Abstract
To investigate the resistance signaling pathways activated by pathogen infection, we previously identified the Arabidopsis thaliana mutant constitutive expresser of PR genes22 (cpr22), which displays constitutive activation of multiple defense responses. Here, we identify the cpr22 mutation as a 3-kb deletion that fuses two cyclic nucleotide-gated ion channel (ATCNGC)–encoding genes, ATCNGC11 and ATCNGC12, to generate a novel chimeric gene, ATCNGC11/12. Genetic, molecular, and complementation analyses suggest that ATCNGC11/12, as well as ATCNGC11 and ATCNGC12, form functional cAMP-activated ATCNGCs and that the phenotype conferred by cpr22 is attributable to the expression of ATCNGC11/12. However, because overexpression of ATCNGC12, but not ATCNGC11, suppressed the phenotype conferred by cpr22, the development of this phenotype appears to be regulated by the ratio between ATCNGC11/12 and ATCNGC12. Analysis of knockout lines revealed that both ATCNGC11 and ATCNGC12 are positive mediators of resistance against an avirulent biotype of Hyaloperonospora parasitica. Through epistatic analyses, cpr22-mediated enhanced resistance to pathogens was found to require NDR1-dependent and EDS1/PAD4-dependent pathways. In striking contrast, none of these pathways was required for cpr22-induced salicylic acid accumulation or PR-1 gene expression. These results demonstrate that NDR1, EDS1, and PAD4 mediate other resistance signaling function(s) in addition to salicylic acid and pathogenesis-related protein accumulation. Moreover, the requirement for both NDR1-dependent and EDS1/PAD4-dependent pathways for cpr22-mediated resistance suggests that these pathways are cross-regulated.
INTRODUCTION
Plants contain a large number of defense systems that can be deployed to prevent pathogen infection. Whether these defense responses are successful appears to be determined largely by the intensity and rapidity of their activation. In certain plant–pathogen interactions, a strong and rapid defense response is triggered by the direct or indirect interaction between the products of a plant R gene and a pathogen avirulence (avr) gene (Flor, 1971; Keen, 1990). After this gene-for-gene–specific recognition, the inoculated leaves usually exhibit increased levels of salicylic acid (SA), the induction of several families of defense genes, including those encoding the pathogenesis-related (PR) proteins, and the development of a hypersensitive response (HR) (Hammond-Kosack and Jones, 1996). The HR is characterized by apoptotic-like cell death at the site(s) of pathogen entry and often the restriction of pathogen to the cells within and immediately surrounding the necrotic lesion(s). At later times after infection, increased SA levels and PR gene expression are detected in the uninoculated portions of the plant, concurrent with the development of systemic acquired resistance, a long-lasting, broad-based resistance to subsequent infection (Ryals et al., 1996; Dempsey et al., 1999).
Many studies have demonstrated that SA is a critical signaling molecule in the pathways leading to local and systemic resistance (Dempsey et al., 1999). To identify other components involved in the resistance pathway, various Arabidopsis thaliana mutants exhibiting altered responses to pathogen infection have been identified. One class displays constitutive PR gene expression, increased SA levels, and heightened resistance to virulent and avirulent pathogens; included in this group are the lsd, cpr, ssi, acd, cim, hlm, cpn, and dnd mutants (Yu et al., 1998; Glazebrook, 1999, 2001; Jambunathan et al., 2001; Balague et al., 2003). A subset of these mutants also spontaneously develop HR-like lesions. By contrast, a second class of mutants exhibit heightened disease susceptibility. Some of these, such as npr1/nim1, sid1/eds5, and sid2/eds16, fail to respond to exogenously supplied SA or to accumulate endogenous SA (Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997; Nawrath and Metraux, 1999). In comparison, other enhanced susceptibility mutants, including pad4, eds1, and ndr1, have lost R gene–mediated resistance to distinct groups of pathogens (Century et al., 1995; Glazebrook et al., 1996; Parker et al., 1996). Comparison of R gene structure with the requirement for EDS1, PAD4, or NDR1 subsequently revealed that those R proteins with a coiled-coil, nucleotide binding site, and leucine-rich repeat (CC-NBS-LRR) structure generally require NDR1 to signal defense responses, whereas those R proteins that have an N-terminal domain homologous with Toll and the interleukin-1 receptor followed by an NBS and LRR (TIR-NBS-LRR) usually require EDS1 and PAD4 (Aarts et al., 1998; Feys et al., 2001).
After R gene-mediated pathogen recognition, one of the most rapidly activated defense responses is a rapid, biphasic increase in reactive oxygen species, known as the oxidative burst. Another very early response is ion fluxes, including an influx of H+ and Ca2+ and an efflux of K+ and Cl− (Atkinson et al., 1996). Of these ions, Ca2+ was shown to play a role in activating the oxidative burst after elicitor treatment (Chandra et al., 1997; Jabs et al., 1997; Sasabe et al., 2000) and in activating pathogen- or elicitor-induced cell death in tobacco (Nicotiana tabacum), cowpea (Vigna unguiculata), and soybean (Glycine max) (Atkinson et al., 1990; Levine et al., 1996; Xu and Heath, 1998; Sasabe et al., 2000). In addition, Ca2+ has been implicated in signaling SA-induced PR gene expression (Raz and Fluhr, 1992; Doke et al., 1996).
Consistent with a critical role for ion fluxes in defense signaling, three Arabidopsis mutants that constitutively express defense responses and spontaneously form HR-like lesions were found to contain defects in Ca2+-related proteins. The cpn1 (bon1) mutation disrupts a copine-encoding gene (Hua et al., 2001; Jambunathan et al., 2001). Copines are a newly identified class of Ca2+-dependent phospholipid binding proteins that have been detected in diverse organisms ranging from ciliates to humans (Creutz et al., 1998). On the other hand, the dnd1 and hlm1/dnd2 mutations disrupt different members of the ATCNGC gene family (Clough et al., 2000; Balague et al., 2003; Jurkowski et al., 2004). ATCNGCs have been studied intensively in animals, in which they play important roles in the perception of light and smell (Zufall et al., 1994; Zagotta and Siegelbaum, 1996). These channels, which are thought to be heterotetrameric in structure (Zhong et al., 2003), facilitate the conductance of Ca2+, K+, Na+, and other cations across the cell membrane (Kaupp and Seifert, 2002). ATCNGC subunits share many similarities with voltage-gated outward rectifying K+-selective ion channel (Shaker) proteins, including a cytoplasmic N terminus, six membrane-spanning regions, a pore domain, and a cytoplasmic C terminus (Zagotta and Siegelbaum, 1996). However, ATCNGCs are gated primarily by the binding of cAMP or cGMP, rather than by voltage.
In Arabidopsis, the ATCNGC gene family consists of 20 members that have been divided into four groups based on sequence similarity (Maser et al., 2001). Cloning of DND1 and HLM1/DND2 revealed that they correspond to ATCNGC2 and ATCNGC4, respectively (Clough et al., 2000; Balague et al., 2003, Jurkowski et al., 2004). These genes are highly related and belong to the same subgroup (Maser et al., 2001; Balague et al., 2003). Intriguingly, although mutations in these genes confer similar constitutive defense response phenotypes, some differences were noted. In particular, the dnd1 mutation in ATCNGC2 did not affect R gene–mediated resistance, whereas the hlm1 mutation in ATCNGC4 suppressed gene-for-gene resistance to certain pathogens but not others (Yu et al., 1998; Balague et al., 2003; Talke et al., 2003). Thus, the role played by ATCNGCs in defense signaling is currently unclear.
To identify the signaling components involved in activating defense responses after pathogen attack, we previously screened 4200 T-DNA–tagged lines from the Wassilewskija (Ws) ecotype for individuals that constitutively expressed PR-1 (Yoshioka et al., 2001). One individual, designated constitutive expresser of PR genes22 (cpr22), was identified. This mutant constitutively activated various defense responses, exhibited stunted growth with curly leaves, and displayed enhanced resistance to the virulent oomycete pathogen Hyaloperonospora parasitica (formerly Peronospora parasitica) Emco5. cpr22 is a semidominant mutation that is lethal in the homozygous state unless the plant is grown under high RH. Here, we demonstrate that the phenotype conferred by cpr22 is attributable to a deletion that generates a chimeric ATCNGC-encoding gene designated ATCNGC11/12. Genetic and molecular analyses suggest that the presence of ATCNGC11/12 leads to the activation of SA-dependent, but NDR1- and EDS1/PAD4-independent, defense responses. By contrast, cpr22-mediated enhanced resistance appears to require both the NDR1- and EDS1/PAD4-mediated signaling pathways.
RESULTS
Isolation of the cpr22 Mutation
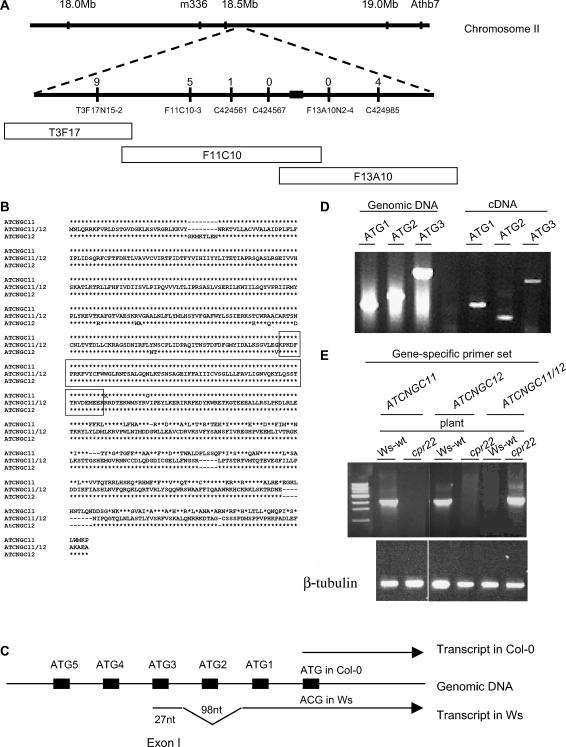
The cpr22 mutation was previously mapped to chromosome 2, ∼2 centimorgan from AthB102 (Yoshioka et al., 2001). However, because of a lack of polymorphisms between the Ws and Columbia (Col-0) ecotypes in this region, a second mapping population of 439 individuals was developed using the F2 progeny of a cross between cpr22 and Landsberg erecta. Markers identified using the CEREON polymorphism database were used to localize CPR22 to a 60-kb region spanned by BAC clones F11C10 and F13A10 (Figure 1A). Of the 16 open reading frames (ORFs) identified within this region, 9 were sequenced from cpr22 and wild-type plants; comparison of these sequences identified a 3-kb deletion in a cluster of putative ATCNGC genes. The deletion generated a chimeric gene derived from ATCNGC11 and ATCNGC12 (Figure 1B).
Figure 1.
cpr22 Is a 3-kb Deletion That Creates a Chimeric ATCNGC-Encoding Gene.
(A) Genetic and physical map of the cpr22 locus on chromosome 2. The top line represents chromosome 2; the bottom line is a larger scale map of the cpr22-containing region, with various markers indicated below the line and the number of recombination events for each marker indicated above it. The location of the 3-kb deletion is indicated as a black box. BAC clones T3F17, F11C10, and F13A10, which span the cpr22-containing region, are represented as open boxes below the map.
(B) Amino acid sequence alignment of ATCNGC11, ATCNGC11/12, and ATCNGC12 from the Ws ecotype. The deduced amino acid sequence for ATCNGC11/12 is in the middle; amino acid residues in ATCNGC11 and/or ATCNGC12 that are identical to those of ATCNGC11/12 are represented as asterisks, whereas residues that differ are indicated above or below the line. The boxed region contains sequences that are identical in all three proteins. Because the predicted ATCNGC11/12 sequence is identical to ATCNGC11 upstream of the boxed sequence and to ATCNGC12 downstream of this sequence, the homologous recombination event that created ATCNGC11/12 likely occurred within this region.
(C) Scheme of the 5′ translated and untranslated regions of the Col-0 and Ws alleles of ATCNGC11. To investigate the position of the translational start codon in the Ws allele of ATCNGC11, RT-PCR analysis was performed with primers corresponding to five ATG codons near the predicted translational start codon of the Col-0 allele. ATG1 to ATG5 are depicted as black boxes on the genomic DNA. This analysis suggested that the transcript for the Ws allele (bottom arrow) contains an extra 98-nucleotide intron and 27-nucleotide exon that are not present in the transcript of the Col-0 allele (top arrow).
(D) PCR analysis of ATCNGC11 (Ws ecotype). Forward primers ATG1, ATG2, and ATG3, which correspond to the first three ATG codons within the predicted 5′ untranslated region of ATCNGC11, were tested for their ability to generate a product using genomic DNA or cDNA from wild-type Ws plants as the template in combination with the ATCNGC11-specific reverse primer UD1R1. ATG1 is closest to the annotated translational start codon, and ATG3 is farther upstream. The products were resolved on an agarose gel and stained with ethidium bromide. See text for additional information.
(E) Semiquantitative RT-PCR analysis of ATCNGC11, ATCNGC12, and ATCNGC11/12 transcript levels in wild-type Ws and cpr22 homozygous plants using gene-specific primers. The products were resolved on an agarose gel and stained with ethidium bromide. β-Tubulin was used as a loading control.
Consistent with the structure of other ATCNGC-encoding genes, ATCNGC11 and ATCNGC12 are predicted to contain a cytoplasmic N terminus, six membrane-spanning segments, a pore domain, and a cytoplasmic C terminus that contains overlapping calmodulin binding and cyclic nucleotide binding domains (Kohler et al., 1999; Leng et al., 1999; Kohler and Neuhaus, 2000). This conserved structure also was observed in the chimeric ATCNGC11/12 gene. Sequence comparison between ATCNGC11/12, ATCNGC11, and ATCNGC12 suggested that a homologous recombination event occurred within the third membrane-spanning region, which fused the 5′ portion of ATCNGC11 with the 3′ portion of ATCNGC12. Because a 222-bp stretch of this region is 100% homologous between ATCNGC11 and ATCNGC12, the actual location of the recombination event cannot be determined.
The predicted ATG start codon for ATCNGC11 in Col-0 corresponds to an ACG codon in the Ws allele. Thus, it was unclear whether ATCNGC11, and by extension the novel chimeric gene, was expressed in Ws plants. Alternatively, these proteins might be translated from an ATG farther 5′ in the predicted untranslated sequence. Analysis of the proposed 5′ region of the Ws ATCNGC11 allele identified several ATG codons; however, without splicing of the RNA transcript, none would be in-frame with the downstream protein-encoding sequence. To assess whether ATCNGC11 was translated from one of these ATG codons, primers corresponding to sequences surrounding the first five upstream ATG codons were used for PCR analysis (Figure 1C). A fragment of the expected size was generated when a primer corresponding to the first ATG (ATG1; closest to the annotated translational start codon) was used with genomic DNA or cDNA as the template (Figure 1D). By contrast, primers corresponding to the second (ATG2) and third (ATG3) ATG codons only generated fragments of the expected size in the presence of genomic DNA; smaller than expected fragments were generated when cDNA was used as the template. Sequence analysis revealed that the fragment generated by the ATG2 primer was attributable to nonspecific amplification, whereas the fragment generated by the ATG3 primer lacked a 98-nucleotide region surrounding ATG2. This result, combined with the inability of primers corresponding to ATG codons upstream of ATG3 to generate product from the cDNA template (data not shown), suggested that ATCNGC11 from Ws contains an extra exon and a 98-nucleotide intron upstream of the predicted start site for the Col-0 allele. The presence of a 27-nucleotide exon that is spliced in-frame with downstream sequences after removal of a 98-nucleotide intron was confirmed by sequencing ATG3-primed PCR products generated from genomic and cDNA templates (Figure 1C; data not shown). To determine whether the chimeric ATCNGC11/12 gene also was expressed, cDNA prepared from cpr22 plants was subjected to sequence analysis using the ATG3 primer. ATCNGC11/12 was found to contain a full-length ORF, and based on semiquantitative RT-PCR analysis, it was expressed in cpr22 to comparable levels as ATCNGC11 and ATCNGC12 in wild-type Ws plants (Figure 1E). Thus, we conclude that ATCNGC11 and ATCNGC11/12 are not pseudogenes.
ATCNGC11 and ATCNGC12 Knockout Lines and Their F1 Progeny Display No Phenotypes Conferred by cpr22 but Exhibit Reduced Resistance to an Avirulent Pathogen
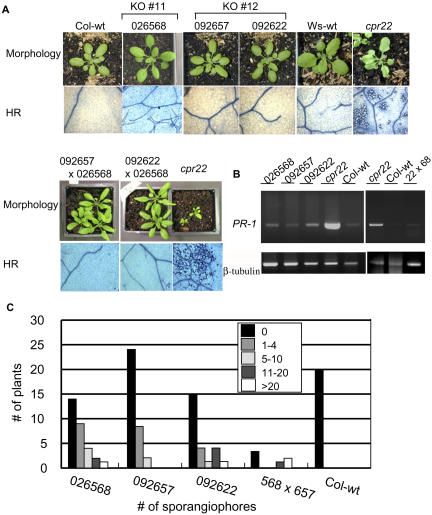
Because ATCNGC11 and ATCNGC12 are expressed in wild-type plants but are combined into a chimeric gene in cpr22, the mutant phenotype could be attributable to the loss of one or both of these genes and/or to the expression of ATCNGC11/12. To begin addressing this question, ATCNGC11 and ATCNGC12 T-DNA insertion lines were obtained from the Salk Institute collection (T-DNA insertion lines Salk_026568 for ATCNGC11 and Salk_092657 and Salk_092622 for ATCNGC12) and analyzed. Homozygous plants were obtained for all three knockout (KO) lines, and RT-PCR was used to confirm the absence of expression of the corresponding genes (see Supplemental Figure 1A online). All of the KO lines exhibited wild-type morphology and failed to spontaneously form lesions or constitutively express PR-1 (Figures 2A and 2B). Their pathogen resistance status was analyzed using H. parasitica biotypes Noco2 and Emwa1. The Col-0 ecotype, which is the background for the Salk Institute T-DNA insertion lines, is susceptible to Noco2 and resistant to Emwa1. None of the three KO lines showed altered susceptibility to Noco2 compared with wild-type Col-0 plants (Table 1). However, resistance to Emwa1 was reduced in all three KO lines; 35 to 55% of the KO plants allowed sporangiophore development, with modest to high levels of sporangiophore development detected on ∼20 to 30% of these plants (Figure 2C, Table 1). By contrast, no sporangiophores were observed on wild-type Col-0 plants. This reduction in resistance suggests that ATCNGC11 and ATCNGC12 are positive mediators of resistance against H. parasitica Emwa1.
Figure 2.
KO Lines of ATCNGC11 (Salk_026568), ATCNGC12 (Salk_ 092657 and Salk_092622), and Their F1 Progeny Display No Phenotypes Conferred by cpr22 but Exhibit Reduced Resistance to H. parasitica Emwa1.
(A) Morphological phenotype and spontaneous cell death in wild-type Col-0 and Ws plants, the cpr22 mutant, ATCNGC11 and ATCNGC12 KO lines, and their F1 progeny. Plants were photographed 3 weeks after planting for single KO lines and 4 weeks after planting for F1 progeny. Lesion formation was monitored microscopically in leaves of 25-d-old plants after trypan blue staining.
(B) RT-PCR analysis for PR-1 gene expression. PR1-F and PR1-R primers were used for the detection of PR-1 gene expression, and β-tubulin-5′ and β-tubulin-3′ primers were used for the detection of β-tubulin gene expression.
(C) Infection with H. parasitica Emwa1. Seven-day-old seedlings were inoculated with H. parasitica Emwa1 (106 spores/mL). At 6 d after inoculation, two cotyledons per plant were analyzed with a microscope and categorized into one of five categories (0, 1 to 4, 5 to 10, 11 to 20, or >20) depending on the number of sporangiophores observed on the two cotyledons. Experiments were performed four times with similar results.
Table 1.
Resistance Status of Single and Double Knockout Plants to H. parasitica
| Isolate | Plant Linea | Total No. of Plants | No. of Infected Plantsb | Infected Plants (%) |
|---|---|---|---|---|
| Noco2 (Vir) | 026568 | 28 | 24 | 85.7 |
| 092657 | 17 | 13 | 76.5 | |
| 092622 | 20 | 17 | 85.0 | |
| 568 × 57 | 7 | 6 | 85.7 | |
| Col wild type | 14 | 12 | 85.7 | |
| Emwal (Avr) | 026568 | 30 | 16 | 53.3 |
| 092657 | 34 | 10 | 29.4 | |
| 092622 | 27 | 10 | 37.0 | |
| 568 × 57 | 6 | 3 | 50.0 | |
| Col wild type | 20 | 0 | 0.0 |
568 × 57; F1 of cross-pollination between Salk_026568 and Salk_092657.
Based on formation of sporangiophores.
Because cpr22 plants lack full-length copies of both ATCNGC11 and ATCNGC12, comparison of their phenotype with that of a double KO plant could be very informative. However, ATCNGC11 and ATCNGC12 are <1.5 kb apart, making the generation of a double KO via recombination extremely difficult. Using RNA interference to create a line specifically silenced for ATCNGC11 and ATCNGC12 also would be problematic, because there are 20 members in the ATCNGC family and some of these genes are highly similar in their DNA sequences (Maser et al., 2001). Given the difficulty involved in obtaining a double KO, we decided to cross the ATCNGC11 and ATCNGC12 KO lines and monitor the phenotype of the F1 progeny. The cpr22 mutation is semidominant and leads to a readily visible phenotype in the heterozygous state (one copy each of ATCNGC11, ATCNGC12, and ATCNGC11/12) when grown under normal RH conditions. Thus, if the phenotype conferred by cpr22 is attributable to the loss of one copy of ATCNGC11 and ATCNGC12, rather than to the expression of ATCNGC11/12, the F1 progeny of crosses between the KO lines should exhibit a phenotype like that conferred by cpr22. F1 plants from crosses between Salk_026568 (ATCNGC11) and Salk_092657 (ATCNGC12) or between Salk_026568 (ATCNGC11) and Salk_092622 (ATCNGC12) were created by reciprocal cross-pollination. To confirm heterozygosity, the T-DNA insertion in each gene was analyzed by PCR using gene-specific primers as well as a primer specific for the T-DNA sequence (see Supplemental Figure 1B online). All of the F1 plants exhibited wild-type morphology and, like the single KO lines, failed to spontaneously form lesions or constitutively express PR-1 (Figures 2A and 2B). Therefore, the phenotype conferred by cpr22 is not attributable simply to the loss of one copy each of these two ATCNGC genes. Analysis of a small number of plants (because of the limited number of F1 seeds) revealed comparable levels of susceptibility to H. parasitica Noco2 and exhibited similarly reduced resistance to Emwa1 as the single KO lines (Table 1, Figure 2C). Because the F1 plants contain a single copy of ATCNGC11 and ATCNGC12, and the parent KO lines contain two copies of either ATCNGC11 or ATCNGC12, these results suggest that these ATCNGCs are at least partially redundant for resistance.
Expression of ATCNGC11/12 Is Required for the Phenotype Conferred by cpr22
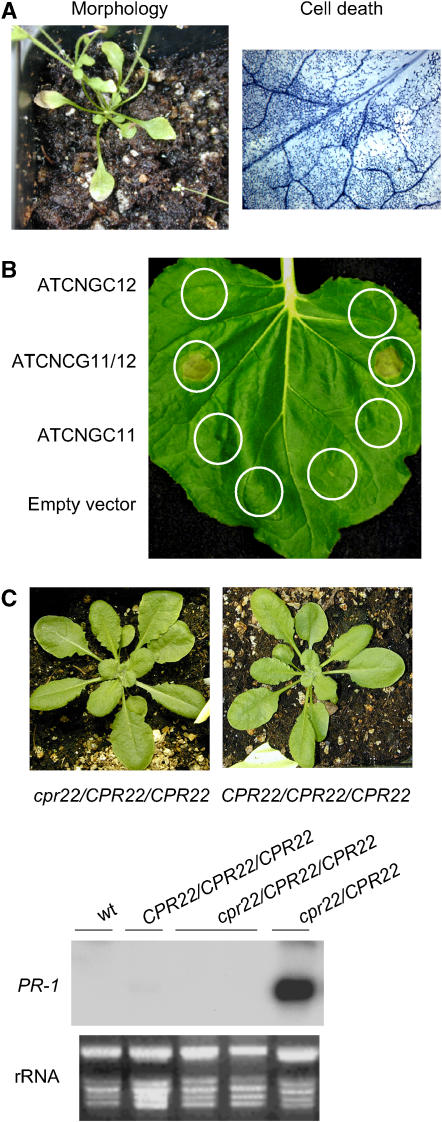
The results with the F1 plants suggest that the phenotype conferred by cpr22, at least in cpr22/CPR22 heterozygous plants, is not attributable to the loss of one functional copy of ATCNGC11 or ATCNGC12. Whether expression of ATCNGC11/12 leads to the phenotype conferred by cpr22 was then tested by transforming diploid wild-type Ws plants with ATCNGC11/12 driven by the powerful cauliflower mosaic virus (CaMV) 35S promoter. By 4 weeks after planting, these transgenic plants developed a phenotype similar to that of cpr22 heterozygous plants, including stunted growth, curly leaves, spontaneous lesion formation and constitutive PR-1 expression, whereas transgenic plants expressing 35S-ATCNGC11 or 35S-ATCNGC12 constructs were indistinguishable from untransformed control plants (Figure 3A; see Supplemental Figure 2 online). Further arguing that expression of ATCNGC11/12 leads to the phenotype conferred by cpr22, transient expression of CaMV 35S-driven ATCNGC11/12, but not ATCNGC11 or ATCNGC12, induced HR-like cell death in Nicotiana benthamiana (Figure 3B). Analysis of cpr22/CPR22/CPR22 triploid plants (created by crossing a cpr22 homozygous mutant with a tetraploid CS3432 plant [Col-0 ecotype]), however, revealed that expression of ATCNGC11/12 does not necessarily lead to the phenotype conferred by cpr22. All 12 F1 progeny from independent crosses displayed wild-type morphology and did not constitutively express PR-1 (Figure 3C). Seed development in the triploid plants, however, was arrested in ambient RH growth conditions and was restored to nearly normal levels by high RH growth conditions. One possible explanation for the different phenotypes of 35S-ATCNGC11/12 transgenic plants and the triploid plants is that the phenotype conferred by cpr22 is determined by the ratio between the expression of ATCNGC11/12 and the wild-type copy(s) of ATCNGC11 and/or ATCNGC12. Because ATCNGC11/12 expression is driven by its endogenous (ATCNGC11) promoter in the triploid plants, its expression level may not be sufficient to result in the phenotype conferred by cpr22 in the presence of two copies of wild-type ATCNGC11 and ATCNGC12. By contrast, stronger expression of ATCNGC11/12 from the 35S promoter in transgenic plants might overcome the presence of the wild-type genes.
Figure 3.
Expression of ATCNGC11/12 Is Required for the Phenotype Conferred by cpr22.
(A) Mutant morphology and spontaneous lesion formation in wild-type Ws plants transformed with ATCNGC11/12 driven by the CaMV 35S promoter. Plants were grown on soil and photographed at 4 weeks after planting . Microscopic analysis of trypan blue–stained leaves from these plants revealed intensely stained areas of dead cells.
(B) Transient expression of ATCNGC12, ATCNGC11/12, or ATCNGC11 in N. benthamiana. Agrobacteria containing an empty vector (pMBP3) or a vector containing ATCNGC12, ATCNGC11, or ATCNGC11/12 driven by the CaMV 35S promoter were infiltrated into N. benthamiana leaves. The infiltrated areas are circled. Leaves were photographed at 3 d after infiltration and showed lesion formation only with ATCNGC11/12 expression.
(C) Phenotypes of wild-type triploid (CPR22/CPR22/CPR22) and a triploid cpr22/CPR22/CPR22 plant. The plants were grown on soil and photographed at 3 weeks after planting (top). RNA gel blot analysis was performed using 8 μg of total RNA harvested from 3-week-old plants (bottom). Ethidium bromide staining of rRNA served as a loading control.
ATCNGC12 Suppresses the Phenotype Conferred by cpr22 in a Dosage-Dependent Manner
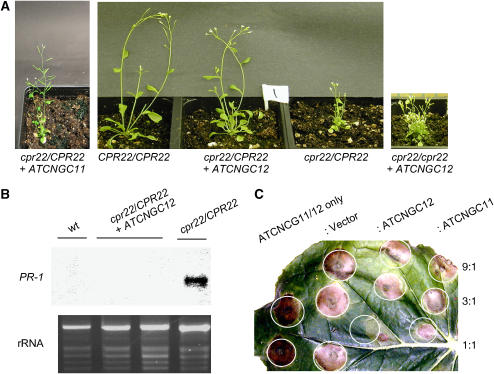
To investigate whether the phenotype conferred by cpr22 is regulated by the ratio between ATCNGC11/12 and ATCNGC11 and/or ATCNGC12, we tested whether the phenotype conferred by cpr22 was affected by increasing the dosage of the wild-type genes. More than 30 independent T1 lines were generated from cpr22 heterozygous plants transformed with an ∼3.3-kb genomic copy of ATCNGC12. Strikingly, the phenotype of many of these plants was nearly identical to that of wild-type plants. Although these T1 plants were slightly stunted, they did not develop spontaneous lesions or constitutively express PR-1 under normal RH growth conditions (Figure 4A). Three independent T1 lines also were generated from cpr22 homozygous plants transformed with the genomic copy of ATCNGC12. Plants from all of these lines grew in low RH conditions without lethality, and the severity of stunting, leaf curling, and spontaneous lesion formation was similar to that of cpr22 heterozygous plants (Figure 4A). Analysis of T2 progeny from all of the T1 lines transformed with ATCNGC12 confirmed that expression of this transgene suppressed cpr22-induced stunting, spontaneous lesion formation, and constitutive PR-1 expression in a dose-dependent manner. Of 25 T2 plants from a self-pollinated cpr22 homozygous line expressing ATCNGC12, 4 progeny were slightly stunted but otherwise wild type in morphology, 15 developed a phenotype similar to that of cpr22 heterozygous plants, and 6 died (consistent with the cpr22 homozygous phenotype); these results conform with the predicted 1:2:1 ratio (χ2 = 1.32; 0.80 > P > 0.50) for a single, semidominant locus. Analysis of 70 T2 progeny from a self-pollinated cpr22 heterozygous line expressing ATCNGC12 identified 43 progeny with the nearly wild-type morphology, 22 with a phenotype similar to that of cpr22 heterozygous plants, and 5 that died. These results are consistent with an 11:4:1 ratio (χ2 = 1.78; 0.5 > P > 0.20), which would be predicted if the transgene segregates independently of the CPR22 locus and two doses of ATCNGC12 are sufficient to confer the wild-type or nearly wild-type morphology.
Figure 4.
The Severity of the Phenotype Conferred by cpr22 Is Determined by the Ratio between ATCNGC11/12 and ATCNGC12.
(A) Morphological phenotypes of wild-type (CPR22/CPR22) and cpr22 (cpr22/CPR22) plants, as well as those of a cpr22 heterozygous plant transformed with an ∼5.5-kb genomic copy of ATCNGC11 (cpr22/CPR22 + ATCNGC11) and cpr22 heterozygous and homozygous plants transformed with an ∼3.3-kb genomic copy of ATCNGC12 (cpr22/CPR22 + ATCNGC12 and cpr22/cpr22 + ATCNGC12, respectively). The plants were grown on soil and photographed at 4 weeks after planting.
(B) PR-1 gene expression in wild-type (CPR22/CPR22) plants, the cpr22/CPR22 mutant, and a cpr22/CPR22 mutant transformed with ATCNGC12. RNA gel blot analysis was performed using 8 μg of total RNA harvested from 3-week-old plants. Ethidium bromide staining of rRNA served as a loading control.
(C) Transient coexpression analysis of ATCNGC11, ATCNGC12, or ATCNGC11/12 in N. benthamiana. Agrobacteria containing a 35S-ATCNGC11/12 construct in the pMBP3 vector were infiltrated alone or coinfiltrated with differing concentrations of agrobacteria containing either an empty vector or a 35S-ATCNGC12 or 35S-ATCNGC11 construct into N. benthamiana leaves. The ratios of ATCNGC11/12 to empty vector, ATCNGC12, and ATCNGC11 are designated at right. The infiltrated leaf was photographed at 3 d after infiltration. The expression level of each gene was confirmed and monitored by protein gel blot analysis using a specific antibody against green fluorescent protein and by fluorescent microscopic observation.
By contrast, transformation of cpr22 heterozygous plants with an ∼5.5-kb genomic copy of ATCNGC11 produced only two independent T1 lines, despite multiple attempts. The T1 lines and their subsequent T2 progeny showed cpr22-like curly leaves, spontaneous HR development, and PR-1 gene expression (Figure 4A; data not shown). Although these results suggested that ATCNGC11 cannot complement the phenotype conferred by cpr22, it should be noted that the level of ATCNGC11 transgene expression, and that of the ATCNGC12 transgene, cannot be monitored because they are indistinguishable from those of the corresponding endogenous genes.
To determine whether increased expression of ATCNGC12 suppresses other aspects of the phenotype conferred by cpr22, we monitored the ability of this transgene to compromise cpr22-mediated disease resistance in Arabidopsis and HR development in N. benthamiana. Analysis of T2 progeny from a cpr22 heterozygous plant transformed with ATCNGC12 revealed that resistance to H. parasitica Emco5 was suppressed (data not shown). Coexpression analysis using green fluorescent protein–tagged ATCNGC11/12, ATCNGC11, and ATCNGC12 in N. benthamiana indicated that ATCNGC12 also reduced lesion formation caused by ATCNGC11/12, but only when both were expressed and/or present at similar levels (Figure 4C). By contrast, neither the empty vector nor ATCNGC11 affected lesion formation when present at a similar (or lesser) level as ATCNGC11/12.
Functional Analysis of ATCNGC11, ATCNGC12, and ATCNGC11/12
Native ATCNGCs in animals are heterotetrameric proteins composed of more than one ATCNGC gene product (Zhong et al., 2003). Some studies have suggested that native plant ATCNGC channel complexes also are composed of multiple subunits expressed from different ATCNGC genes. In addition, Arazi et al. (1999) found that overexpression of a plant ATCNGC could enhance the uptake of one cation and reduce the uptake of another in transgenic plants. Together, these results are consistent with a model positing that native plant ATCNGC channels are heteromeric and that altered expression of a specific ATCNGC subunit could alter the assembly of the native channel complex and change its conductance profile. Thus, provided that ATCNGC11/12 is capable of forming channels, the phenotype conferred by cpr22 might be caused by altered cation concentrations attributable to the presence of heteromeric channels containing ATCNGC11/12 and ATCNGC12 or other ATCNGC family members.
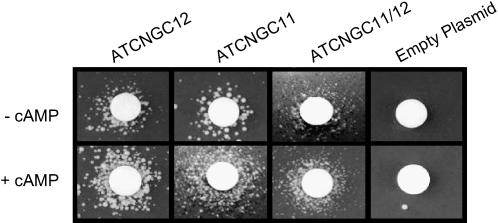
To determine whether ATCNGC12, ATCNGC11, and ATCNGC11/12 encode functional cation channels, heterologous expression assays were performed in the trk1,2 yeast mutant. This mutant lacks the functional K+ uptake proteins TRK1 and TRK2 (Gaber et al., 1988; Ko and Gaber, 1991). Reduced cation uptake in this mutant results in a hyperpolarized (inside negative) membrane potential and concomitant hypersensitivity to the cationic antibiotic hygromycin B. However, the trk1,2 yeast mutant can grow in medium containing hygromycin B plus K+ if a cation transport protein capable of conducting K+ is expressed in the mutant (Madrid et al., 1998; Bihler et al., 2002). Moreover, Mercier et al. (2004) recently showed that expression of plant ATCNGCs complemented the hypersensitivity of trk1,2 growth (around a filter disk containing K+) to hygromycin B.
We used the functional assay developed by Mercier et al. (2004) to test the cyclic nucleotide-dependent inward cation conductance (i.e., ATCNGC function) of ATCNGC11, ATCNGC12, and ATCNGC11/12. The trk1,2 mutant transformed with the empty plasmid (control) did not grow around a K+-containing filter disk on medium containing hygromycin B either in the absence or presence of cAMP (Figure 5). By contrast, ATCNGC11, ATCNGC12, and the chimeric ATCNGC11/12 complemented the growth of trk1,2. However, the growth of the mutant yeast expressing ATCNGC11/12 was less vigorous than that of mutants expressing either ATCNGC11 or ATCNGC12.
Figure 5.
ATCNGC11, ATCNGC12, and ATCNGC11/12 Encode Functional Cyclic Nucleotide-Gated Cation Channels.
Complementation of hygromycin hypersensitivity of trk1,2 yeast by transfection with empty plasmid, ATCNGC11, ATCNGC12, or ATCNGC11/12. After transformation, the ability of yeast cells to grow around a filter disk containing 3 M KCl (center of each panel) on solid YPGal medium containing 0.07 mg/mL hygromycin B in the presence or absence of 100 μM cAMP was monitored. Photographs were taken after 3 d of growth. Data shown are representative results obtained with this assay in four independent experiments.
One of the defining attributes of members of the ATCNGC family is the (direct) activation of currents by the ligand cAMP (Kaupp and Seifert, 2002). As shown most clearly by mutant yeast expressing ATCNGC11 and ATCNGC12, addition of a lipophilic analog of cAMP to the growth medium increased colony growth around the filter disk (Figure 5), thus demonstrating that these proteins are ATCNGCs. Although cAMP clearly stimulated the growth of mutant yeast expressing chimeric ATCNGC11/12, again, growth was less vigorous than in yeast expressing the wild-type ATCNGCs. Addition of dibutyryl-cGMP did not enhance the growth of yeast expressing ATCNGC11, ATCNGC12, or ATCNGC11/12 (data not shown); thus, it appears that these channels are activated solely by cAMP and not by cGMP. Interestingly, Lemtiri-Chlieh and Berkowitz (2004) recently documented the presence of native ATCNGCs in mesophyll and guard cells of Arabidopsis plants activated by cAMP but not by cGMP.
Lesion Formation Is Not Impaired in cpr22 Plants Inoculated with an Avirulent Pathogen
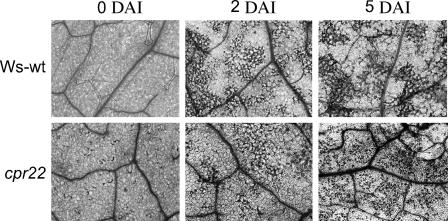
Previously, three Arabidopsis ATCNGC mutants were isolated, dnd1 and hlm1/dnd2 (Clough et al., 2000; Balague et al., 2003; Jurkowski et al., 2004). Although there are several differences in their phenotypes, all three mutants display delayed and/or suppressed HR cell death upon infection with various avirulent pathogens (Yu et al., 1998; Balague et al., 2003). Therefore, we tested whether HR cell death is altered in cpr22 plants. After inoculation with the avirulent pathogen Pseudomonas syringae pv tomato DC3000 expressing avrRpt2, both visual inspection and microscopic analysis with trypan blue revealed that the kinetics of HR cell death were comparable in wild-type Ws and cpr22 plants. As reported previously (Yoshioka et al., 2001), cpr22 is humidity-sensitive. High RH (>95%) suppresses the phenotype conferred by cpr22/CPR22 almost completely, and relatively high RH (70%) causes a milder phenotype, including less stunting and lesion formation. To facilitate the observation of pathogen-induced HR in cpr22 plants, all plants were grown under 70% RH. Therefore, spontaneous lesion formation in cpr22 at 0 d after inoculation was milder than that under normal RH conditions (RH of 55%; compare Figure 6, 0 d after inoculation, with Figure 7B). Cell death was first detected ∼24 h after inoculation and became more pronounced by 2 d after inoculation in both wild-type and cpr22 plants (Figure 6; data not shown). Thus, cpr22, unlike dnd1 and hlm1/dnd2, is capable of developing a normal HR in response to infection with an avirulent pathogen.
Figure 6.
The cpr22 Mutant Develops a Normal HR after Infection with an Avirulent Pathogen.
Microscopic analysis of trypan blue–stained leaves from ∼20-d-old wild-type Ws and cpr22/CPR22 plants. After inoculation with P. syringae pv tomato carrying avrRpt2, leaves were photographed at 0, 2, or 5 d after inoculation (DAI).
Figure 7.
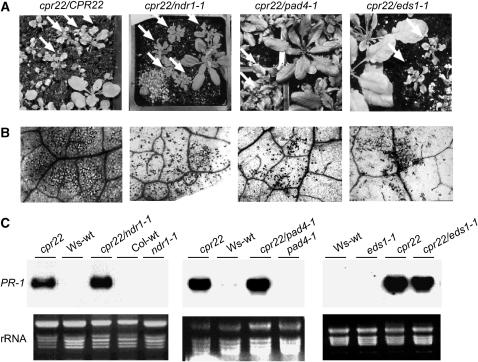
PAD4, EDS1, and NDR1 Are Not Required for cpr22-Mediated Stunting, Spontaneous Lesion Formation, or PR-1 Gene Expression.
(A) Comparison of the morphology displayed by segregating F3 progeny from a self-pollinated cpr22/CPR22 mutant plant and those of self-pollinated cpr22/CPR22 plants homozygous for ndr1-1, pad4-1, or eds1-1 (cpr22 ndr1-1, cpr22 pad4-1, and cpr22 eds1-1, respectively). Arrows indicate plants containing the cpr22/CPR22 genotype (based on molecular and progeny analyses); regardless of the allele at the NDR1, PAD4, or EDS1 locus, these plants displayed stunted growth and curly leaves. Plants were photographed at 4 weeks after planting.
(B) Microscopic analysis of trypan blue–stained leaves from 3-week-old F3 progeny heterozygous for cpr22 as well as from cpr22/CPR22 plants homozygous for ndr1-1, pad4-1, or eds1-1. Darkly stained areas indicative of cell death were detected in all sets of progeny.
(C) Expression of the PR-1 gene in CPR22/CPR22 (Ws-wt and Col-wt) plants, heterozygous cpr22/CPR22 (cpr22) and homozygous ndr1-1, pad4-1, or eds1-1 single mutant plants, and cpr22 heterozygous and ndr1-1, pad4-1, or eds1-1 homozygous double mutants. RNA gel blot analysis was performed using 8 μg of total RNA harvested from 3-week-old soil-grown plants. Ethidium bromide staining of rRNA was used as a loading control.
Many cpr22-Associated Phenotypes Are Independent of EDS1, PAD4, and NDR1
To determine whether cpr22 constitutively activates PR-1 expression, spontaneous lesion formation, SA accumulation, and enhanced resistance to H. parasitica Emco5 via defense signaling pathways used during R gene–mediated resistance, we previously crossed cpr22 plants with the ndr1-1 mutant (Yoshioka et al., 2001). Analysis of the resultant progeny indicated that NDR1 is required for cpr22-mediated resistance to H. parasitica. To expand this analysis and also to determine whether any of the other cpr22-associated phenotypes are dependent on PAD4 or EDS1, we crossed a cpr22/CPR22 plant with homozygous pad4-1 and ndr1-1 plants. In addition, a cpr22 homozygous plant grown under high RH was crossed with a homozygous eds1-1 mutant. In the F1 generation, ∼50% of the progeny from the pad4-1 and ndr1-1 crosses exhibited stunted growth, curly leaves, and constitutive PR-1 expression, consistent with the semidominant nature of the cpr22 mutation (Table 2). By contrast, the remaining ∼50% of the plants were morphologically wild type. All F1 progeny from the cross between cpr22 homozygous plants and the eds1-1 mutant were stunted and constitutively expressed PR-1, as expected.
Table 2.
Morphological Phenotype and PR-1 Gene Expression in Progeny of Crosses between cpr22 and ndr1-1, pad4-1, or eds1-1 Plants
| Total No. of Plants
|
Morphology/PR-1 Expression
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Crossa | Generation | Nb | PR-1+/cpr22c | PR-1−/wtd | Hypothesise | χ2f | P | |
| cpr22/CPR22 × cpr22/CPR22 | F1 | 20 | 5 | 9 | 6 | 1:2:1 | 0.30 | 0.95 > P > 0.8 |
| cpr22/CPR22 × ndr1-1/ndr1-1 | F1 | 7 | 0 | 3 | 4 | 0:1:1 | – | |
| F2 | 43 | 10 | 22 | 11 | 1:2:1 | 0.43 | 0.95 > P > 0.8 | |
| cpr22/CPR22 × pad4-1/pad4-1 | F1 | 24 | 0 | 10 | 14 | 0:1:1 | – | |
| F2 | 76 | 15 | 41 | 20 | 1:2:1 | 1.32 | 0.8 > P > 0.5 | |
| cpr22/cpr22 × eds1-1/eds1-1 | F1 | 5 | 0 | 5 | 0 | 0:1:0 | – | |
| F2 | 47 | 10 | 26 | 11 | 1:2:1 | 0.59 | 0.95 > P > 0.8 | |
The pollen donor is indicated first and the accepting plant second.
Nonviable plants.
All plants showed typical cpr22 morphology and constitutive expression of the PR-1 gene.
All plants showed morphology similar to wild-type plants and did not exhibit constitutive PR-1 expression.
Hypothesis: cpr22 is inherited as a semidominant mutation that is lethal in the homozygous condition. The NDR1, PAD4, and EDS1 genes are not required for cpr22-induced morphology or PR-1 gene expression.
Two degrees of freedom.
After allowing the stunted F1 progeny from each cross to self-pollinate, the F2 progeny were analyzed. For each cross, ∼25% of the F2 progeny died at ambient (low) RH, ∼50% exhibited the phenotype conferred by cpr22, as indicated by stunted growth, curly leaves, and constitutive PR-1 expression, and ∼25% were morphologically wild type (Table 2). This 1:2:1 ratio suggests that development of these cpr22-associated phenotypes is independent of NDR1, PAD4, and EDS1. These results were confirmed using F3 progeny analysis and cleaved-amplified polymorphic sequence (CAPS) markers to identify F2 plants heterozygous for cpr22 and homozygous for ndr1-1, pad4-1, or eds1-1. As expected, the cpr22 ndr1-1, cpr22 eds1-1, and cpr22 pad4-1 double mutants were all stunted, although the phenotype of cpr22 pad4-1 plants was somewhat less severe (Figure 7A). Trypan blue staining revealed that these double mutants developed similar numbers of spontaneous HR-like lesions as cpr22 single mutants (Figure 7B). They also constitutively expressed PR-1 to comparable levels as cpr22 single mutant plants (Figure 7C). Together, these results demonstrate that PAD4, EDS1, and NDR1 are not required for cpr22-associated lethality, stunting, spontaneous lesion formation, or constitutive PR-1 expression.
Enhanced Disease Resistance Conferred by cpr22 Is Dependent on EDS1, PAD4, and NDR1
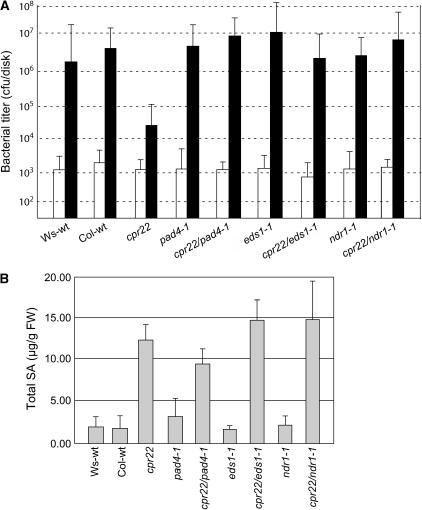
Whether cpr22-induced disease resistance requires EDS1, PAD4, or NDR1 was then assessed by monitoring symptom development after inoculation with an oomycete or bacterial pathogen. Wild-type Ws and Col-0 plants were susceptible to H. parasitica biotype Emco5, with ∼95% developing sporangiophores by 7 d after inoculation, whereas cpr22/CPR22 plants were completely resistant (Table 3). Mutations in PAD4, EDS1, or NDR1 suppressed cpr22-mediated enhanced resistance. cpr22 heterozygous pad4-1 or eds1-1 homozygous plants exhibited similar susceptibility as wild-type plants or the pad4-1 or eds1-1 single mutant plants, respectively. cpr22 heterozygous ndr1-1 homozygous plants, like ndr1-1 single mutants, displayed somewhat heightened susceptibility to Emco5, as reported previously (Yoshioka et al., 2001). cpr22-mediated resistance to the virulent bacterial pathogen P. syringae pv maculicola ES4326 also was suppressed by mutations in EDS1, PAD4, or NDR1. Although the cpr22 single mutant displayed ∼100-fold less bacterial growth than wild-type Ws or Col-0 plants, this resistance was lost in all three double mutants (Figure 8A). Thus, cpr22-induced resistance to virulent oomycete and bacterial pathogens appears to be mediated by one or more signaling pathway(s) that use both the CC-NBS-LRR signal transducer NDR1 and the TIR-NBS-LRR transducers EDS1 and PAD4.
Table 3.
Resistance Status of cpr22 Single and Double Mutants to H. parasitica Isolate Emco5
| Total No. of Plants
|
No. of Plantsa
|
|||
|---|---|---|---|---|
| Genotype | R | S | Plants (%)b | |
| CPR22/CPR22 (Ws) | 70 | 5 | 65 | 92.9 |
| CPR22/CPR22 (Col-0) | 32 | 2 | 30 | 93.7 |
| cpr22/CPR22 | 76 | 76 | 0 | 0.0 |
| CPR22/CPR22/pad4-1/pad4-1 | 63 | 8 | 55 | 87.3 |
| cpr22/CPR22/pad4-1/pad4-1 | 93 | 8 | 85 | 82.7 |
| CPR22/CPR22/eds1-1/eds1-1 | 42 | 0 | 42 | 100.0 |
| cpr22/CPR22/eds1-1/eds1-1 | 35 | 5 | 30 | 85.7 |
| CPR22/CPR22/ndr1-1/ndr1-1 | 20 | 0 | 20 | 100.0 |
| cpr22/CPR22/ndr1-1/ndr1-1 | 28 | 0 | 28 | 100.0 |
Based on formation of sporangiophores. R, resistance; S, susceptible.
Percentage of plants that show formation of sporangiophores.
Figure 8.
EDS1, PAD4, and NDR1 Are Required for cpr22-Mediated Resistance to P. syringae pv maculicola ES4326 but Not for SA Accumulation.
(A) After infiltration with P. syringae pv maculicola ES4326, bacterial growth was monitored in the leaves of 3-week-old wild-type Ws and Col-0 plants, cpr22/CPR22, eds1-1, pad4-1, and ndr1-1 single mutants, and cpr22 heterozygous, ndr1-1, pad4-1, or eds1-1 homozygous double mutant plants by collecting three leaf discs at 0 d after inoculation (open bars) and 3 d after inoculation (closed bars). Colony-forming units (cfu) are expressed ±sd and represent averages of four independent samples.
(B) Total SA levels were assayed in leaves of 3-week-old, soil-grown wild-type Ws and Col-0 plants, cpr22/CPR22 (cpr22), eds1-1, pad4-1, and ndr1-1 single mutants, and cpr22 heterozygous, ndr1-1, pad4-1, or eds1-1 homozygous double mutant plants. The values are presented in micrograms per gram fresh weight (FW) and represent averages ± sd of three samples consisting of leaves from three plants per line.
Mutations in EDS1, PAD4, and NDR1 Do Not Suppress cpr22-Induced SA Accumulation
SA is a critical signal for R gene–mediated resistance, and mutations in EDS1, PAD4, and NDR1 have been shown to suppress its accumulation in certain Arabidopsis–pathogen interactions (Zhou et al., 1998; Feys et al., 2001; Shapiro and Zhang, 2001). Because cpr22-mediated resistance to H. parasitica is SA-dependent (Yoshioka et al., 2001), we tested whether the loss of resistance in cpr22 eds1-1, cpr22 pad4-1, or cpr22 ndr1-1 double mutants was attributable to reduced SA production. Similar to the cpr22/CPR22 mutant, all three double mutants contained threefold to sevenfold higher levels of SA than wild-type plants or the eds1-1, pad4-1, or ndr1-1 single mutants (Figure 8B). Thus, cpr22-induced SA accumulation is largely independent of EDS1, PAD4, and NDR1.
DISCUSSION
Here, we report the cloning of cpr22, an ∼3-kb deletion mutant that generates a chimeric gene, ATCNGC11/12, by linking the putative ATCNGC-encoding genes ATCNGC11 and ATCNGC12. Complementation studies and analysis of KO lines revealed that expression of ATCNGC11/12 is required for the phenotype conferred by cpr22. However, because expression of a ATCNGC12 transgene suppressed the phenotype conferred by cpr22 in a dosage-dependent manner, the ratio between ATCNGC11/12 and ATCNGC12 also appears to be critical for phenotype development. Structural and functional analyses indicated that ATCNGC11/12, ATCNGC11, and ATCNGC12 are all functional cation channels. Based on these combined results, several mechanisms through which ATCNGC11/12 induces the phenotype conferred by cpr22 can be envisioned. The first possibility is that ATCNGC11/12 activates the phenotype conferred by cpr22 by destroying the function of CNCG12-containing channels. However, because ATCNGC12 single KO plants are almost indistinguishable from wild-type plants, this possibility seems unlikely. Instead, we suggest that ATCNGC11/12 competes with ATCNGC12 to form tetramer channels. In this scenario, there are two possibilities: the channels containing ATCNGC11/12 either gain an altered function or function as a constitutively active form of the original channels. Consistent with the latter possibility, ATCNGC12 single KO plants exhibit reduced levels of resistance against an avirulent isolate of H. parasitica, compared with wild-type plants. Because this finding suggests that wild-type ATCNGC12 is a positive regulator of R gene–mediated resistance, expression of a constitutively active form of ATCNGC12 (in the form of ATCNGC11/12) would likely confer enhanced pathogen resistance.
The demonstration that the resistance to H. parasitica Emwa1 was comparably suppressed in ATCNGC11 and ATCNGC12 single KO plants as well as in their F1 progeny suggests that ATCNGC11, like ATCNGC12, is a positive regulator of resistance and that these proteins may work cooperatively. Although others (Zhong et al., 2003) have suggested that plant ATCNGCs form heterotetramers and may cooperate based on results in nonplant systems, our results indicate that such cooperation exists in plants. Although ATCNGC11 and ATCNGC12 are similar and may cooperate, their functions do not appear to be identical. Expression of a ATCNGC12 transgene driven by its endogenous promoter suppressed cpr22-related phenotypes, whereas expression of a ATCNGC11 transgene did not. Unfortunately, because the expression levels of the ATCNGC11 and ATCNGC12 transgenes cannot be determined because of the presence of their endogenous copies, it remains unclear whether this result reflects a functional difference between these ATCNGCs or is attributable simply to different expression levels. Stronger evidence that ATCNGC11 and ATCNGC12 have at least some functional differences comes from the demonstration that ATCNGC11/12-induced lesion formation in N. benthamiana was suppressed by ATCNGC12 but not by ATCNGC11.
In addition to this study, there is growing evidence that ATCNGCs play a role in regulating defense response activation. In particular, mutations in ATCNGC2 (DND1) and ATCNGC4 (HLM1, DND2) confer constitutive PR gene expression, increased SA levels, and enhanced resistance to pathogens (Yu et al., 1998; Clough et al., 2000; Balague et al., 2003; Jurkowski et al., 2004). Despite these similarities with the phenotype conferred by cpr22, however, ATCNGC11/12, ATCNGC2, and ATCNGC4 do not appear to have identical roles in the resistance pathway. The dnd1 mutant displays gene-for-gene disease resistance to avirulent pathogens in the absence of cell death (Yu et al., 2000), and the HR in hlm1/dnd2 mutants is strongly reduced and/or delayed (Balague et al., 2003; Jurkowski et al., 2004). By contrast, HR cell death in cpr22 was comparable to that displayed by wild-type plants after inoculation with P. syringae pv tomato carrying avrRpt2. In addition, our preliminary studies suggest that the expression patterns of ATCNGC11/12, ATCNGC2, and ATCNGC4 and the patterns of spontaneous cell death exhibited by their respective mutants differ (K. Yoshioka and D.F. Klessig, unpublished data).
The ATCNGC gene family of Arabidopsis consists of 20 members whose overall genomic sequence similarity ranges from 55 to 83% (Maser et al., 2001). Based on sequence similarity, these genes have been divided into four groups, with group IV the most distantly related to the other ATCNGCs. ATCNGC2 and ATCNGC4, which are closely related to each other, are the sole members of subgroup IVB. By contrast, both ATCNGC11 and ATCNGC12 are only distantly related to these genes and are two of six genes belonging to group I. Interestingly, the calmodulin binding domain sequence in ATCNGC12 is significantly divergent from those of other ATCNGCs. This observation raises the possibility that ATCNGC12 activity is regulated via a specific calmodulin that does not bind to other ATCNGCs.
In addition to cpr22, dnd1, and hlm1, several other Arabidopsis mutants that develop spontaneous lesions, constitutively express defense responses, and display enhanced disease resistance have been identified (Dempsey et al., 1999; Silva et al., 1999; Clough et al., 2000; Jambunathan et al., 2001; Yoshioka et al., 2001; Balague et al., 2003). Whether these resistance responses are activated via a signaling pathway(s) also used by some R genes or simply by perturbing critical cellular processes is unclear at present. However, several lines of evidence suggest that cpr22 exerts at least some of its effects by stimulating pathways used by certain R genes. For example, the ssi4 mutant, which contains a defect in a TIR-NBS-LRR–type gene, and transgenic plants overexpressing the resistance genes RPW8.1 and RPW8.2 also display humidity-sensitive, constitutive activation of defense responses and disease resistance (Shirano et al., 2002; Xiao et al., 2003; Zhou et al., 2004). Additionally, the demonstration that KO mutants lacking ATCNGC11 or ATCNGC12 display suppressed R gene–mediated resistance strongly suggests that cpr22-mediated resistance is activated by defense signaling pathways rather than by metabolic perturbation. Further supporting this hypothesis, genetic analysis revealed that the resistance signal transducers EDS1, PAD4, and NDR1 are all required for cpr22-mediated enhanced disease resistance, although they are not required for cpr22-induced PR gene expression, SA accumulation, or spontaneous lesion formation. It is interesting that dnd1- and dnd2-induced resistance to virulent bacteria requires PAD4, whereas PR gene expression, SA accumulation, and disease resistance do not (Jirage et al., 2001). Because PAD4 interacts with EDS1 (Feys et al., 2001), it is likely that dnd1- and dnd2-induced resistance, like that of cpr22, requires EDS1; whether NDR1 also is involved will require further epistatic analysis.
The finding that cpr22-mediated resistance requires not only NDR1 but also EDS1 and PAD4 was unexpected. EDS1/PAD4 and NDR1 are generally thought to function in mutually exclusive pathways regulated by TIR-NBS-LRR or CC-NBS-LRR R genes, respectively (Aarts et al., 1998; Feys et al., 2001).
Other exceptions to this rule are RPP4 in the Col-0 ecotype (an ortholog of RPP5 in the Landsberg erecta ecotype) (van der Biezen et al., 2002) and RPP5. In cotyledons, these R genes mediate partial resistance to H. parasitica via a pathway that is fully dependent on EDS1 and partially dependent on NDR1 (Aarts et al., 1998; McDowell et al., 2000; van der Biezen et al., 2002). The discovery that cpr22 induces strong resistance to virulent bacterial and oomycete pathogens in an EDS1-, PAD4-, and NDR1-dependent manner supports the idea that these distinct signaling pathways undergo cross-regulation. Moreover, because RPP4 confers resistance to H. parasitica Emwa1 in the Col-0 ecotype and this resistance is compromised in ATCNGC11 and ATCNGC12 KO lines, it is tempting to speculate that these two ATCNGCs play a role in RPP4/5-mediated resistance. Whether their role in defense signaling is specific for RPP4/5-mediated resistance or is also part of other R gene–triggered defense signaling cascades remains to be determined.
Compared with enhanced disease resistance, cpr22-induced PR-1 expression and SA accumulation were not affected by mutations in EDS1, PAD4, or NDR1. Because these three signal transducers are believed to mediate signaling through SA (Zhou et al., 1998), this result was surprising. Together, these results suggest that one or more SA-independent branches, in addition to the SA-dependent branch(es), functions downstream of these three signal transducers and is required for disease resistance, but not for some defense responses, such as PR-1 expression.
In summary, our results suggest that ATCNGC11/12 induces pathogen resistance through multiple defense signaling pathways, whereas ATCNGC11 and ATCNGC12 function as positive components in one or more of these pathways. Although the mechanism by which ATCNGC11/12 activates resistance is currently unclear, ion fluxes are important early events in the defense signaling process. Further characterization of ATCNGC11/12, ATCNGC11, and ATCNGC12 should provide important insights into the role(s) that ATCNGCs plays in transducing different signals after pathogen infection.
METHODS
Plant Growth Conditions
Arabidopsis thaliana plants were grown on Pro-Mix soil (Premier Horticulture) in a growth chamber under ambient humidity as described by Silva et al. (1999). For the high RH condition, 95% RH was applied.
Map-Based Cloning of cpr22
The CPR22 locus was mapped using the F2 progeny from crosses between cpr22/CPR22 (Ws ecotype) and either wild-type Col-0 or wild-type Landsberg erecta plants. Novel CAPS (Konieczny and Ausubel, 1993) and simple sequence length polymorphism (Bell and Ecker, 1994) markers were generated based on information from the CEREON polymorphism collection. Of 1355 F2 plants examined using these markers, one recombination event was identified using marker C424561 and four recombination events were identified using C424985. No recombination events were identified using additional markers within this 60-kb region. Thus, CPR22 is located within a region spanning the end of BAC clone F11C10 and the beginning of F13A10. Specific PCR primers to amplify the putative ORFs in this 60-kb region were designed using the F11C10 and F13A10 sequences. The PCR products, generated using genomic DNA from wild-type WS or cpr22 homozygous plants as the template, were sequenced directly using an automatic sequencer (ABI Prism model 3100).
PCR Analysis
To determine whether ATCNGC11 is translated from an alternative start codon, PCR analysis was performed using genomic DNA from wild-type Ws plants and primers corresponding to several upstream ATG codons. The primers used were ATG1F (5′-ATGGAAAATTGAAAAGTGTTAGAGGA-3′), ATG2F (5′-ATGAACAAGTTTAACATAGCTTC-3′), and ATG3F (5′-ATGAATCTTCAGAGGAAAT-3′) as forward primers and UD1R1 (5′-GGTCTTCCTCCAGTAAACCT-3′) as the reverse primer. RT-PCR also was performed with these primers using cDNA generated by SuperScript reverse transcriptase (Invitrogen Life Technologies) according to the manufacturer's instructions. Both PCR and RT-PCR products were sequenced. To isolate a cDNA of ATCNGC11/12, RT-PCR was performed using total RNA from cpr22 homozygous plants and the primers Bam-ATG3F (5′-GGGATCCCATGAATCTTCAGAGGAGAAA-3′) and ORF14-R1-24 (5′-CTATGCTTCAGCCTTTGCAAACTC-3′). This cDNA was cloned into plasmid pGEM-T Easy (Promega) and sequenced by the same method described above.
Homozygous plants of KO lines were created by self-pollination and confirmed by PCR according to suggestions from the Salk Institute (http://signal.salk.edu/tdnaprimers.html) using primers LBb1 (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′), 22RP (5′-CAGGCAAGAGAATGAAACCTTCAA-3′), 57RP (5′-TCACCCGGTAGGACATCCATTT-3′), 568RP2 (5′-TCAGTATCGTCGCCTCCATAG-3′), 22LP (5′-ATTGCTTTTGGTGGGGTCTCC-3′), 57LP (5′-CGTCGCTGGTGTCAGAGAGAA-3′), and 568LP (5′-GGTTGAGGTGGTGAGTATTCG-3′).
Expression of ATCNGC11, ATCNGC12, ATCNGC11/12, PR-1, and β-tubulin was monitored by RT-PCR using the following gene-specific primers: for ATCNGC11, cDNA-13-F1 (5′-AGGTTTACTGGAGGAAGACC-3′) and cDNA-13-R1 (5′-CTAGCGATCACATTACATAGAG-3′); for ATCNGC12, cDNA-14-F1 (5′-CGGGAAGATGAAAACACTGG-3′) and cDNA-14-R1 (5′-CACTATGCTTCAGCCTTTGC-3′); for ATCNGC11/12, cDNA-13-F1 and cDNA-14-R1; for PR-1, PR1-F (5′-AGCCTATCGTCACACTCCCGCTCAA-3′) and PR1-R (5′-GCTACAAATCACCCAAATATACTCA-3′); and for β-tubulin, β-tubulin-5′ (5′-CGTGGATCACAGCAATACAGAGCC-3′) and β-tubulin-3′ (5′-CCTCCTGCACTTCCACTTCGTCTTC-3′). cDNAs were generated by the same method described above.
Plant Transformation
A genomic ATCNGC12 clone, containing 0.8 kb of 5′ promoter sequence and 0.5 kb of the 3′ untranslated region, and a genomic ATCNGC11 clone, containing 1.0 kb of 5′ promoter sequence and 0.5 kb of untranslated region, were generated by PCR using primers NEWCLONE-F1BAM (5′-CTAGGATCCTAGCGAGAATCTCACCACCACAACCAC-3′) and NEWCLONE-R1ECO (5′-GCGAATTCGGTCTCTGAGCAGCAAAGGATCATAGCG-3′) for ATCNGC12 and primers CLONE13G-F1 (5′-GTTTCTTCAATTCTGTTGAC-3′) and D1R1 (5′-CCAGTGTTTTCATCTTC-CCG-3′) for ATCNGC11. PCR was performed using Pfu DNA polymerase (Stratagene). The PCR product was cloned into pGEM-T Easy (Promega) and sequenced. This genomic ATCNGC12 clone was then subcloned into pBIN19 (Clontech) and transformed into both cpr22 homozygous and heterozygous plants using the vacuum infiltration method (Bechtold and Pelletier, 1998). Transformants were selected on Murashige and Skoog (1962) medium containing 50 μg/mL kanamycin and then transferred to soil. The presence of the introduced ATCNGC12 or ATCNGC11 clone and T-DNA region in the T1 and T2 progeny of cpr22 homozygous transgenic plants was confirmed by PCR analysis. For the T1 and T2 progeny of cpr22 heterozygous transformants, the presence of the introduced ATCNGC12 or ATCNGC11 clone and the endogenous ATCNGC11/12 chimeric gene was determined by performing PCR analysis for full-length ATCNGC12 or ATCNGC11, the T-DNA, full-length ATCNGC11/12, and the genomic region between ATCNGC11 and ATCNGC12. To detect the T-DNA, the following specific primers were used: LBc-F (5′-ACGGGCAACAGCYGATT-3′) and LBc-R (5′-AGTTGCGCAGCCTGAAT-3′).
For stable transformation and transient expression, the cDNAs ATCNGC11, ATCNGC12, and ATCNGC11/12 were subcloned into pMBP3 (Cambia). Stable transformation was performed as described above. Agrobacterium tumefaciens–mediated transient expression in Nicotiana benthamiana was performed as described by Sessa et al. (2000). A. tumefaciens strain GV2260 (final density of 0.2 OD600) was used to syringe-infiltrate N. benthamiana leaves, and cell death was monitored at 3 d after infiltration.
Trypan Blue Staining
Leaf samples were taken from 2- to 3-week-old plants grown on soil. Trypan blue staining was performed as described previously (Bowling et al., 1994).
RNA Extraction and Gel Analysis
Small-scale RNA extraction was performed using the Trizol reagent (Gibco BRL) according to the manufacturer's instructions. RNA gel blot analysis and synthesis of random primed probes were performed as described previously (Shah et al., 1999). RNA gel blot hybridization was performed as described previously (Kachroo et al., 1995).
Functional Characterization of ATCNGCs in Yeast
ATCNGC11 cDNA in the pGEM plasmid (see above) was amplified by PCR using primers BamAtc11 (5′-CGGGATCCCGATGAATCTTCAGAGGAG-3′) and EcoAtc11 (5′-GGAATTCCCTAGGTTTCATCCATAGG-3′) and then subcloned into the pYES (Invitrogen) plasmid BamHI and EcoRI sites. ATCNGC12 and ATCNGC11/12 cDNAs in the pGEM plasmid (see above) were restriction-digested with BamHI and NotI and subcloned into the pYES plasmid. The K+ uptake–deficient yeast (trk1,2) strain WD3 (Mat a; leu2-3, 2-112; trp1-1; ura3-3; ade2-1; his3-11; can1-100; trk1-LEU2; trk2-HIS3) was transformed with the empty plasmids pYES, pYES-ATCNGC11, pYES-ATCNGC12, and pYES-ATCNGC11/12 using the Qbiogene/Bio 101 EZ-Yeast transformation kit according to the manufacturer's protocol. Positive transformant colonies were selected on solid YNB-uracil (ura) medium (Qbiogene) supplemented with complete synthetic medium without ura (Qbiogene), (NH4)2SO4 as a nitrogen source, glucose as a carbon source, and 100 mM KCl. Transformed yeast strains were maintained (−80°C) as 25% (w/v) glycerol stocks, and freshly streaked cultures were generated from the glycerol stocks on solid YNB-ura. For each experiment, liquid YNB-ura cultures were inoculated from solid YNB-ura medium plates. Colonies on solid plates were never >4 weeks old, to prevent suppressor and revertant mutations. The trk1,2 complementation assay followed the method described by Mercier et al. (2004). YNB-ura liquid cultures of transformants were grown with shaking at 30°C for 24 h. Aliquots were pelleted by microcentrifuge and rinsed with sterile water. The concentrations of aliquots (100 μl) of the yeast cultures were adjusted, and the aliquots were then applied to assay plates containing YPGal (1% yeast extract, 2% peptone, and 2% galactose as a carbon source) with 0.07 mg/mL hygromycin B using glass beads to facilitate equal distribution across the surface. Sterile 3 M KCl (300 μl) was added to 1-cm-diameter filter disks (740-E; Schleicher & Schuell) under sterile conditions, and the filter disks were added to the assay plates. Assay plates were incubated in the dark at room temperature for 3 to 4 d. Complementation of K+ uptake–deficient WD3 yeast strain growth was evaluated as colony formation around the 3 M KCl filter disks on each assay plate. When added to the YPGal assay medium, cAMP and/or cGMP were provided as dibutyryl (lipophilic) analogs at 100 μM final concentration.
Double Mutant Isolation
Double mutant lines were generated using the pollen from a cpr22/CPR22 plant to fertilize a pad4-1 homozygous (Col-0 ecotype) plant and the pollen from a cpr22/cpr22 plant grown in high RH to fertilize an eds1-1 (Ws ecotype) homozygous plant. Construction of cpr22/CPR22; ndr1-1/ndr1-1 plants was as described by Yoshioka et al. (2001). Simple sequence length polymorphism and CAPS marker analyses were done in the F1 generation to score for a successful cross between the cpr22 mutant and pad4-1 or eds1-1. The genotypes of F1, F2, and F3 progeny at the PAD4 locus were determined by BsmF1 digestion of a 1620-bp PCR fragment using the CAPS primers Pad4F (5′-ATGGACGATTGTCGATTCG-3′) and Pad4R (5′-CTCCATTGCGTCACTCTCAT-3′). The genotype at the EDS1 locus was determined by digesting a 710-bp fragment amplified using the primers EDS3f (5′-GGATAGAAGATGAATACAAGCC-3′) and EDS1r (5′-ACCTAAGGTTCAGGTATCTGT-3′) with MseI.
Pathogen Infection
Infection with Hyaloperonospora parasitica isolates Emco5, Emwa1, or Noco2 was performed by applying a single drop of asexual inoculum suspension (106 conidiosporangia/mL) per cotyledon of 7-d-old seedlings. The seedlings were grown at 16°C and >90% RH with an 8-h photoperiod. Plants whose leaves displayed sporangiophores were scored as susceptible at 7 d after inoculation. Infection with Pseudomonas syringae pv maculicola ES4326 was performed on 4-week-old soil-grown plants as described previously (Shah et al., 1997). Leaves were infiltrated with a bacterial suspension (OD600 = 0.002) in 10 mM MgCl2, and colony-forming units were counted at 3 d after inoculation.
Extraction and Analysis of SA
Total SA was extracted from ∼0.5 g of leaf tissue, and levels were determined as described by Bowling et al. (1994).
Accession Numbers
Sequence data for ATCNGC11 and ATCNGC12 can be found in the GenBank/EMBL data libraries under the following accession numbers: H84902 (At2g46440) and A84902 (At2g46450), respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of T-DNA Insertion Lines and Their F1 Progeny.
Supplemental Figure 2. Morphology of Wild-Type Ws Plants Transformed with ATCNGC11 and ATCNGC12 Driven by the CaMV 35S Promoter.
Supplementary Material
Acknowledgments
We thank J.E. Parker for eds1-1 seeds, P. Repetti and B.J. Staskawicz for ndr1-1 seeds and marker information, CEREON for information on the polymorphism, and the Salk Institute for T-DNA insertion lines. We gratefully acknowledge D'Maris Dempsey for critical reading of the manuscript and fruitful discussions. We also acknowledge Yumiko Shirano, Hui-ju Wu, and Fasong Zhou for helpful advice. This work was supported by Grant MCB-0110404 from the National Science Foundation to D.F.K. and by a Discovery Grant from the Science and Engineering Research Council of Canada to K.Y.
Current address: Department of Plant Pathology, University of Kentucky, Lexington, KY 40546.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Keiko Yoshioka (yoshioka@botany.utoronto.ca) and Daniel F. Klessig (dfk8@cornell.edu).
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.038786.
References
- Aarts, N., Metz, M., Holub, E., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arazi, T., Sunkar, R., Kaplan, B., and Fromm, H. (1999). A tobacco plasma membrane calmodulin-binding transporter confers Ni2+ tolerance and Pb2+ hypersensitivity in transgenic plants. Plant J. 20 171–182. [DOI] [PubMed] [Google Scholar]
- Atkinson, M.M., Keppler, L.D., Orlandi, E.W., Baker, C.J., and Mischke, C.F. (1990). Involvement of plasma membrane calcium influx in bacterial induction of the K+/H+ and hypersensitive responses in tobacco. Plant Physiol. 92 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson, M.M., Midland, S.L., Sims, J.J., and Keen, N.T. (1996). Syringolide 1 triggers Ca2+ influx, K+ efflux, and extracellular alkalization in soybean cells carrying the disease-resistance gene Rpg4. Plant Physiol. 112 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balague, C., Lin, B., Alcon, C., Flottes, G., Malmstrom, S., Kohler, C., Neuhaus, G., Pelletier, G., Gaymard, F., and Roby, D. (2003). HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82 259–266. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Bihler, H., Slayman, C.L., and Bertl, A. (2002). Low-affinity potassium uptake by Saccharomyces cerevisiae is mediated by NSC1, a calcium-blocked non-specific cation channel. Biochim. Biophys. Acta 1558 109–118. [DOI] [PubMed] [Google Scholar]
- Bowling, S.A., Guo, A., Cao, H., Gordon, A.S., Klessig, D.F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S., Holub, E.B., and Staskawicz, B.J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 92 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra, S., Stennis, M., and Low, P.S. (1997). Measurement of Ca2+ fluxes during elicitation of the oxidative burst in aequorin-transformed tobacco cells. J. Biol. Chem. 272 28274–28280. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., Fengler, K.A., Yu, I.C., Lippok, B., Smith, R.K., Jr., and Bent, A.F. (2000). The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 97 9323–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creutz, C.E., Tomsig, J.L., Snyder, S.L., Gautier, M.C., Skouri, F., Beisson, J., and Cohen, J.J. (1998). The copines, a novel class of C2domain-containing, calcium-dependent, phospholipid-binding proteins conserved from Paramecium to humans. J. Biol. Chem. 273 1393–1402. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 2 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey, D., Shah, J., and Klessig, D.F. (1999). Salicylic acid and disease resistance in plants. Crit. Rev. Plant Sci. 18 547–575. [Google Scholar]
- Doke, N., Miura, Y., Sanchez, L.M., Park, H.J., Noritake, T., Yoshioka, H., and Kawakita, K. (1996). The oxidative burst protects plants against pathogen attack: Mechanism and role as an emergency signal for plant bio-defence. Gene 179 45–51. [DOI] [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.-A., and Parker, J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H. (1971). Current status of gene-for-gene concept. Annu. Rev. Phytopathol. 9 275–296. [Google Scholar]
- Gaber, R.F., Styles, C.A., and Fink, G.R. (1988). TRK1 encodes a plasma membrane protein required for high-affinity potassium transport in Saccharomyces cerevisiae. Mol. Cell. Biol. 8 2848–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (1999). Genes controlling expression of defense responses in Arabidopsis. Curr. Opin. Plant Biol. 2 280–286. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr. Opin. Plant Biol. 4 301–308. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, J., Grisafi, P., Cheng, S.-H., and Flink, G.R. (2001). Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev. 15 2263–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Jabs, T., Tschöpe, M., Colling, C., Hahlbrock, K., and Scheel, D. (1997). Elicitor-stimulated ion fluxes and
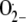 from the oxidative burst are essential components in triggering defense gene activation and phytoalexin synthesis in parsley. Proc. Natl. Acad. Sci. USA 94 4800–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
from the oxidative burst are essential components in triggering defense gene activation and phytoalexin synthesis in parsley. Proc. Natl. Acad. Sci. USA 94 4800–4805. [DOI] [PMC free article] [PubMed] [Google Scholar] - Jambunathan, N., Siani, J.M., and McNellis, T.W. (2001). A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13 2225–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D., Zhou, N., Cooper, B., Clarke, J.D., Dong, X., and Glazebrook, J. (2001). Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J. 26 395–407. [DOI] [PubMed] [Google Scholar]
- Jurkowski, G.I., Smith, R.K., Jr., Yu, I.C., Ham, J.H., Sharma, S.B., Klessig, D.F., Fengler, K.A., and Bent, A.F. (2004). Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol. Plant Microbe Interact. 17 511–520. [DOI] [PubMed] [Google Scholar]
- Kachroo, P., Leong, S.A., and Chattoo, B.B. (1995). Mg-SINE: A short interspersed nuclear element from the rice blast fungus, Magnaporthe grisea. Proc. Natl. Acad. Sci. USA 92 11125–11129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp, U.B., and Seifert, R. (2002). Cyclic nucleotide-gated ion channels. Physiol. Rev. 82 769–824. [DOI] [PubMed] [Google Scholar]
- Keen, N.T. (1990). Gene-for-gene complementarity in plant-pathogen interactions. Annu. Rev. Genet. 24 447–463. [DOI] [PubMed] [Google Scholar]
- Ko, C.H., and Gaber, R.F. (1991). TRK1 and TRK2 encode structurally related K+ transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 11 4266–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, C., Merkle, T., and Neuhaus, G. (1999). Characterization of a novel gene family of putative cyclic nucleotide- and calmodulin-regulated ion channels in Arabidopsis thaliana. Plant J. 18 97–104. [DOI] [PubMed] [Google Scholar]
- Kohler, C., and Neuhaus, G. (2000). Characterization of calmodulin binding to cyclic nucleotide-gated ion channels from Arabidopsis thaliana. FEBS Lett. 14 133–136. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Lemtiri-Chlieh, F., and Berkowitz, G.A. (2004). Cyclic adenosine monophosphate regulates calcium channels in the plasma membrane of Arabidopsis leaf guard and mesophyll cells. J. Biol. Chem. 279 35306–35312. [DOI] [PubMed] [Google Scholar]
- Leng, Q., Mercier, R.W., Yao, W., and Berkowitz, G.A. (1999). Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiol. 121 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, A., Pennell, R.I., Alvarez, M.E., Palmer, R., and Lamb, C. (1996). Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr. Biol. 6 427–437. [DOI] [PubMed] [Google Scholar]
- Madrid, R., Gomez, M.J., Ramos, J., and Rodriquez-Navarro, A. (1998). Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J. Biol. Chem. 273 14838–14844. [DOI] [PubMed] [Google Scholar]
- Maser, P., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126 1646–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M., Cuzick, A., Can, C., Beynon, J., Dangl, J.L., and Holub, E.B. (2000). Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J. 22 523–529. [DOI] [PubMed] [Google Scholar]
- Mercier, R.W., Rabinowitz, N.M., Ali, R., Gaxiola, R.A., and Berkowitz, G.A. (2004). Yeast hygromycin sensitivity as a functional assay of cyclic nucleotide gated cation channels. Plant Physiol. Biochem. 42 529–536. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Nawrath, C., and Metraux, J.P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Holub, E.B., Frost, L.N., Falk, A., Gunn, N.D., and Daniels, M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz, V., and Fluhr, R. (1992). Calcium requirement for ethylene-dependent responses. Plant Cell 4 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.-Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe, M., Takeuchi, K., Kamoun, S., Ichinose, Y., Govers, F., Toyoda, K., Shiraishi, T., and Yamada, T. (2000). Independent pathways leading to apoptotic cell death, oxidative burst and defense gene expression in response to elicitin in tobacco cell suspension culture. Eur. J. Biochem. 267 5005–5013. [DOI] [PubMed] [Google Scholar]
- Sessa, G., D'Ascenzo, M., and Martin, G.B. (2000). Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto-Pto-mediated hypersensitive response. EMBO J. 19 2257–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., and Klessig, D.F. (1999). The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10 69–78. [DOI] [PubMed] [Google Scholar]
- Shapiro, A.D., and Zhang, C. (2001). The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 127 1089–1101. [PMC free article] [PubMed] [Google Scholar]
- Shirano, Y., Kachroo, P., Shah, J., and Klessig, D.F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, H., Yoshioka, K., Dooner, H.K., and Klessig, D.F. (1999). Characterization of a new Arabidopsis mutant exhibiting enhanced disease resistance. Mol. Plant Microbe Interact. 12 1053–1063. [DOI] [PubMed] [Google Scholar]
- Talke, I.N., Blaudez, D., Maathuis, F.J.M., and Sanders, D. (2003). ATCNGCs: Prime targets of plant cyclic nucleotide signaling? Trends Plant Sci. 8 286–293. [DOI] [PubMed] [Google Scholar]
- van der Biezen, E.A., Freddie, C.T., Kahn, K., Parker, J.E., and Jones, J.D. (2002). Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29 439–451. [DOI] [PubMed] [Google Scholar]
- Xiao, S., Brown, S., Patrick, E., Brearley, C., and Turner, J.G. (2003). Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H., and Heath, M.C. (1998). Role of calcium in signal transduction during the hypersensitive response caused by basidiospore-derived infection of the cowpea rust fungus. Plant Cell 10 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka, K., Kachroo, P., Tsui, F., Sharma, S.B., Shah, J., and Klessig, D.F. (2001). Environmentally-sensitive, SA-dependent defense response in the cpr22 mutant of Arabidopsis. Plant J. 26 447–459. [DOI] [PubMed] [Google Scholar]
- Yu, I.C., Fengler, K.A., Clough, S.J., and Bent, A.F. (2000). Identification of Arabidopsis mutants exhibiting an altered hypersensitive response in gene-for-gene disease resistance. Mol. Plant Microbe Interact. 13 227–286. [DOI] [PubMed] [Google Scholar]
- Yu, I.C., Parker, J., and Bent, A.F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. USA 95 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta, W.N., and Siegelbaum, S.A. (1996). Structure and function of cyclic nucleotide-gated channels. Annu. Rev. Neurosci. 19 235–263. [DOI] [PubMed] [Google Scholar]
- Zhong, H., Lai, J., and Yau, K.W. (2003). Selective heteromeric assembly of cyclic nucleotide-gated channels. Proc. Natl. Acad. Sci. USA 100 5509–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F., Menke, F.L., Yoshioka, K., Moeder, W., Shirano, Y., and Klessig, D.F. (2004). High humidity suppresses ssi4-mediated cell death and disease resistance upstream of MAP kinase activation, H2O2 production and defense gene expression. Plant J. 39 920–932. [DOI] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., Tsui, F., Klessig, D.F., and Glazebrook, J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufall, F., Firestein, S., and Shepherd, G.M. (1994). Cyclic nucleotide-gated ion channels and sensory transduction in olfactory receptor neurons. Annu. Rev. Biophys. Biomol. Struct. 23 577–607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.