genomic piece fused in-frame to GFP. The vector was transformed into Agrobacterium tumefaciens (strain GV3101), and flowering pen3-1 plants were dipped into A. tumefaciens–containing infiltration medium as described (Cutler et al., 2000). T1 seeds were surface-disinfected and plated on Murashige and Skoog agar plates containing 50 μg kanamycin/mL (Murashige and Skoog, 1962). Two-week-old healthy seedlings were screened for GFP fluorescence with a Leica dissecting microscope equipped with an epifluorescence filter set. GFP-expressing seedlings were transferred to soil and inoculated with E. cichoracearum or B. g. hordei 2 weeks later. Single inoculated leaves from T1 plants were mounted on microscope cover slips in a 0.01 mg/mL propidium iodide solution in water (Ramonell et al., 2005). Imaging was done using a Nikon inverted fluorescence microscope equipped with a Bio-Rad MRC 1024 confocal head. Images were processed using the software ImageJ.
Accession Numbers
Microarray data sets were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; Barrett et al., 2005) under accession number GSE3220. The Arabidopsis Genome Initiative locus identifier for PEN3/PDR8 is At1g59870.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Expression of the 12 Members of the Arabidopsis PDR Family in Various Tissues.
Supplemental Table 2. PEN3 Transcript Levels Are Increased under Different Biotic and Abiotic Stresses.
Supplemental Table 3. Changes in Transcript Levels of PDR Family Members upon Powdery Mildew Infection in Wild-Type and pen3-1 Plants.
Supplemental Table 4. Host and Nonhost Resistance Phenotypes of Mutants Resistant to E. cichoracearum.
Supplemental Table 5. Primer Combinations Used to Genotype Mutations by PCR.
Supplemental Figure 1. Transcript Profiling of pen3-1 and Col-0 after Inoculation with the Arabidopsis and Barley Powdery Mildew Pathogens.
Supplementary Material
Acknowledgments
We thank Alex Paredez (Carnegie Institute), David Ehrhardt (Carnegie Institute), and Serry Koh (Carnegie Institute) for their help with confocal imaging; Marc Nishimura (Carnegie Institute) for sharing CAPS markers; Matt Humphry (Carnegie Institute) for assistance with GeneSpring; Melisa Lim (Carnegie Institute) for monitoring the expression of At3g30720; and Gemma López (Universidad Politécnica de Madrid) for technical assistance. Thanks to X. Dong (Duke University; npr1-1), C. Nawrath (Fribourg University; sid2-1), J. Glazebrook (University of Minnesota; pad4-1), J. Turner (University of East Anglia; coi1-1), J. Parker (Max Planck Institute for Plant Breeding Research; eds1-1), J. Ryals (Novartis; NahG), Jeff Dangl (University of North Carolina; sgt1b-1 rar1-10), and the ABRC (eds1-1, Etr1-1, jar1-1) for providing seeds. M.S. was supported in part by a Stanford Graduate Fellowship and by funding from the National Science Foundation to S.S. C.S.-R. was the recipient of a PhD fellowship from the Ministerio de Educación y Ciencia of Spain. This work was also supported in part by the Carnegie Institute (S.S.) and the Ministerio de Educacion y Ciencia of Spain (Grant BIO2003-4424 to A.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Shauna Somerville (ssomerville@stanford.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.038372.
References
- Abramoff, M.D., Magelhaes, P.J., and Ram, S.J. (2004). Image processing with ImageJ. Biophotonics International 11 36–42. [Google Scholar]
- Adam, L., and Somerville, S.C. (1996). Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J. 9 341–356. [DOI] [PubMed] [Google Scholar]
- Assaad, F.F., Qiu, J.L., Youngs, H., Ehrhardt, D., Zimmerli, L., Kalde, M., Wanner, G., Peck, S.C., Edwards, H., Ramonell, K., Somerville, C.R., and Thordal-Christensen, H. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15 5118–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, M.J., Muskett, P., Kahn, K., Feys, B.J., Jones, J.D.G., and Parker, J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295 2077–2080. [DOI] [PubMed] [Google Scholar]
- Barrett, T., Suzek, T.O., Troup, D.B., Wilhite, S.E., Ngau, W.C., Ledoux, P., Rudnev, D., Lash, A.E., Fujibuchi, W., and Edgar, R. (2005). NCBI GEO: Mining millions of expression profiles—Database and tools. Nucleic Acids Res. 33 D562–D566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo, M., and Molina, A. (2004). ETHYLENE RESPONSE FACTOR 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol. Plant Microbe Interact. 17 763–770. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo, M., Molina, A., and Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29 23–32. [DOI] [PubMed] [Google Scholar]
- Bhat, R.A., Miklis, M., Schmelzer, E., Schulze-Lefert, P., and Panstruga, R. (2005). Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. USA 102 3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker, A.B., Estelle, M., Somerville, C., and Kende, H. (1988). Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241 1086–1089. [DOI] [PubMed] [Google Scholar]
- Braun, U., Cook, R.T.A., Inman, A.J., and Shin, H.-D. (2002). The taxonomy of the powdery mildew fungi. In The Powdery Mildews, A Comprehensive Treatise, R.R. Belanger, W.R. Bushnell, A.J. Dik, and T.L.W. Carver, eds (St. Paul, MN: APS Press), pp. 13–55.
- Brugiere, S., Kowalski, S., Ferro, M., Seigneurin-Berny, D., Miras, S., Salvi, D., Ravanel, S., d'Herin, P., Garin, J., Bourguignon, J., Joyard, J., and Rolland, N. (2004). The hydrophobic proteome of mitochondrial membranes from Arabidopsis cell suspensions. Phytochemistry 65 1693–1707. [DOI] [PubMed] [Google Scholar]
- Busch, W., Wunderlich, M., and Schoffl, F. (2005). Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J. 41 1–14. [DOI] [PubMed] [Google Scholar]
- Campbell, E.J., Schenk, P.M., Kazan, K., Penninckx, I.A.M.A., Anderson, J.P., Maclean, D.J., Cammue, B.P.A., Ebert, P.R., and Manners, J.M. (2003). Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiol. 133 1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X.N. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162 156–159. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D., Aarts, N., Feys, B.J., Dong, X.N., and Parker, J.E. (2001). Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J. 26 409–420. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., Fengler, K.A., Yu, I.C., Lippok, B., Smith, R.K., and Bent, A.F. (2000). The Arabidopsis DND1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 97 9323–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, N.C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qiu, J.L., Hückelhoven, R., Stein, M., Freialdenhoven, A., Somerville, S.C., and Schulze-Lefert, P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425 973–977. [DOI] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., and Somerville, C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos, M., Van Oosten, V.R., Van Poecke, R.M.P., Van Pelt, J.A., Pozo, M.J., Mueller, M.J., Buchala, A.J., Métraux, J.P., Van Loon, L.C., Dicke, M., and Pieterse, C.M.J. (2005). Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 18 923–937. [DOI] [PubMed] [Google Scholar]
- Ecker, J.R. (2002). A sequence-indexed library of insertion mutations in the Arabidopsis genome. Plant Physiol. 129 405–406. [Google Scholar]
- Ellingboe, A.H. (1972). Genetics and physiology of primary infection by Erysiphe graminis. Phytopathology 62 401–406. [Google Scholar]
- Fernandez, M.R., and Heath, M.C. (1991). Interactions of the nonhost French bean plant Phaseolus vulgaris with parasitic and saprophytic fungi. 4. Effect of preinoculation with the bean rust fungus on growth of parasitic fungi nonpathogenic on beans. Can. J. Bot. 69 1642–1646. [Google Scholar]
- Feys, B.J., Benedetti, C., Penfold, C., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.A., and Parker, J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins EDS1 and PAD4. EMBO J. 20 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye, C.A., and Innes, R.W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., and Ausubel, F.M. (1994). Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA 91 8955–8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glombitza, S., et al. (2004). Crosstalk and differential response to abiotic and biotic stressors reflected at the transcriptional level of effector genes from secondary metabolism. Plant Mol. Biol. 54 817–835. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5 1003–1011. [DOI] [PubMed] [Google Scholar]
- Goodman, C.D., Casati, P., and Walbot, V. (2004). A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell 16 1812–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J.T., and Ausubel, F.M. (1993). Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. Plant J. 4 327–341. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., and Yao, N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6 201–211. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (2000). Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol. 3 315–319. [DOI] [PubMed] [Google Scholar]
- Heck, S., Grau, T., Buchala, A., Métraux, J.P., and Nawrath, C. (2003). Genetic evidence that expression of NahG modifies defense pathways independent of salicylic acid biosynthesis in the Arabidopsis-Pseudomonas syringae pv. tomato interaction. Plant J. 36 342–352. [DOI] [PubMed] [Google Scholar]
- Hernan, R., Fasheh, R., Calabrese, C., Frank, A.J., Maclean, K.H., Allard, D., Barraclough, R., and Gilbertson, R.J. (2003). ERBB2 up-regulates S100A4 and several other prometastatic genes in medulloblastoma. Cancer Res. 63 140–148. [PubMed] [Google Scholar]
- Holt, B.F., Belkhadir, Y., and Dangl, J.L. (2005). Antagonistic control of disease resistance protein stability in the plant immune system. Science 309 929–932. [DOI] [PubMed] [Google Scholar]
- Hua, J., and Meyerowitz, E.M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94 261–271. [DOI] [PubMed] [Google Scholar]
- Huitema, E., Vleeshouwers, V.G.A.A., Francis, D.M., and Kamoun, S. (2003). Active defense responses associated with non-host resistance of Arabidopsis thaliana to the oomycete pathogen Phytophthora infestans. Mol. Plant. Pathol. 4 487–500. [DOI] [PubMed] [Google Scholar]
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski, M., Stukkens, Y., Degand, H., Purnelle, B., Marchand-Brynaert, J., and Boutry, M. (2001). A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 13 1095–1107. [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., and Takemoto, D. (2004). Plant innate immunity: Direct and indirect recognition of general and specific pathogen-associated molecules. Curr. Opin. Immunol. 16 48–62. [DOI] [PubMed] [Google Scholar]
- Kang, L., Li, J.X., Zhao, T.H., Xiao, F.M., Tang, X.Y., Thilmony, R., He, S.Y., and Zhou, J.M. (2003). Interplay of the Arabidopsis nonhost resistance gene NHO1 with bacterial virulence. Proc. Natl. Acad. Sci. USA 100 3519–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki, H., Saitoh, H., Ito, A., Fujisawa, S., Kamoun, S., Katou, S., Yoshioka, H., and Terauchi, R. (2003). Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol. Plant. Pathol. 4 383–391. [DOI] [PubMed] [Google Scholar]
- Kleffmann, T., Russenberger, D., von Zychlinski, A., Christopher, W., Sjolander, K., Gruissem, W., and Baginsky, S. (2004). The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14 354–362. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Yamada, M., Kobayashi, I., and Kunoh, H. (1997). Actin microfilaments are required for the expression of nonhost resistance in higher plants. Plant Cell Physiol. 38 725–733. [Google Scholar]
- Kolling, R. (2002). Mutations affecting phosphorylation, ubiquitination and turnover of the ABC-transporter Ste6. FEBS Lett. 531 548–552. [DOI] [PubMed] [Google Scholar]
- Kuchler, K., Sterne, R., and Thorner, J. (1989). Saccharomyces cerevisae STE6 gene product, a novel pathway for protein export in eukaryotic cells. EMBO J. 8 3973–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir, S., Babiychuk, E., Storozhenko, S., Davey, M.W., Papenbrock, J., De Rycke, R., Engler, G., Stephan, U.W., Lange, H., Kispal, G., Lill, R., and Van Montagu, M. (2001). A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell 13 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton, K., Weymann, K., Friedrich, L., Vernooij, B., Uknes, S., and Ryals, J. (1995). Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol. Plant Microbe Interact. 8 863–870. [DOI] [PubMed] [Google Scholar]
- Lee, H., Lee, K., Lee, J., Noh, E.W., and Lee, Y. (2005). AtPDR12 contributes to lead resistance in Arabidopsis. Plant Physiol. 138 827–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, H., Yao, N., Song, L.T., Luo, S., Lu, H., and Greenberg, L.T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17 2636–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka, V., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310 1180–1183. [DOI] [PubMed] [Google Scholar]
- Llorente, F., Alonso-Blanco, C., Sánchez-Rodríguez, C., Jorda, L., and Molina, A. (2005). ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 43 165–180. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Gillmor, C.S., and Scheible, W.R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a Genome Initiative working for you. Plant Physiol. 123 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach, J.M., Castillo, A.R., Hoogstraten, R., and Greenberg, J.T. (2001). The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl. Acad. Sci. USA 98 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck, K., Neuenschwander, U., Cade, R.M., Dietrich, R.A., Dangl, J.L., and Ryals, J.A. (2002). Isolation and characterization of broad-spectrum disease-resistant Arabidopsis mutants. Genetics 160 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia, E., Klein, M., Geisler, M., Bovet, L., Forestier, C., Kolukisaoglu, U., Muller-Rober, B., and Schulz, B. (2002). Multifunctionality of plant ABC transporters—More than just detoxifiers. Planta 214 345–355. [DOI] [PubMed] [Google Scholar]
- Masri, S.S., and Ellingboe, A.H. (1966). Primary infection of wheat and barley by Erysiphe graminis. Phytopathology 56 389–395. [PubMed] [Google Scholar]
- Mellersh, D.G., and Heath, M.C. (2003). An investigation into the involvement of defense signaling pathways in components of the nonhost resistance of Arabidopsis thaliana to rust fungi also reveals a model system for studying rust fungal compatibility. Mol. Plant Microbe Interact. 16 398–404. [DOI] [PubMed] [Google Scholar]
- Meyer, S.L.F., and Heath, M.C. (1988). A comparison of the death induced by fungal invasion or toxic chemicals in cowpea epidermal cells. II. Responses induced by Erysiphe cichoracearum. Can. J. Bot. 66 624–634. [Google Scholar]
- Moller, S.G., Kunkel, T., and Chua, N.H. (2001). A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev. 15 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, K., Mackerness, S.A.H., Page, T., John, C.F., Murphy, A.M., Carr, J.P., and Buchanan-Wollaston, V. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23 677–685. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Muskett, P.R., Kahn, K., Austin, M.J., Moisan, L.J., Sadanandom, A., Shirasu, K., Jones, J.D.G., and Parker, J.E. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell 14 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., and Métraux, J.P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Neff, J.D., Chory, J., and Pepper, A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 14 387–392. [DOI] [PubMed] [Google Scholar]
- Nishimura, M.T., Stein, M., Hou, B.H., Vogel, J.P., Edwards, H., and Somerville, S.C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301 969–972. [DOI] [PubMed] [Google Scholar]
- Nühse, T.S., Stensballe, A., Jensen, O.N., and Peck, S.C. (2004). Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16 2394–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Holub, E.B., Frost, L.N., Falk, A., Gunn, N.D., and Daniels, M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart, J.R., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120. [DOI] [PubMed] [Google Scholar]
- Pighin, J.A., Zheng, H.Q., Balakshin, L.J., Goodman, I.P., Western, T.L., Jetter, R., Kunst, L., and Samuels, A.L. (2004). Plant cuticular lipid export requires an ABC transporter. Science 306 702–704. [DOI] [PubMed] [Google Scholar]
- Ramonell, K., Berrocal-Lobo, M., Koh, S., Wan, J.R., Edwards, H., Stacey, G., and Somerville, S. (2005). Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol. 138 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonell, K.M., Zhang, B., Ewing, R.M., Chen, Y., Xu, D., Stacey, G., and Somerville, S. (2002). Microarray analysis of chitin elicitation in Arabidopsis thaliana. Mol. Plant. Pathol. 3 301–311. [DOI] [PubMed] [Google Scholar]
- Rate, D.N., Cuenca, J.V., Bowman, G.R., Guttman, D.S., and Greenberg, J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate, D.N., and Greenberg, J.T. (2001). The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J. 27 203–211. [DOI] [PubMed] [Google Scholar]
- Rhee, S.Y., Osborne, E., Poindexter, P.D., and Somerville, C.R. (2003). Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol. 133 1170–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., and Skaletsky, H.J. (2000). Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols: Methods in Molecular Biology, S. Krawetz and S. Misener, eds (Totowa, NJ: Humana Press), pp. 365–386. [DOI] [PubMed]
- Saenz, G.S., and Taylor, J.W. (1999). Phylogeny of the Erysiphales (powdery mildews) inferred from internal transcribed spacer ribosomal DNA sequences. Can. J. Bot. 77 150–168. [Google Scholar]
- Sasabe, M., Toyoda, K., Shiraishi, T., Inagaki, Y., and Ichinose, Y. (2002). cDNA cloning and characterization of tobacco ABC transporter: NtPDR1 is a novel elicitor-responsive gene. FEBS Lett. 518 164–168. [DOI] [PubMed] [Google Scholar]
- Schaller, G.E., and Keiber, J.J. (1September2001). Ethylene. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0071, http://www.aspb.org/publications/arabidopsis/.
- Shah, J. (2003). The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 6 365–371. [DOI] [PubMed] [Google Scholar]
- Shih, M., Heinrich, P., and Goodman, H. (1991). Cloning and chromosomal mapping of nuclear genes encoding chloroplast and cytosolic glyceraldehyde-3-phosphate-dehydrogenase from Arabidopsis thaliana. Gene 104 133–138. [DOI] [PubMed] [Google Scholar]
- Staswick, P., Su, W., and Howell, S. (1992). Methyl-jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukkens, Y., Bultreys, A., Grec, S., Trombik, T., Vanham, D., and Boutry, M. (2005). NpPDR1, a pleiotropic drug resistance-type ATP-binding cassette transporter from Nicotiana plumbaginifolia, plays a major role in plant pathogen defense. Plant Physiol. 139 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturaro, M., Hartings, H., Schmelzer, E., Velasco, R., Salamini, F., and Motto, M. (2005). Cloning and characterization of GLOSSY1, a maize gene involved in cuticle membrane and wax production. Plant Physiol. 138 478–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen, H. (2003). Fresh insights into processes of nonhost resistance. Curr. Opin. Plant Biol. 6 351–357. [DOI] [PubMed] [Google Scholar]
- Tsuji, J., Jackson, E.P., Gage, D.A., Hammerschmidt, R., and Somerville, S.C. (1992). Phytoalexin accumulation in Arabidopsis thaliana during the hypersensitive reaction to Pseudomonas syringae pv. syringae. Plant Physiol. 98 1304–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brule, S., and Smart, C.C. (2002). The plant PDR family of ABC transporters. Planta 216 95–106. [DOI] [PubMed] [Google Scholar]
- van Wees, S.C.M., and Glazebrook, J. (2003). Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J. 33 733–742. [DOI] [PubMed] [Google Scholar]
- Vogel, J., and Somerville, S. (2000). Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 97 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J.P., Raab, T.K., Schiff, C., and Somerville, S.C. (2002). PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14 2095–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J.P., Raab, T.K., Somerville, C.R., and Somerville, S.C. (2004). Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 40 968–978. [DOI] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yun, B.W., Atkinson, H.A., Gaborit, C., Greenland, A., Read, N.D., Pallas, J.A., and Loake, G.J. (2003). Loss of actin cytoskeletal function and EDS1 activity, in combination, severely compromises non-host resistance in Arabidopsis against wheat powdery mildew. Plant J. 34 768–777. [DOI] [PubMed] [Google Scholar]
- Zimmerli, L., Jakab, C., Métraux, J.P., and Mauch-Mani, B. (2000). Potentiation of pathogen-specific defense mechanisms in Arabidopsis by beta-aminobutyric acid. Proc. Natl. Acad. Sci. USA 97 12920–12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli, L., Stein, M., Lipka, V., Schulze-Lefert, P., and Somerville, S. (2004). Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J. 40 633–646. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- Kobae et al. (2006) report that pdr8/pen3 mutants are compromised in nonhost resistance to Phytophthora infestans but show increased resistance to Pseudomonas syringae pv tomato DC3000.
- Kobae, Y., Sekino, T., Yoshioka, H., Nakagawa, T., Martinoia, E., and Maeshima, M. (13January2006). Loss of AtPDR8, a plasma membrane ABC transporter of Arabidopsis thaliana, causes hypersensitive cell death upon pathogen infection. Plant Cell Physiol. http://dx.doi.org/10.1093/pcp/pcj001. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.