Abstract
The hnRNPs (heterogeneous nuclear ribonucleoproteins) F and H2 share a similar protein structure. Both have been implicated as regulating polyadenylation, but hnRNP H2 had a positive effect, whereas hnRNP F acted negatively. We therefore carried out side-by-side comparisons of their RNA-binding and in vivo actions. The binding of the CstF2 (64 kDa cleavage stimulatory factor) to SV40 (simian virus 40) late pre-mRNA substrates containing a downstream GRS (guanine-rich sequence) was reduced by hnRNP F, but not by hnRNP H2, in a UV-cross-linking assay. Point mutations of the 14-nt GRS influenced the binding of purified hnRNP F or H2 in parallel. Co-operative binding of the individual proteins to RNA was lost with mutations of the GRS in the G1−5 or G12−14 regions; both regions seem to be necessary for optimal interactions. Using a reporter green fluorescent protein assay with the GRS inserted downstream of the poly(A) (polyadenine) signal, expression in vivo was diminished by a mutant G1−5 sequence which decreased binding of both hnRNPs (SAA20) and was enhanced by a 12–14-nt mutant that showed enhanced hnRNP F or H2 binding (SAA10). Using small interfering RNA, down-regulation of hnRNP H2 levels diminished reporter expression, confirming that hnRNP H2 confers a positive influence; in contrast, decreasing hnRNP F levels had a negligible influence on reporter expression with the intact GRS. A pronounced diminution in reporter expression was seen with the SAA20 mutant for both. Thus the relative levels of hnRNP F and H2 in cells, as well as the target sequences in the downstream GRS on pre-mRNA, influence gene expression.
Keywords: electrophoretic mobility-shift assay (EMSA), heterogeneous nuclear ribonucleoprotein (hnRNP), polyadenylation, RNA interference (RNAi), UV-cross-linking
Abbreviations: CstF-64, cleavage stimulatory factor 64 kDa subunit, also known as CstF2; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; GFP, green fluorescent protein; GRS, guanine-rich sequence; GST, glutathione S-transferase; HEK-293T, human embryonic kidney; hnRNP, heterogeneous nuclear ribonucleoprotein; IPTG, isopropyl β-D-thiogalactoside; ORF, open reading frame; RNAi, RNA interference; RRM, RNA-recognition motif; siRNA, small interfering RNA; SV40, simian virus 40; SVL, SV40 late; wtSVL, wild-type SVL
INTRODUCTION
hnRNPs (heterogeneous nuclear ribonucleoproteins) display sequence-specific RNA-binding activity, while associating predominantly with pol II (RNA polymerase II) transcripts [1]. Most of them are located exclusively in the nucleus, while others shuttle between cytoplasm and nucleus. The RNA-binding activities of these proteins are conferred by either RRMs (RNA-recognition motifs) or arginine, glycine and glycine-rich domains or both [2]. The RRM-containing hnRNPs typically have multiple RRM domains; one or more of the RRMs is responsible for sequence-specific binding [3]. The binding of multi-RRM-containing proteins to RNA is co-operative and displays complex kinetics. Individual RRMs may bridge multiple RNAs binding in trans, while other RRMs can bind with the same RNA to increase binding affinity in cis [4]. Among the many proposed functions for hnRNP proteins are mRNA trafficking, splicing, telomere length control, mRNA stability, transcription and polyadenylation [1].
The hnRNP H protein family consists of hnRNP H1, H2 (DSEF or H′), H3 (2H9) and hnRNP F [5]. The 346-amino-acid-long hnRNP H3 is a member of the family that has been shown to have various alternatively spliced forms whose functions are not well characterized [6]. Meanwhile, hnRNP H1 and H2 are 449-amino-acid proteins with three RRMs, the first two of which are separated by only 14 amino acids, while the third RRM is 100 amino acids downstream of the cluster of two [7]. The 96% identity between H1 and H2 may ensure similar function. In contrast, hnRNP F is 415-amino-acids long, with three RRMs arrayed in the same spacing, but lacks the very C-terminal region that is present in hnRNPs H1 and H2. While hnRNP F shows an overall 72 and 70% identity with hnRNP H1 and H2 respectively, there is more than 90% identity in the corresponding RRMs [8]. The roles of hnRNP H and F in splicing have been studied extensively with multiple different substrates. They can act in either a positive or a negative way, since the proteins bind to both splicing silencers or enhancers [9,10].
The hnRNP H2 protein, also known as DSEF-1, binds to a 14-nt GRS (guanine-rich sequence) region in SVL [SV40 (simian virus 40) late] pre-mRNA, downstream of the poly(A) (polyadenine) addition site [11], see Table 1 for the sequence. Binding of recombinant hnRNP H2 to the GRS downstream of the SVL poly(A) site was shown to activate 3′ processing in vitro. Although several GRS mutants were made and analysed in vitro [11], a conclusive mutation analysis of the GRS region on SVL pre-mRNA had not been performed to determine the minimal optimal binding site. Diverse pre-mRNAs can be influenced by hnRNP H1/H2 [12], but the question of the exact sequence specificity important for modulating polyadenylation remained.
Table 1. Sequences of GRS oligoribonucleotides.
Mutagenic residues are indicated in bold.
| Oligoribonucleotide | Sequence |
|---|---|
| wtGRS | 5′-GGGGGAGGUGUGGG-3′ |
| SAA1 | 5′-AGGGGAGGUGUGGG-3′ |
| SAA2 | 5′-GAGGGAGGUGUGGG-3′ |
| SAA3 | 5′-GGAGGAGGUGUGGG-3′ |
| SAA4 | 5′-GGGAGAGGUGUGGG-3′ |
| SAA5 | 5′-GGGGAAGGUGUGGG-3′ |
| SAA6 | 5′-GGGGGAAGUGUGGG-3′ |
| SAA7 | 5′-GGGGGAGAUGUGGG-3′ |
| SAA8 | 5′-GGGGGAGGUAUGGG-3′ |
| SAA9 | 5′-GGGGGAGGUGUAGG-3′ |
| SAA10 | 5′-GGGGGAGGUGUGAG-3′ |
| SAA11 | 5′-GGGGGAGGUGUGGA-3′ |
| SAA12 | 5′-GGGGGAGGUGUGAA-3′ |
| SAA13 | 5′-GGGGGAGGUGUAAG-3′ |
| SAA14 | 5′-GGGGGAAAUGUGGG-3′ |
| SAA15 | 5′-GGGAAAGGUGUGGG-3′ |
| SAA16 | 5′-GGAAGAGGUGUGGG-3′ |
| SAA17 | 5′-GAAGGAGGUGUGGG-3′ |
| SAA18 | 5′-AAGGGAGGUGUGGG-3′ |
| SAA19 | 5′-GGAGAAGGUGUGGG-3′ |
| SAA20 | 5′-GGAAAAGGUGUGGG-3′ |
| SAA21 | 5′-GGGGGAGGUGUCCC-3′ |
The hnRNP F protein was implicated in poly(A) site choice within the immunoglobulin heavy chain [13]. Whereas hnRNP F blocked the binding of CstF-64 (cleavage stimulatory factor 64 kDa subunit, also known as CstF2) to its GU-rich target sequences downstream of the immunoglobulin secretory and membrane poly(A) sites in a plasma cell extract, the precise binding site of hnRNP F was not mapped.
To better understand the mechanism of action of these two hnRNP proteins, F and H2, we wanted to evaluate their RNA sequence-selectivity in a side-by-side comparison and determine their relative affinities for RNA. In the present study, we have shown that hnRNP F and H2 both bind to a five-guanine stretch (G1−5) downstream of the poly(A) site on SVL pre-mRNA, in the 14-nt GRS. The binding of both hnRNP F and H2 was enhanced by an adenine residue in the 12–14 positions downstream of the G1−5 cluster. While the proteins differ slightly in their RNA affinities, they both display co-operative binding to the wild-type GRS and non-co-operative binding to the substrate in which the G1−5 region is severely interrupted. The binding of CstF-64 is inhibited by purified hnRNP F, but purified hnRNP H2 still allows CstF-64 to bind to RNA. Diminishing the binding of hnRNP F and H2 by mutating the GRS in RNA inhibited gene expression in vivo. Decreasing the levels of hnRNP F and H2 by RNAi (RNA interference) in mammalian cells influenced gene expression in an RNA-sequence-dependent manner. Multiple lines of evidence therefore indicate that hnRNP F and H2 proteins bind to the same RNA sequence, but can act differently to modulate gene expression.
EXPERIMENTAL
Protein purification of hnRNP F and H2
Plasmid pET-15b with the human hnRNP F ORF (open reading frame) [14] and the plasmid pGEX-2T, with human hnRNP H2 ORF [15], were gifts from Dr Douglas Black (University of California Los Angeles, Los Angeles, CA, U.S.A.) and Dr Jeff Wilusz (Colorado State University, Fort Collins, CO, U.S.A.) respectively. Histidine-tagged hnRNP F for UV-cross-linking assays and EMSAs (electophoretic mobility-shift assays) was purified as described [16]. To obtain highly pure and non-denatured hnRNP F and H2 for the filter binding assay, two-step purification methods were developed for each. Escherichia coli (Rosetta™, Novagen) cells were used to express either protein. These cells have a plasmid coding for six tRNAs for mammalian codons that are rarely used in bacteria and improved the yield of the hnRNPs. Cells were grown at 37 °C until the D600 reached 0.5. Then they were induced with 1 mM IPTG (isopropyl β-D-thiogalactoside) and allowed to grow at 28 °C for 3 h until the final D600 reached approx. 2.0.
In the case of the His–hnRNP F purification, cells were resuspended in 50 mM Tris/HCl, pH 8.0, 1% (v/v) Triton X-100 and 5 mM ATP. Three bursts of sonication for 20 s at 10 W (RMS) were performed on ice to lyse the cells. The tube was spun at 15000 g for 15 min, and the pellet was discarded. The supernatant was loaded on to a gravity-operated 10 ml DEAE-Sepharose plastic column (15 mm diameter; Pierce) that was equilibrated with 50 mM Tris/HCl, pH 8.0. Flowthrough was collected and purified over a 2 ml metal-affinity Talon (Clontech) plastic column (7 mm diameter; Pierce) according to the manufacturer's protocol. Two-step elutions were performed with 10 mM and 300 mM imidizole; the latter eluted sample contained the pure His–hnRNP F.
For GST (glutathione S-transferase)–hnRNP H2 purification, the induction was performed with 0.1 mM IPTG to enhance the solubility of the protein. The cells were resuspended in PBS, pH 7.3, with 1% (v/v) Triton X-100 and 5 mM ATP. The sonication and spin were performed as described above for hnRNP F. The hnRNP H2 recombinant protein was first enriched by using a 2 ml glutathione–Sepharose column (Amersham Biosciences) according to the manufacturer's protocol. A two-step elution with 1 and 100 mM glutathione in 50 mM Tris/HCl, pH 8.0, was performed, and the latter eluted sample contained the most enriched fraction. Glutathione was eliminated by exchanging buffers on a 10 ml medium G-25 Sephadex column. The final fractions in PBS, pH 7.3, were loaded on to a 10 ml DEAE-Sepharose column, and a two-step elution was performed with 250 and 750 mM NaCl; the latter eluted sample contained the pure GST–hnRNP H2.
Finally, both protein samples were buffer-exchanged with Buffer D [20 mM Hepes, pH 7.9, 100 mM KCl, 1 mM DTT, 20% (v/v) glycerol and 0.2 mM EDTA with protease inhibitor (Cocktail Set I™; Calbiochem)] by using a 10 ml medium G-25 Sephadex column. Each fusion protein is a single band on a protein gel following Coomassie Blue staining.
CstF-64 RNA-binding domain
Plasmid pZ64-18 [17] containing the entire cDNA for CstF-64 was digested with EcoRV and XhoI and then inserted in-frame into the XhoI and SmaI sites of pGEX 4T-2 (Amersham Biosciences). The resulting plasmid made a protein containing the RNA-binding domain of CstF-64, the hinge region and the proline/glycine-rich domain but lacking the MEARA (Met-Glu-Ala-Arg-Ala) repeat region. This GST-tagged protein was purified according to the glutathione–Sepharose 4B protocol provided by Amersham Biosciences. Briefly, E. coli XL1-Blue (Invitrogen) cells were grown until the D600 was 0.5. Then IPTG was added to 0.1 mM, and the culture was grown at 30 °C for an additional 2–6 h. The pellet was resuspended in ice-cold PBS plus protease inhibitors (Cocktail Set I™), and sonicated on ice for six blasts of 10 s at 50% power. Triton X-100 was added to a final concentration of 1%, and the lysate was incubated on a rocker for 20 min at 4 °C. After pelleting the debris, the clarified supernatant was incubated with glutathione–Sepharose 4B resin for 2 h at 4 °C. After washing four times with PBS and, eluting with glutathione elution buffer (3.1 mg/ml reduced glutathione in 50 mM Tris/HCl, pH 8.0), the purified protein was dialysed against 20 mM Hepes, 0.2 mM EDTA, 10% glycerol, 80 mM potassium glutamate and 1 mM DTT (dithiothreitol).
UV-cross-linking studies
These studies were conducted as previously described [13,18]. Briefly, 40 nM, 20 nM, 10 nM or 5 nM His-tagged hnRNP F or 50 nM GST-tagged hnRNP H2 were incubated in 25 μl reaction mixtures containing 40 μg/ml yeast tRNA, 1 mM ATP, 0.7 mM MgCl2, 0.2 mM EDTA, 2 mM DTT, 10% (v/v) glycerol and 230 mM potassium glutamate in 20 mM Hepes, pH 7.9, and 60 fmol of [32P]GTP-labelled RNA. After an incubation of 5 min at 30 °C, 250 nM GST CstF-64 RBD was added, followed by an additional incubation for 5 min at 30 °C. Reactions were transferred to the wells of 96-well plates on ice and irradiated with 1.8 J/cm2 of UV light, then digested with 2.5 μg of RNase A for 15 min at 37 °C. Radiolabelled proteins were resolved by SDS/10% PAGE and visualized by autoradiography.
Probes for EMSAs, filter-binding and UV-cross-linking
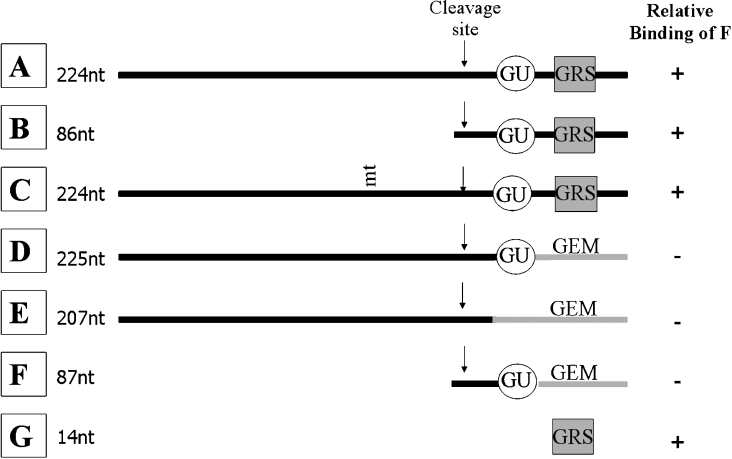
Plasmids that were used to create RNA probes A, B, C and E (Figure 1) for UV-cross-linking and EMSAs were a gift from Dr Jeff Wilusz and are described in [19]. The pSVL plasmid had the 224-nt wtSVL (wild-type SVL) poly(A) region with the upstream sequences, AAUAAA, cleavage site and downstream region, whereas pSVL-5, which was used to produce the 86-nt probe B, had only the cleavage site and downstream sequences. For probe C, a point mutation in pSVL-G3 changed the poly(A) signal from AAUAAA to AAGAAA. Probe E, also called pSVL-GEM, had sequences downstream of the cleavage site replaced with GEM vector sequence. DNA templates that were used to create RNA probes D and F (Figure 1) were synthesized by PCR as described in [20]. These lacked the GRS, but still contained the GU sequence for CstF-64 binding. Probe G was in vitro transcribed from a DNA oligonucleotide template containing the 14-nt GRS and the T7 promoter sequence.
Figure 1. SVL pre-mRNA based probes used in the present study.
The name and length of each RNA probe is indicated on the left. The relative binding affinities to hnRNP F as determined from data summarized in Figure 3 are indicated in the right column. Probe A is wtSVL; GU, GU-rich binding site for CstF-64; GEM, vector pGem sequence. mt indicates the site of mutation.
Probes for filter binding assays in Figures 5(A) and 5(B) were transcribed from PCR-derived templates. Using 248-5′CC (5′-TA-ATACGACTCACTATAGAATACAC CGAGCT-3′), containing the T7 promoter sequence (in italics) and an EcoRI site, boxed in grey, as a common forward primer and 248wt-3′ (5′-AAAAAACCTCCCACACCTCCCCCTGAACCTGAAACATAAA- ATGAAT-3′) and 248SAA20-3′ (5′-AAAAAACCTCCCACA- CCTTTTCCTGAACCTGAAACATAAAATGAAT-3′) as reverse primers, the 86-nt wtSVL and SAA20 probes were created. The SAA20 mutation is underlined and in bold. The PCR protocol consisted of 30 cycles of 94 °C for 1 min, 58 °C for 30 s and 72 °C for 30 s, and used Accupol DNA polymerase (Gene-Choice). For subsequent filter binding assays (Figure 5C) oligonucleotides were synthesized to contain the wild-type GRS (in bold type), SAA10 or SAA21 mutations (underlined) in a 67-nt sequence which when transcribed by T7 (primer complement in italics) would produce a 50-nt sense strand: 5′-CAUUCAUUUUAUGUUUCAGGUUCAGGGGGAGGUGUGGGAGGUUUUUU-3′. The templates were: wild-type, 5′-AAAAAACCTCCCACACCTCCCCCTGAACCTGAAACATAAAATGAATGCCCTATAGTGAGTCGTATTA-3′; SAA10 template, 5′-AAAAAACCTCTCACACCTCCCCCTGAACCTGAAACATAAAATGAATGCCCTATAGTGAGTCGTATTA-3′; and SAA21 template, 5′-AAAAAACCTGGGACACCTCCCCCTGAACCTGAAACATAAAATGAATGCCCTATAGTGAGTCGTATTA-3′.
CGAGCT-3′), containing the T7 promoter sequence (in italics) and an EcoRI site, boxed in grey, as a common forward primer and 248wt-3′ (5′-AAAAAACCTCCCACACCTCCCCCTGAACCTGAAACATAAA- ATGAAT-3′) and 248SAA20-3′ (5′-AAAAAACCTCCCACA- CCTTTTCCTGAACCTGAAACATAAAATGAAT-3′) as reverse primers, the 86-nt wtSVL and SAA20 probes were created. The SAA20 mutation is underlined and in bold. The PCR protocol consisted of 30 cycles of 94 °C for 1 min, 58 °C for 30 s and 72 °C for 30 s, and used Accupol DNA polymerase (Gene-Choice). For subsequent filter binding assays (Figure 5C) oligonucleotides were synthesized to contain the wild-type GRS (in bold type), SAA10 or SAA21 mutations (underlined) in a 67-nt sequence which when transcribed by T7 (primer complement in italics) would produce a 50-nt sense strand: 5′-CAUUCAUUUUAUGUUUCAGGUUCAGGGGGAGGUGUGGGAGGUUUUUU-3′. The templates were: wild-type, 5′-AAAAAACCTCCCACACCTCCCCCTGAACCTGAAACATAAAATGAATGCCCTATAGTGAGTCGTATTA-3′; SAA10 template, 5′-AAAAAACCTCTCACACCTCCCCCTGAACCTGAAACATAAAATGAATGCCCTATAGTGAGTCGTATTA-3′; and SAA21 template, 5′-AAAAAACCTGGGACACCTCCCCCTGAACCTGAAACATAAAATGAATGCCCTATAGTGAGTCGTATTA-3′.
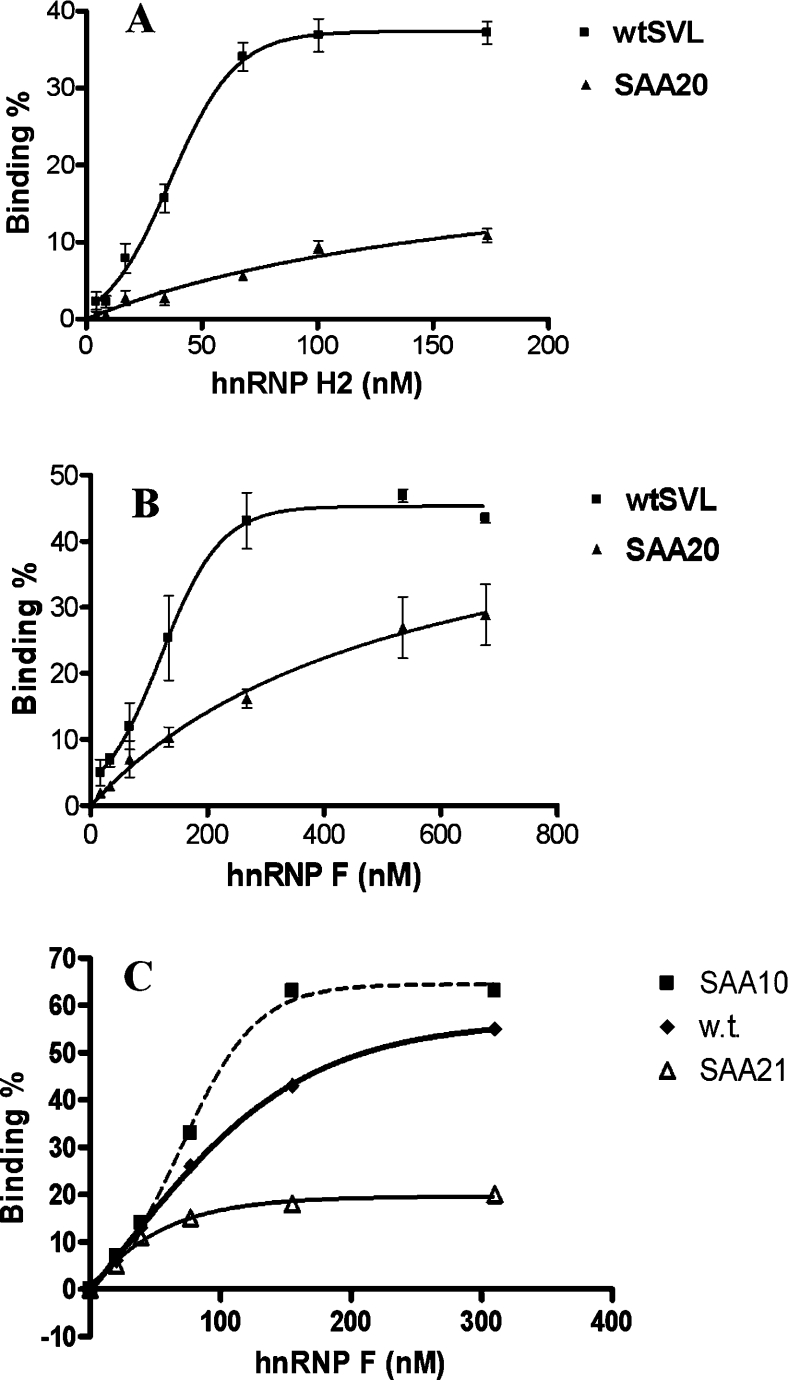
Figure 5. Filter binding assay with hnRNP H2 and hnRNP F.
(A) Binding curves of GST–hnRNP H2 to 250 pM wild-type probe B in Figure 1 or probe B having the SAA20 mutation, as determined by filter binding assay. Each reaction was performed three times, and results are means±S.E.M. (B) Binding curves of His–hnRNP F to the same RNA molecules were obtained as described in (A). Each reaction was performed three times, and results are means±S.E.M. (C) Binding was performed with His–hnRNP F and 250 pM RNA probes (50 nt) containing wild-type (w.t.), SAA10 or SAA21 GRS regions. Results are the means of at least two determinations.
pEGFP-based SVL plasmid constructs
To clone the regions into pEGFP, a 5′ PCR primer with an EcoRI overhang and a 3′ primer with mutant GRS overhang was used. We then synthesized PCR products having the 224-nt SVL poly(A) region with the mutant GRS. The 248-5′CC primer (above) is the common forward primer. The individual 3′ primers with the MluI sites boxed in grey and mutations underlined were: WT-3′CLONING, 5′-TACCG AAAAAACCTCCCACACCTCCCCCTGAACCTGAAACATAAAATGAAT-3′; SAA-20-3′CLONING, 5′-TACCG
AAAAAACCTCCCACACCTCCCCCTGAACCTGAAACATAAAATGAAT-3′; SAA-20-3′CLONING, 5′-TACCG AAAAAACCTCCCACACCTTTTCCTGAACCTGAAACATAAAATGAAT-3′; and SA-A10-3′CLONING, 5′-TACCG
AAAAAACCTCCCACACCTTTTCCTGAACCTGAAACATAAAATGAAT-3′; and SA-A10-3′CLONING, 5′-TACCG AAAAAACCTCTCACACCTCCCCCTGAACCTGAAACATAAAATGAAT-3′. The GFP (green fluorescent protein) ORF on pEGFP-C1 (Clontech) has an SV40 early poly(A) site. By using EcoRI and MluI sites, this default site was removed and then the inserts produced by PCR were cloned into the same sites. Each PCR product was double-digested with EcoRI and MluI for 4 h at 37 °C. All cloning experiments were performed as described in [21].
AAAAAACCTCTCACACCTCCCCCTGAACCTGAAACATAAAATGAAT-3′. The GFP (green fluorescent protein) ORF on pEGFP-C1 (Clontech) has an SV40 early poly(A) site. By using EcoRI and MluI sites, this default site was removed and then the inserts produced by PCR were cloned into the same sites. Each PCR product was double-digested with EcoRI and MluI for 4 h at 37 °C. All cloning experiments were performed as described in [21].
To create pEGFP-nopoly(A), the default poly(A) site of GFP ORF was removed from pEGFP-C1 plasmid by using EcoRI and MluI sites, boxed in grey, and two oligonucleotides were inserted to the same position. The sequences of the oligonucleotides were: EcoRI, 5′- GATCGGATATCAGTACTA-3′; and MluI, 5′-
GATCGGATATCAGTACTA-3′; and MluI, 5′- AGTACTGATATCCGATCG-3′. Every cloned plasmid was verified by DNA sequencing. All the pEGFP and pDsRed-Express plasmids were linearized by SspI digestion for transfection. This only disrupts the f1 origin in all the plasmids.
AGTACTGATATCCGATCG-3′. Every cloned plasmid was verified by DNA sequencing. All the pEGFP and pDsRed-Express plasmids were linearized by SspI digestion for transfection. This only disrupts the f1 origin in all the plasmids.
DNA oligonucleotides and in vitro transcription
All oligonucleotides were synthesized at the DNA Synthesis Facility, University of Pittsburgh. The 14-nt oligoribonucleotides given in Table 1 were produced by in vitro transcription reactions of longer DNA oligonucleotides with T7 RNA polymerase, essentially as described previously [22] with the exception that no cap analogue was used.
EMSAs and filter binding assays
Each EMSA used 12.5 μg of purified yeast tRNA, 3 mM DTT, 1.2 mM ATP, 16.7 mM creatine phosphate, 0.75 mM MgCl2, 2.7% (v/v) poly(vinyl alcohol), RNA and protein (dialysed against Buffer D). To each tube, 1 fmol of radiolabelled RNA (5 fmol if RNA probe was <50 nt) and the indicated amount of protein were added. Each tube was made up to 20 μl in total; volumes were equalized with Buffer D resulting in the same amount of Buffer D, hence identical salt concentrations in each tube. All of the EMSA reaction mixtures were incubated for 10–20 min at 30 °C.
Acrylamide gels of 4, 6 or 8% acrylamide with a 20:0.25 acrylamide/bisacrylamide ratio were run at 30 mA, 300 V and 15 W for approx. 1–2 h. Gel running buffer contained 25 mM Tris base, 25 mM boric acid and 1 mM EDTA. Gels were also prepared in this buffer. Gel runs were always started with both a cold gel and cold buffer and were performed at room temperature (25 °C).
The EMSAs with competitors were performed with a 2 min pre-incubation of protein with RNA competitor followed by 10 min incubation of the 32P-labelled probe B. The specific competitor was a tritium-labelled 86-nt probe B, whereas the non-specific competitor was poly(A) having the same length.
Filter binding assays were carried out essentially as described in [3] at 30 °C for 45–60 min. Each 50 μl reaction mixture contained 250 pM [32P]RNA and 0.1M KCl and each assay was performed two to three times and error bars (where shown) represent the S.E.M. The curves were fitted to the data points using Prism 4 software (version 4.01) from Graphpad Software.
Transfection
Cells were transfected with linearized DNA plasmids by using Fugene 6 (Roche). The pDsRedExpress plasmid (Clontech) coding for DsRedExpress protein, a variant of the original DsRed protein [23], was used as a transfection marker. Cells [(1.5–3.0)×105] were plated on six-well plates 12–18 h before transfection. Approx. 2 μg of equimolar plasmid, serum-free medium and Fugene 6 transfection agent with a Fugene 6/plasmid ratio of 3:2 (v/w) were combined in a 100 μl volume. This mixture was incubated for 15 min at room temperature and added to the wells having 2 ml of serum-positive medium.
RNAi transfection
Approx. 1.5×105 cells were seeded on each well of six-well plates 1 day before the transfection. A total of 200 μl of transfection mixture, including 0.1 nmol of duplex siRNA (small interfering RNA), 8 μl of amine transfection reagent (Ambion) and serum-free Dulbecco's modified Eagle's medium was added to each well having 800 μl of medium with serum, essentially according to the manufacturer's instructions. After 20–24 h of incubation, the medium was exchanged with 2 ml of fresh medium with serum. After a total of 2 days of incubation, cells were transfected with pEGFP-based plasmids. They were then harvested after 24 h of incubation for flow cytometry. The siRNA sequences are given in Table 2. The region targeted at the hnRNP H2 mRNA was identical in hnRNP H1 and H2. Mismatches between the hnRNP F and H transcripts in this region ensured that there was no cross-reactivity.
Table 2. siRNAs used in RNAi transfection.
Mismatches between the hnRNP F and H transcripts are highlighted.
| siRNA to hnRNP F (Ambion siRNA lot # 044786siB) | siRNA to hnRNP H1 (Ambion siRNA lot # 048945si) |
|---|---|
| 5′-GGUGUCCAUUUCAUCUACAtt-3′ | 5′-GGUAUUCGUUUCAUCUACAtt-3′ |
| 5′-UGUAGAUGAAAUGGACACCtg-3′ | 5′-UGUAGAUGAAACGAAUACCtt-3′ |
Flow cytometry
After incubating for 18–41 h in medium, cells were washed with PBS and trypsinized for 1 min. Cells were pelleted in 5 ml Falcon tubes. Cell pellets were sequentially washed with PBS and then with FACS buffer [5% (v/v) foetal calf serum and 0.01% (w/v) sodium azide in PBS]. Finally, cells were fixed in paraformaldehyde in PBS.
All the flow-cytometry experiments were performed with the Coulter Epics XL flow cytometer operating with a single 488 nm laser. Both GFP and DsRedExpress proteins were excited with a 488 nm laser, and emissions were recorded in the FL1 (green) or FL3 (red) channels. A modest compensation was made by subtracting 3.7% of the FL1 channel from FL3. A total of (5–20)×103 events were recorded for each sample. The analysis was done with the WinMdi software available at http://facs.scripps.edu/software.html (November 2004).
Western blots
Western blots were performed as described in [13,24]. Anti-GFP and anti-DsRedExpress antibodies were purchased from Clontech, whereas anti-neomycin antibody was obtained from Upstate Biochemical Cell Signaling Solutions. Antibodies against GAPDH (glyceraldehyde-3-phosphate dehydrogenase), a loading control, hnRNP H2 and hnRNP F were described previously [13].
RESULTS
UV-cross-linking studies show differences in hnRNP F and H2 binding
We had shown previously using RNA EMSA that purified hnRNP F could bind to a 224-nt sequence of the SVL poly(A) region (probe A represented in Figure 1), but that binding was inhibited by replacing the region downstream of the poly(A) site with irrelevant pGEM sequences (probe E) [13]. In order to explore the binding of hnRNP F, H2 and CstF-64 further, we performed UV-cross-linking experiments. When exposed to UV light, radio-labelled RNA is covalently bound to protein with which it is in very close contact. The cross-linked portion of the RNA is protected from subsequent digestion by RNase A by virtue of the protein that shields it. Meanwhile, the protein(s) bound to the RNA can be identified by SDS/PAGE because of its newly acquired radioactivity. As shown in Figure 2, hnRNP F and H2 proteins, either alone or when mixed together, bound to probe A and acquired radioactivity. When either protein was added to SLV-GEM (probe E) where the region downstream of the poly(A) addition site was substituted with vector (pGEM) sequences, no binding was observed, indicating that the binding seen in the intact SVL was in the region downstream of the cleavage site. These results are consistent with previous EMSA data.
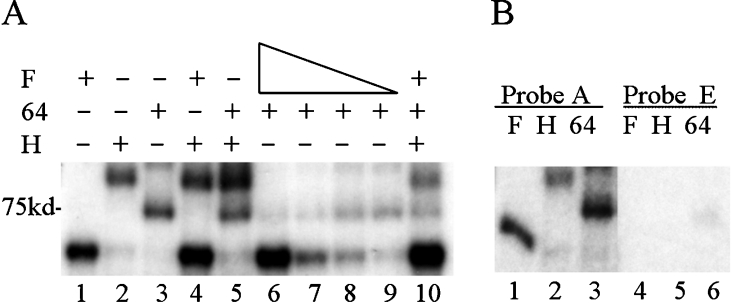
Figure 2. hnRNP F inhibits CstF-64 binding.
Purified hnRNP proteins and/or the RNA-binding domain of CstF-64 were incubated with 32P-labelled RNA under UV-cross-linking conditions (see the Experimental section) and subjected to UV-irradiation on ice. Following RNase digestion, proteins were separated by SDS/10% PAGE. Dried gels were exposed to film. In the indicated lanes, 50 nM GST-tagged hnRNP H2 or 260 nM GST–CstF-64 RBD were added. (A) Lane 1, 40 nM His-hnRNP F; lane 2, GST-hnRNP H2; lane 3, GST–CstF-64 RBD; lane 4, 40 nM His–hnRNP F plus GST–hnRNP H2; lane 5, GST–hnRNP H2 plus GST–CstF-64 RBD; lane 6, 40 nM His–hnRNP F plus GST–CstF-64 RBD; lane 7, 20 nM His–hnRNP F plus GST–CstF-64 RBD; lane 8, 10 nM His–hnRNP F plus GST–CstF-64 RBD; lane 9, 5 nM His–hnRNP F plus GST–CstF-64 RBD; lane 10, 40 nM His–hnRNP F, GST–CstF-64 RBD and GST–hnRNP H2. (B) SVL or SVL-GEM, probes A or E incubated with purified proteins; lanes 1 and 4, 40 nM His–hnRNP F; lanes 2 and 5, 50 nM GST–hnRNP H2; lanes 3 and 6, 260 nM GST–CstF-64 RBD. The position of the 75 kDa protein is indicated. F, hnRNP F; H, hnRNP H; 64, CstF-64.
Recombinant CstF-64, containing only the RNA-binding/N-terminal half of the molecule, bound to SVL (probe A), but less well to probe E, which lacks the GU-rich target for CstF-64 as well as the GRS. We observed that, when a mixture of hnRNP H2 and recombinant CstF-64 were incubated with probe A, both bound (Figure 2). In contrast, when hnRNP F and CstF-64 were added to probe A, CstF-64 binding was inhibited at high concentrations of hnRNP F (Figure 2A). Based on these results, we conclude that hnRNP F and H2 either might bind to the same site with different consequences or perhaps recognize different sites downstream of the poly(A) site in the SVL region.
However, when hnRNP F, H2 and CstF-64 were all present, the negative effect of hnRNP F on CstF-64 binding was partially abolished (see Figure 2A, lane 10). This is consistent with an earlier observation that hnRNPs H and F can exist as heterodimers in cells [16] and that H and F are found in a slower migrating complex by EMSAs [13]. These hnRNP H–hnRNP F heterodimers apparently still allow CstF-64 to bind to RNA, while F alone does not.
The binding of hnRNP H2 to pre-mRNA had been shown previously to stimulate cleavage, while the binding of hnRNP F had been shown to block cleavage [12,13], consistent with changes in the consequences of CstF-64 binding in extracts and the present UV-cross-linking studies. However, the binding characteristics of both proteins had not been determined rigorously in a side-by-side comparison. Therefore we explored the binding specificities of both proteins to determine their sites of action.
Mapping the binding site of hnRNP F on SVL pre-mRNA
The three RNA-binding domains of hnRNP F show extensive similarity to hnRNP H2, but, since hnRNP F behaved differently from hnRNP H2 in the UV-cross-linking studies and differ outside the RRMs, we wanted to map the binding region of hnRNP F on SVL. We made use of several SVL pre-mRNA-based constructs [20] (Figure 1). The EMSAs were performed with these RNA samples labelled with [32P]GTP and histidine-tagged recombinant hnRNP F as shown in Figures 3 and 4. Since hnRNP F was shown to bind the cap-binding complex, all of the in vitro transcription reactions were performed without the cap analogue. Despite the fact that the transcripts were uncapped, RNA stability was maintained.
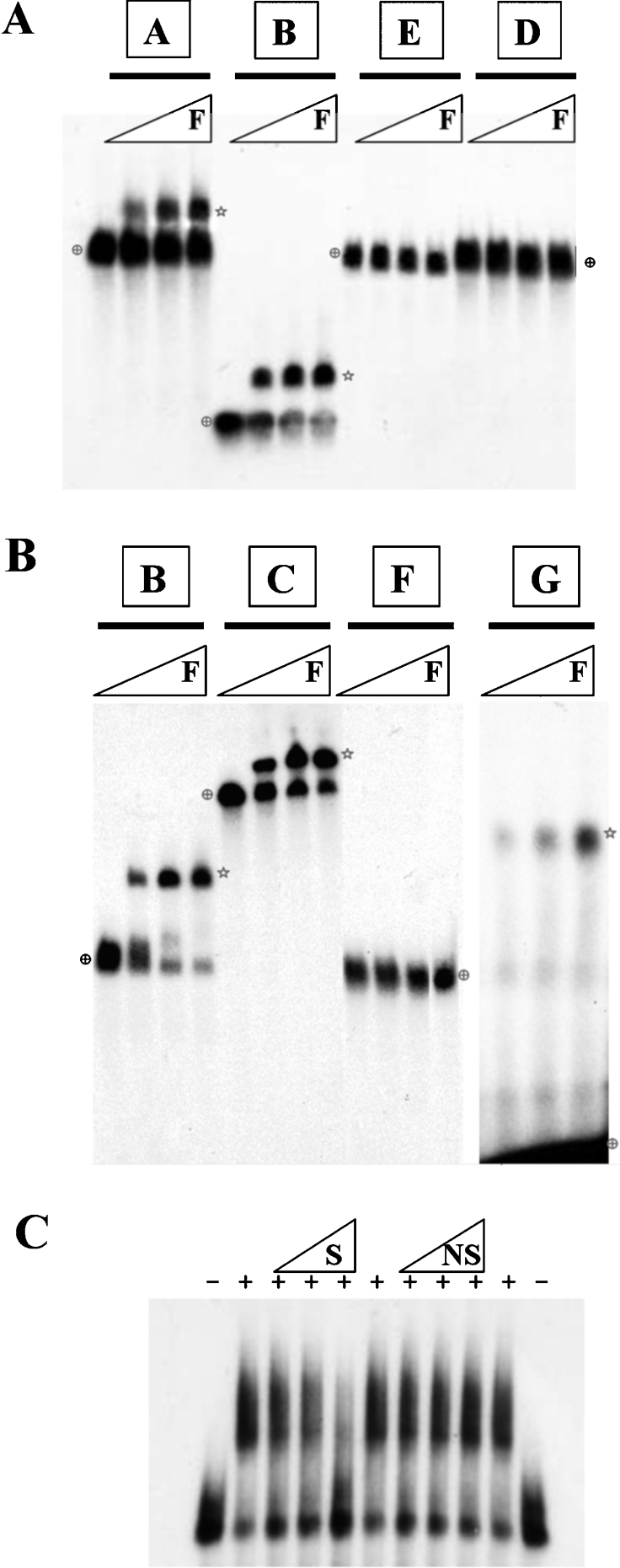
Figure 3. hnRNP F binds to the GRS in SVL pre-mRNA in EMSAs.
(A) EMSAs were performed with 50 pM [32P]GTP-labelled SVL pre-mRNAs in 20 μl reaction volumes. Recombinant His-hnRNP F (0, 55, 110 or 220 nM) was used to shift each RNA probe. Samples were run on a 4% (20:0.25) native acrylamide gel containing 0.5% agarose. Stars indicate the positions of hnRNP F-RNA complexes. Cross-hairs show the positions of unshifted RNA probes. (B) EMSAs were performed as described in (A). Samples were run on 8% (20:0.25) native acrylamide gels containing 0.5% agarose. Probe G, 250 pM, was used in the last four lanes. (C) EMSAs were performed with 50 pM probe B and 0.5, 5 or 50 nM specific (S) or non-specific (NS) competitors in the presence (+) or absence (−) of 165 nM His-hnRNP F.
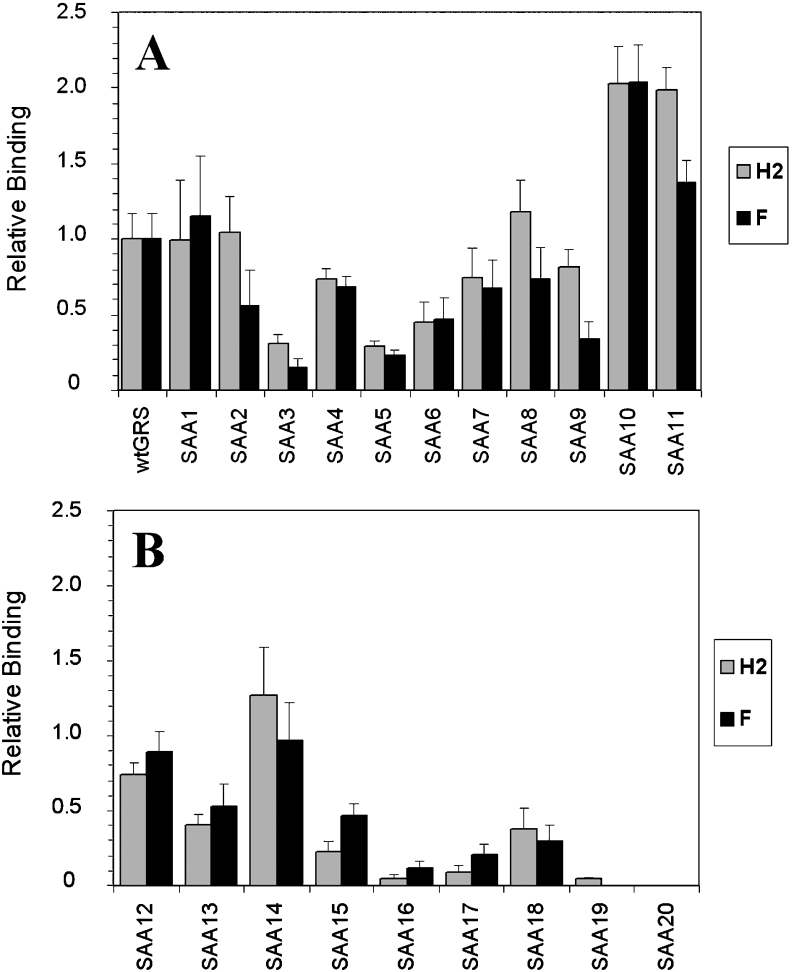
Figure 4. hnRNP F and H2 show similar binding to mutations in the GRS.
(A, B) Relative binding of wild-type and mutant GRS oligoribonucleotides whose sequences are shown in Table 1 to hnRNP F (F) and hnRNP H2 (H2) proteins. EMSAs were performed with 250 pM of each probe and 330 nM His–hnRNP F or 210 nM GST–hnRNP H2. Relative binding of each mutant to wild-type GRS was set as 1.0 as shown in the histogram. Results are means±S.E.M. for at least three determinations.
We found that hnRNP F protein was able to bind to the first three of the RNA samples: wtSVL (probe A), SVL having a poly(A) signal mutation AAGAAA (probe C) and a portion of the SVL having only the downstream guanine-rich signal (probe B), see Figures 3(A) and 3(B). These data confirm the UV-cross-linking result that hnRNP F binds somewhere in the region at or downstream of the cleavage site. Note that, even at the highest concentrations of hnRNP F, some unbound RNA remained. Further increases in the amount of hnRNP F did not result in a complete shift of the RNA to the bound fraction (results not shown). In an EMSA, once the RNA and probe dissociate, a process which can be accelerated by physical forces upon entering the gel matrix, they cannot re-associate, since the unbound RNA is rapidly separated from the RNA–protein complex.
Replacement of the GU-rich and guanine-rich regions downstream of the cleavage site (see probe E) or just the region downstream of the GU-rich CstF-64-binding site with vector pGEM sequence, in probe D, abolished the binding of hnRNP F. Replacement of the 14-nt GRS with vector pGEM sequences in the smaller probe F also eliminated the hnRNP F binding. The 14-nt GRS of SVL alone, probe G (see Table 1 for sequence), was transcribed, and hnRNP F was able to shift its mobility (Figure 3B). Competing RNA was used to determine whether the interaction observed between hnRNP F and GRS was a result of specific RNA–protein binding (Figure 3C). The binding of hnRNP F to GRS was competed by a specific (S) RNA that is GRS-positive, but not by a non-specific RNA, demonstrating the specificity of binding. Together, these data strongly suggest that hnRNP F binds to the GRS, just like hnRNP H2, but perhaps with altered specificity in sequence or orientation which could possibly influence CstF-64 binding.
Comparing the binding site(s) for hnRNP F and H2
After narrowing the binding region of hnRNP F to 14 nt, we wanted to determine the RNA sequence characteristics important for both hnRNP F and H2 binding within that interval. For this purpose, a serial single G→A mutation study was performed to detect which guanine residues are essential for the interaction of hnRNP F and H2 with the RNA. There are 11 guanine residues in the GRS and each was mutated individually to adenine for a total of 11 single mutants. The exact sequence of each mutant region is given in Table 1. The concentrations of hnRNP F or hnRNP H2 were increased until no further binding with the wild-type sequence was observed; this was 330 nM for hnRNP F and 210 nM for hnRNP H2 with 250 pM probe (results not shown). The binding of the wild-type sequence in the probe was arbitrarily set at 1.0, as summarized in Figure 4, even though some probe remained unbound (see Figure 3) because of the nature of the EMSA. The binding of the mutant probes was normalized to the amount of radioactivity seen with the wild-type probe in the bound fraction.
The data for single substitution mutants using EMSA are summarized in Figure 4(A). The most potent single-point mutants for diminishing binding to hnRNP F and H2 are SAA3 or SAA5, which have mutation at the third or fifth positions of the GRS respectively. These mutants show approx. 15–30% of wild-type GRS binding to hnRNP F and H2. Mutants SAA10 and SAA11 in the G12−14 region bind to hnRNP F and H2 more efficiently than wild-type GRS does. The effect of each of the mutations is similar on both hnRNP F and H2 binding, although SAA3 and SAA9 have a slightly more pronounced effect on hnRNP F than on H2. In summary, EMSA results with single mutations indicate that the third and fifth guanine residues are the most important for both hnRNP F and hnRNP H2 binding. The SAA3 mutation disrupts the longest guanine stretch in the GRS exactly in the middle.
We then made every possible two consecutive G→A mutations in GRS (Table 1) and performed EMSAs with them (results are summarized in Figure 4B). Mutants SAA16 and SAA17 were the most effective in reducing binding. Whereas SAA16 has mutations at the third and fourth positions, SAA17 has substitutions at the second and third nucleotides (Table 1). With this set of experiments, it was clear that the first five guanine residues are very important for both hnRNP F and H2 binding. Mutants SAA12 and SAA13 showed less binding than the wild-type GRS so, while one adenine in the G12−14 region of the GRS aids binding, two adenines reduce it.
We made two other mutants: one with mutations at the third and fifth positions (SAA19) the other with triple G→A mutations between the third and fifth residues (SAA20) (Table 1). The latter completely knocked out both hnRNP F and H2 binding, whereas the former showed some residual binding of hnRNP H2 in EMSAs (Figure 4B).
Thus the G1−5 cluster of the GRS is a major determinant for both hnRNP F and H2 binding. Meanwhile, increased binding with mutations in the 13th or 14th position in the GRS (SAA10 and SAA11) indicate the positive influence of one adenine within the purine triplet downstream of the G1−5 cluster, a feature of several hnRNP H-binding sequences (see Figure 8). This may imply a second point of contact with the RRMs of these hnRNPs, a line of reasoning which is examined further below.
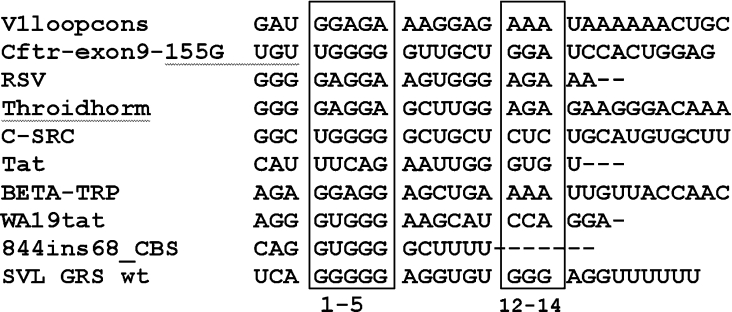
Figure 8. Alignment of hnRNP F- and hnRNP H-binding sites.
Sequences from the indicated genes implicated as binding to hnRNP F or hnRNP H/H2 were taken from the references indicated in the Discussion. The genes are: V1loopcons, consensus for Exon 6D splicing regulatory region of divergent HIV-1 isolates; Cftr-exon9-155G, natural mutation 155G of cystic fibrosis transmembrane regulator Exon 9; RSV, negative regulator of splicing element from rous sarcoma virus; Throidhorm, TRα2 isoform of thyroid hormone receptor gene c-erbAα; C-SRC, intronic splicing enhancer downstream of the N1 5′ splice site of the c-src N1 exon; Tat, a second exonic splicing silencer of tat exon 2 within HIV type I; BETA-TRP, exonic splicing silencer of the rat β-tropomyosin gene; WA19tat, high-affinity hnRNP A1-binding sequence identified by SELEX (systematic evolution of ligands by exponential enrichment); 844 ins68_CBS, 3′ splice site in 884ins68 polymorphism of the cystathionine β-synthase gene. The sequences between nucleotides 1–5 and 12–14 of the GRS are in the boxes.
Next we determined the apparent dissociation constants (Kd) of hnRNP F and H2 to SVL pre-mRNA using a filter binding assay as described previously for other hnRNPs [3]. Either the 86-nt wild-type probe B (see Figure 1) or probe B but having the SAA20 mutation inserted was used for the binding assays shown in Figures 5(A) and 5(B). Proteins were purified extensively as described in the Experimental section. We found that hnRNP H2 binds to wtSVL with an affinity (37 nM) that is apparently higher than that of hnRNP F (121 nM; see Figure 5B). This may ensure that, physiologically, when both are present in equal amounts in the nucleus, the binding sites are preferentially occupied by hnRNP H2. The SAA20 mutation, with three consecutive G→A mutations (see Table 1), which reduced binding almost to zero on EMSAs, decreased the affinity of both proteins (Figures 5A and 5B) in the filter binding assay.
While this difference in protein affinity was reproducible, we cannot rule out the possibility that it may be attributable to the difference in the affinity tags used for the purification of the proteins. There is a histidine tag on hnRNP F and a GST tag on hnRNP H2; each tag is at the N-terminal end. In addition, even though both purifications used ion-exchange and affinity chromatography, the procedures were not identical, and changes in specific activity may contribute to the apparent differences in affinity.
In order to confirm the EMSA data with SAA10 where we saw increased binding relative to wild-type GRS, we used 50-nt 32P-labelled RNA probes generated by T7 RNA polymerase from oligonucleotides (Figure 5C) and examined the binding to hnRNP F with the wild-type, SAA10 mutation or another mutant SAA21 (Table 1). We observed more binding of the SAA10 probe than of the wild-type probe in the lower concentration range of hnRNP F, indicating that SAA10 binds more efficiently than the wild-type probe, consistent with our EMSA data in Figure 4. Meanwhile mutant SAA21, where the G12−14 region was replaced with cytosines, showed only 15–20% binding of the probe to hnRNP F, even at the highest hnRNP F concentrations. Therefore, although the G1−5 cluster in the GRS is important for hnRNP F binding, it is not sufficient to produce an interaction strong enough to withstand the filter binding assay. We conclude that the region at nucleotides 12–14 also plays a role in binding.
Taken together, the data show that hnRNP F and H2 have parallel sequence selectivity. From the data in Figure 5, the sigmoidal nature of the binding of both proteins with the wild-type substrate is apparent, whereas with the SAA20 mutant co-operativity is lost with both hnRNPs. The implications of the sequence selectivity observed in Figures 3, 4 and 5 on the co-operative binding properties seen in the shapes of the curves in Figure 5 for hnRNP F and H2 are addressed in the Discussion.
We see approx. 50% of the wild-type probe in Figure 5 remaining bound to protein after filtration, even at the highest concentrations of hnRNPs. Similarly, we were unable to shift all the RNA in an EMSA. While heterogeneity in the RNA preparation could account for this, a more likely explanation is, as discussed previously [25], that some complexes are not able to withstand the rigors of the assays and are not adequately retained on the filter, just as during electrophoresis they are not able to remain together when travelling through the pores of a gel. In the filter binding assay in Figure 5(C), in contrast with the EMSA data, where we saw a 2-fold higher binding of SAA10 than with wild-type GRS with hnRNP F, the difference from wild-type is less pronounced. This may be a reflection of the different requirements for complex stability in the two assays and/or the fact that the probe in Figure 4 was significantly shorter and less likely to be subject to shear forces. Although the substitution of cytosines for guanine residues in SAA21 could possibly lead to binding with the guanine residues in the G1−5 region, the Tm (melting temperature) of such a hairpin would be <12 °C and, since the binding assays were conducted at 30 °C, this is highly unlikely.
Mutations that decrease hnRNP H2/F affinities decrease poly(A) site use
When nine out of 14 nucleotides of the SVL GRS were deleted in a previous study, the observed in vitro 3′ processing efficiency fell approx. 2.5-fold [26]. In addition, the amount of specific polyadenylation complexes formed on the same mutant fell significantly [27], supporting the idea that binding of hnRNP H2 or F to the GRS increases the rate of complex formation, hence overall 3′ processing efficiency. Since hnRNP F/H depletion and add-back in vitro polyadenylation experiments had shown that hnRNP H2 acted positively, while hnRNP F acts in a negative fashion [13,15], we wanted to extend the findings of sequence specificity to the in vivo, cell-culture level, which had not been done before. The 224-nt wild-type, SAA20 and SAA10 SVL poly(A) regions were cloned into the regions 3′ of the SVL poly(A) site in the mammalian GFP expression plasmid pEGFP-C1 (Clontech). When GFP pre-mRNA was efficiently cleaved and polyadenylated, we expected to observe higher expression and more intense GFP fluorescence. Before testing the mutated GRS in the pEGFP plasmids, we wanted to make sure that the poly(A) site had a measurable effect in our gene expression assay. Because the pEGFP parent plasmid has a very strong CMV (cytomegalovirus) immediate-early promoter, it was possible that the overall expression of GFP in HEK-293T (human embryonic kidney) cells might have been so high that a single strong or weak poly(A) site might not have made a significant difference in expression.
To construct the baseline vector, pEGFP with no poly(A), the default poly(A) site of the GFP ORF was removed from the pEGFP-C1 plasmid. The 224-nt wtSVL poly(A) site was synthesized by PCR and inserted to replace the default poly(A) site of pEGFP-C1, creating pEGFP-wtSVL (see the Experimental section for details). Both plasmids were linearized before transfection and lack downstream non-consensus poly(A) signals.
HEK-293T cells were transfected, and, after 18–41 h of incubation with vectors, cells were fixed for quantitative flow-cytometry analysis. Numerous replications of this experiment showed a reproducible 9–10-fold induction of GFP fluorescence with the wtSVL samples compared with the nopoly(A) construct, as quantified by flow cytometry and summarized in Figure 6(A). Since >10000 events are measured for each flow-cytometric experiment, these differences are extremely significant.
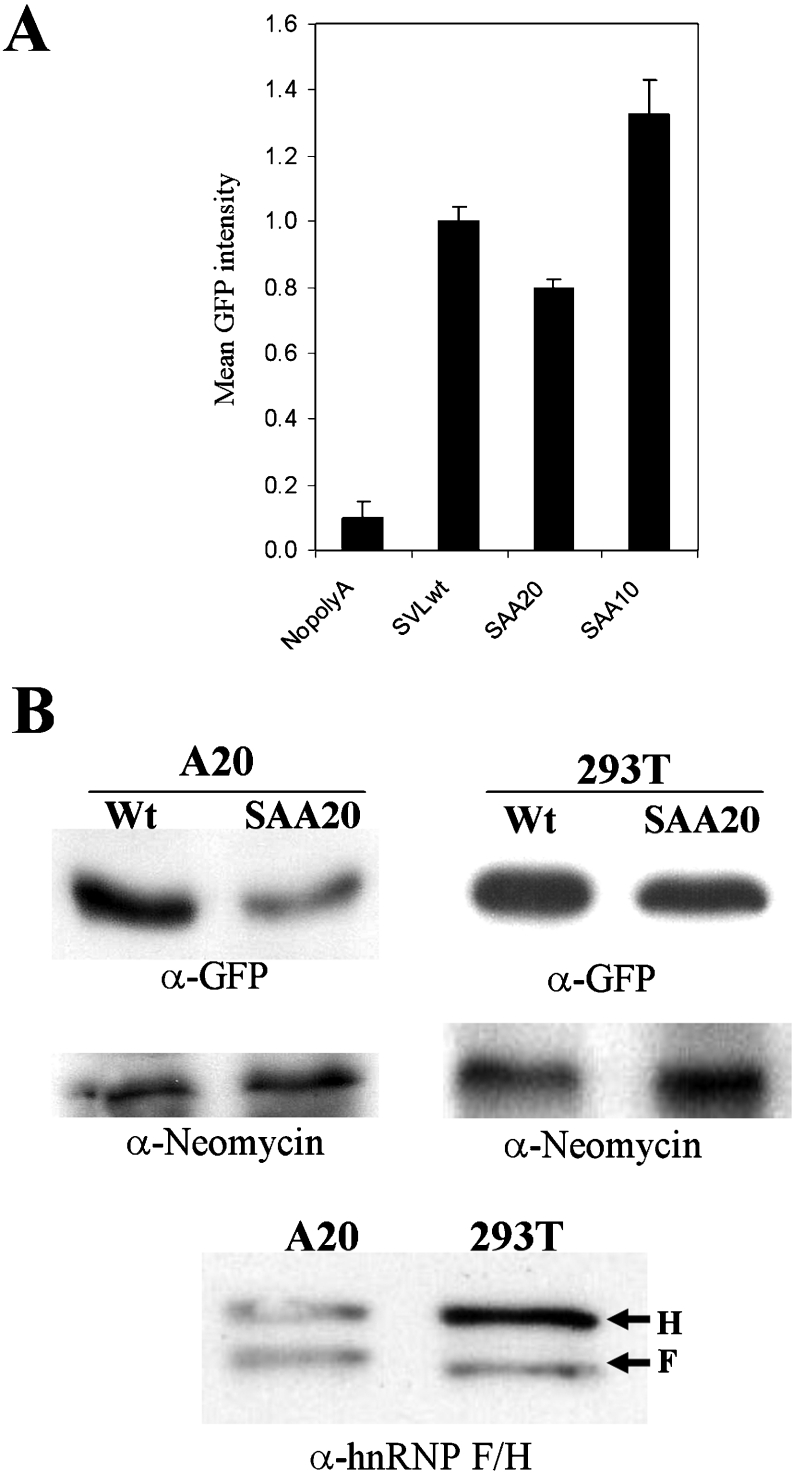
Figure 6. Decreasing the binding of hnRNP F or hnRNP H2 to the downstream region of the GFP transcript reduces the GFP expression in vivo.
(A) Mean GFP fluorescence intensities corresponding to pEGFP-nopoly(A) (NopolyA), pEGFP-wtSVL (SVLwt), pEGFP-SAA20 (SAA20) and pEGFP-SAA10 (SAA10) transfected HEK-293T cells were quantified by flow cytometry. The DsRed fluorescence from a separate plasmid served as a transfection control for normalization (results not shown). Each transfection was performed at least three times, and results are means±S.E.M. (B) GFP levels in pEGFP-wtSVL (Wt) or pEGFP-SAA20 (SAA20)-transfected A20 or HEK-293T cells were determined by Western blots with anti-GFP (α-GFP) antibody. Neomycin encoded on the GFP–plasmid was used for the protein blot loading control (α-Neomycin). Relative hnRNP H and hnRNP F levels in A20 and HEK-293T extracts were detected using a Western blot assay with anti-(hnRNP H) (H) and anti-(hnRNP F) (F) antibodies.
Mutations corresponding to the SAA20 and SAA10 GRS regions were inserted into the context of the 224-nt SVL construct by PCR and then cloned into the pEGFP vector. Final plasmids contain a single poly(A) site having either SVL with wild-type GRS or SVL plus GRS with the substitution of the SAA20 or SAA10 mutations.
Quantitative flow-cytometry results with the HEK-293T cell line showed a statistically significant decrease in GFP mean intensity of pEGFP-SAA20 and a significant increase with the SAA10-transfected cells (Figure 6A) relative to wtSVL respectively. Whenever there is more hnRNP H2/F binding (SAA10), more GFP is expressed, and, when there is less binding (SAA20), the opposite is true.
Level of GFP is related to GRS and levels of hnRNP H2/F
The effect of the SAA20 mutation on GFP expression was examined by Western blot analysis on the HEK-293T and A20 transfected cells. While transfected HEK-293T cells showed a less than 2-fold decrease in GFP, transfection of A20 cells, a B-cell lymphoma line, showed approx. 2–3-fold repression of GFP expression in the case of pEGFP-SAA20 (Figure 6B). The neomycin phosphotransferase gene is present in all our pEGFP plasmids; Western blot analysis of this protein serves as an internal transfection control (Figure 6B). Since GFP compared with control neomycin phosphotransferase levels are reduced with the SAA20 mutant relative to wtSVL, we concluded that weakening the hnRNP H2/F binding site decreases GFP expression significantly.
In view of the fact that the difference in expression of GFP between wtSVL and SAA20 was greater in the A20 cell line than in HEK-293T cells in the Western blot assay (Figure 6B), we speculated that the ratios of hnRNP H2 and hnRNP F might differ between the cell lines. We determined the relative levels of the two hnRNP proteins in these cell lines by Western blot analysis using an antibody that recognizes both proteins (Figure 6B). The HEK-293T cells have more hnRNP H than hnRNP F, whereas the A20 cells have comparable levels of both. In HEK-293T cells, hnRNP H may be present at high enough levels to allow some binding to our SAA20 mutant, thus raising GFP levels relative to that seen in the A20 cell line.
Down-regulation of hnRNP H2 decreases GFP gene expression
If hnRNP H2 acts in a positive manner on the wtSVL GRS, then down-modulating its levels in HEK-293T cells should result in decreased gene expression on a GFP reporter plasmid with an SVL poly(A) site and GRS downstream region. The hnRNP H-specific siRNA was targeted to a region where hnRNP H1 and H2 mRNAs are identical, but differ from hnRNP F mRNA (see the Experimental section). A scrambled oligonucleotide with no similarity to either hnRNP F or hnRNP H was used as a transfection control.
As predicted, down-regulation of hnRNP H protein expression resulted in decreased hnRNP H as assayed by Western blot (Figure 7A) and decreased GFP expression with the pEGFP-wtSVL plasmid in HEK-293T cells as assayed by flow cytometry (Figure 7B). Interestingly, whenever the pEGFP-SAA20 plasmid was assayed, the effect of down-regulation by the anti-hnRNP H siRNA was much larger (Figure 7C). Not only are the hnRNP H levels down 4–5-fold by virtue of the siRNA, but also, with the SAA20 mutation, hnRNP H2 binds with reduced affinity to the GRS. Therefore we conclude that both the GRS and the hnRNP H protein levels affect GFP gene expression.
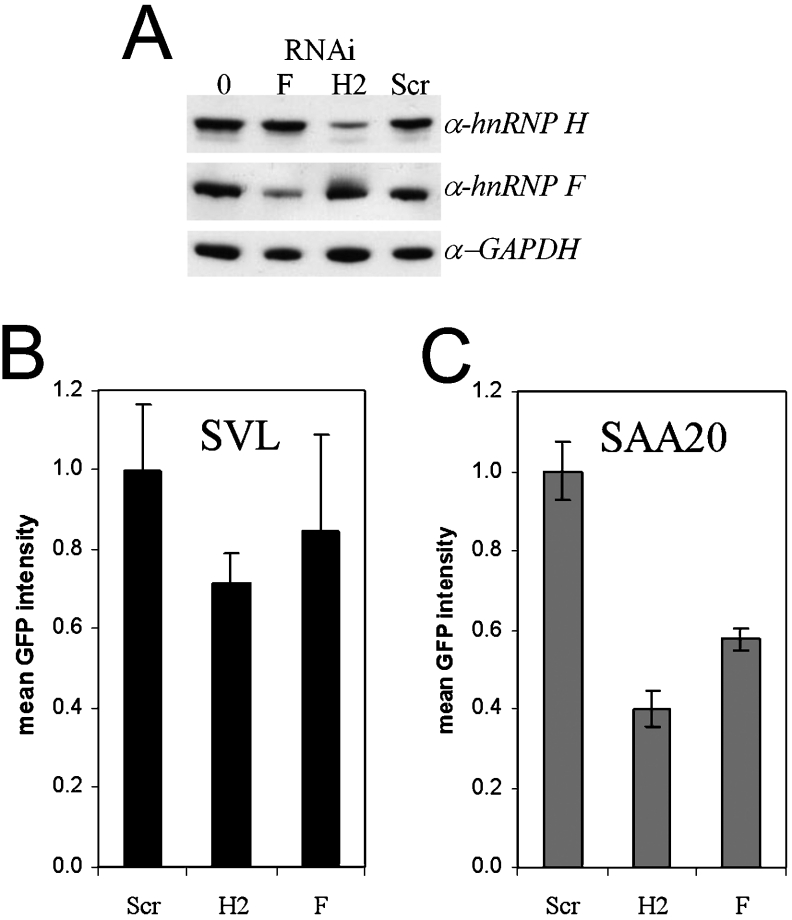
Figure 7. Effects of down-regulation of hnRNP F and hnRNP H by siRNAs on GFP expression.
(A) Western blots performed with anti-hnRNP H1/H2 (α-hnRNP H), anti-hnRNP F (α-hnRNP F) and anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase; α-GAPDH) antibodies. The lane marked 0 shows mock-transfected cells, F is hnRNP F siRNA, H2 is hnRNP H2 siRNA, and Scr (scrambled) is a negative-control siRNA that does not hybridize any known mRNA. (B, C) Quantification of GFP levels by flow cytometry. (B) shows the data from HEK-293T cells co-transfected with pEGFP-wtSVL, whereas, in (C), the results were obtained from cells transfected with pEGFP-SAA20 and the indicated siRNAs. Results are means±S.E.M.
Using siRNA that specifically knocks down hnRNP F protein expression (Figure 7A) and the GRS from the wtSVL sequence, GFP expression was not significantly affected (Figure 7B). However, when the siRNA to hnRNP F was used in conjunction with the SAA20 mutation that reduces hnRNP F binding, GFP expression was significantly reduced (Figure 7C). Therefore mutating the GRS to a lowered affinity for both hnRNPs had a more profound effect on GFP expression than did diminishing the amount of hnRNP F alone.
DISCUSSION
The two proteins hnRNP F and H2 share a high degree of similarity in organization, amino acid composition and, as we show in the present study, RNA sequence specificity; however, our results show that they differ from each other in at least two ways. First, hnRNP F inhibits the binding of CstF-64 to its substrate while hnRNP H2 does not. Secondly, the depletion of hnRNP F by RNAi, even in cells where F is already low, does not influence GFP expression and poly(A) site use substantially whereas depletion of hnRNP H2 does. When the hnRNPs are depleted and the GRS-binding site is weakened, there is a significant fall in GFP expression with blocking either protein.
The lack of a systematic mutational analysis had prevented a determination of the optimal RNA-binding sequence for hnRNP F or H2. By performing extensive mutational analyses, we conclude that a run of five guanine residues and a purine doublet/triplet starting approx. 5–6 nt downstream of the guanine core (G12−14) make up the consensus hnRNP F/H2-binding site(s). Multiple guanine residues form quaternary guanine structures consistent with our observation that it was necessary to disrupt the G1−5 run by introducing three substitutions in order to decrease binding to a point where it was undetectable in the EMSAs. The decreased binding of mutants SAA9, SAA12 and SAA13 relative to the enhanced binding in SAA10 and SAA11 indicate that a downstream adenine can enhance binding, but it should be near a short run of guanines for maximal binding in the G12−14 cluster. Changing the guanine residues to cytosine in SAA21 had a significant effect in decreasing binding. In Figure 8 various hnRNP F- or hnRNP H-binding sites are shown in individual genes that were published previously [16,28–35]. We found the G1−5 region and the downstream purine-rich cluster at nucleotides 12–14 in a majority of the reported hnRNP F or H sites by aligning them using the ClustalW software (http:www.ebi.ac.uk/clustalw, cited December 2004).
The hnRNPs are very abundant in the nucleus and there are no competing histones binding to RNA preventing the protein–RNA interactions. That is perhaps why the dissociation constants of hnRNP F and H2 proteins from the substrate (estimated at 121 and 37 nM respectively) are higher as compared with values with a typical transcription factor, generally in the single digit nanomolar range. The extensive similarities of the three RRMs of hnRNP F and H2 proteins led us to think that any differences in affinity between them may be due to the most C-terminal sequences present in hnRNP H2, but absent in hnRNP F. This C-terminal hnRNP H2-specific region may also be responsible for the differences we see between hnRNP F and H2 in their interactions with CstF-64 binding to RNA. While hnRNP F inhibits CstF-64 binding, either as a monomer or dimer, hnRNP H2, hnRNP H2–hNRP F or hnRNP H2–hnRNP H2 dimers allow the binding to occur. Heterodimers of hnRNP H and F have been implicated in other studies [16]. If the RRMs of hnRNP F were held in a different configuration because the C-terminal domain was truncated relative to hnRNP H2, this might block the adjacent GU-rich region of SVL which has been shown to be the CstF-64-binding site. The binding of CstF-64 to the GU-rich region involves a conformational change while the protein–RNA interface remains mobile, allowing it to bind to many variations of the uracil- or GU-rich sequence [36]. The hnRNP F nearby may impede this conformational change by binding to the substrate. Alternatively, hnRNP F at higher concentrations may multimerize and take up a bigger footprint on the RNA substrate.
Our data show that both hnRNP F and H2 appear to bind co-operatively to the wtSVL RNA substrate. We cannot rigorously distinguish between effects of protein dimerization or of two RNA sites in producing the sigmoidal shape of the curve. On EMSAs, as shown in Figure 3, we typically see a single band with either hnRNP F or hnRNP H2, which might imply only one protein per RNA. The fact that the binding of multi-RRM-containing proteins is co-operative and displays complex kinetics has been appreciated previously [4]. The crystal structure of PTB (polypyrimidine-tract-binding protein), another RRM-containing protein, indicates that all four of its RRMs bind RNA [37]. The hnRNP F and H2 proteins each have three RRMs; the first two of which are separated by only 14 amino acids, while the last is 100 amino acids downstream. In other studies, RRM sites separated by >60 amino acids were seen to have segregated affinities, while those closer than 60 amino acids showed affinities which do not add up to the product of the two affinities [4]. The SAA19 and SAA20 mutations have an impact on the G1−5 cluster of the GRS, resulting in a severe defect in binding to the upstream site with a loss in co-operativity, hence the hyperbolic curve with the SAA20 mutant in filter binding. The SAA21 mutation impacts on the guanine region at nucleotides 12–14 and substitution of cytosines for those purines reduces binding. The enhanced binding with mutants SAA10 and SAA11 is also consistent with a binding site at the end of the 14-nt GRS in the wild-type sequence that is enhanced by the presence of an alanine substitution. Taken together, these results suggest that strong binding requires co-operative interactions of either hnRNP H2 or hnRNP F to at least two or more sites on the GRS, G1−5 and G12−14.
We have verified the effect of sequence-specific binding of hnRNP F and H2 in vivo on polyadenylation and reporter expression by independent lines of evidence. Mutations in the GRS were placed in the region downstream of the SVL poly(A) site of a GFP reporter plasmid; transfection-based experiments showed that mutations in GRS can have positive or negative effects on GFP expression mediated through poly(A) site use. With the SAA20 mutation, which interrupts the G1−5 of GRS and reduces binding to hnRNP F and H2 in vitro, we saw reduced GFP expression in vivo. With SAA10, where in vitro binding of hnRNP F and H2 was increased, we saw more GFP expression in vivo. The complementary data supporting the role of sequence-specific hnRNP binding that influences poly(A) site use in vivo are the experiments in which RNAi mediated a down-regulation of hnRNP protein expression. Reducing hnRNP H2 expression with siRNA reduces GFP production slightly, but reducing the affinity of the RNA site for hnRNP H2 while simultaneously reducing the H2 levels with RNAi results in a significant decrease in GFP intensity. Reducing hnRNP F expression with siRNA does not increase expression with the wild-type GRS, which may seem paradoxical. However, while hnRNP F blocks CstF-64 binding, hnRNP F in the presence of hnRNP H2 allows it to occur, perhaps through dimerization of hnRNP H and F. When we reduce hnRNP F in these experiments, we are doing so in a HEK-293T cell which already has a higher amount of hnRNP H than hnRNP F; so the effects of further hnRNP F depletions may be negligible because the concentration of negatively acting hnRNP F is so low initially. Meanwhile, decreasing hnRNP F may reduce the number of positively acting hnRNP H–hnRNP F dimers. Thus blocking hnRNP F could, in fact, have a negative effect on gene expression. Only when both the concentration of hnRNP F and the affinity of the GRS site are reduced do we see a significant decrease in GFP expression. Not only do hnRNP F and H2 have a lower affinity for the SAA20 mutant than the wild-type, but also there is a 4–5-fold knockdown of hnRNP F and H protein in these siRNA experiments. The severely reduced hnRNP F and H2 binding to RNA resulted in a clear reduction of GFP expression. The GFP expression was not reduced to zero since the hnRNP proteins modulate polyadenylation but are not constitutively required for it to occur.
We have shown that hnRNP F and H2 proteins are modulators of gene expression through their sequence-specific effect on polyadenylation. We and others have shown that hnRNP F and H proteins are differentially expressed in normal and cancer tissues [13,38]. Almost 35% of the 219 genes surveyed in a random sample of mammalian poly(A) signals showed guanine-rich elements that were potential binding sites for hnRNP H or hnRNP F within 100 nt downstream [12]. The implications of our current results are that the hnRNP F or hnRNP H amounts in the cell and the binding affinity of the RNA sequence of a target gene itself may modulate the use of a poly(A) site in any gene and, in genes with multiple tandem poly(A) sites, such as the immunoglobulin heavy chains, may alter the choice of poly(A) sites.
Acknowledgments
This work was supported by grant # CA86433 to C. M. and DAMD #179919352 Training grant to S. A. A. We thank Dr Paula Grabowski for stimulating discussions.
References
- 1.Krecic A. M., Swanson M. S. HnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 2.Birney E., Kumar S., Krainer A. R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu H., Zhang W., Reed R. B., Liu W., Grabowski P. J. Mutations in RRM4 uncouple the splicing repression and RNA-binding activities of polypyrimidine tract binding protein. RNA. 2002;8:137–149. doi: 10.1017/s1355838202015029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shamoo Y., Abdul-Manan N., Williams K. R. Multiple RNA binding domains (RBDs) just don’t add up. Nucleic Acids Res. 1995;23:725–728. doi: 10.1093/nar/23.5.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen H. B., Vorum H. H., Dejgaard K., Liu X., Gromov P., Madsen P., Gesser B., Tommerup N., Celis J. E. Heterogeneous nuclear ribonucleoproteins (hnRNP) H, H′, and F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J. Biol. Chem. 1995;270:28780–28789. doi: 10.1074/jbc.270.48.28780. [DOI] [PubMed] [Google Scholar]
- 6.Honore B. The hnRNP 2H9 gene, which is involved in the splicing reaction, is a multiply spliced gene. Biochim. Biophys. Acta. 2000;1492:108–119. doi: 10.1016/s0167-4781(00)00092-0. [DOI] [PubMed] [Google Scholar]
- 7.Honore B., Rasmussen H. H., Vorum H., Dejgaard K., Liu X. D., Gromov P., Madsen P., Gesser B., Tommerup N., Celis J. E. Heterogeneous nuclear ribonucleoprotein-H, ribonucleoprotein-H′, and ribonucleoprotein-F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J. Biol. Chem. 1995;270:28780–28789. doi: 10.1074/jbc.270.48.28780. [DOI] [PubMed] [Google Scholar]
- 8.Matunis M. J., Xing J., Dreyfuss G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22:1059–1067. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grabowski P. J. A molecular code for splicing silencing: configurations of guanosine-rich motifs. Biochem. Soc. Trans. 2004;32:924–927. doi: 10.1042/BST0320924. [DOI] [PubMed] [Google Scholar]
- 10.Sironi M., Menozzi G., Riva L., Cagliani R., Comi G. P., Bresolin N., Giorda R., Pozzoli U. Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res. 2004;32:1783–1791. doi: 10.1093/nar/gkh341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagga P. S., Ford L. P., Chen F., Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arhin G. K., Boots M., Bagga P. S., Milcarek C., Wilusz J. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 2002;30:1842–1850. doi: 10.1093/nar/30.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veraldi K. L., Arhin G. K., Martincic K., Chung-Ganster L. H., Wilusz J., Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 2001;21:1228–1238. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen F., MacDonald C., Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bagga P., Arhin G., Wilusz J. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 1998;26:5343–5350. doi: 10.1093/nar/26.23.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou M. Y., Rooke N., Turck C. W., Black D. L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takagaki Y., MacDonald C. C., Shenk T., Manley J. L. The human 64-kDa polyadenylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1403–1407. doi: 10.1073/pnas.89.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veraldi K., Edwalds-Gilbert G., MacDonald C., Wallace A., Milcarek C. Isolation and characterization of polyadenylation complexes assembled in vitro. RNA. 2000;6:768–777. doi: 10.1017/s135583820099246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian Z., Wilusz J. An RNA binding protein specifically interacts with a functionally important domain of the downstream element of the SV40 late polyadenylation signal. Mol. Cell. Biol. 1991;11:5312–5320. doi: 10.1128/mcb.11.10.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilusz J., Felig D. I., Shenk T. The C proteins of heterogeneous nuclear ribonucleoprotein complexes interact with RNA sequences downstream of polyadenylation cleavage sites. Mol. Cell. Biol. 1988;8:4477–4483. doi: 10.1128/mcb.8.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spear M. A. Efficient DNA subcloning through selective restriction endonuclease digestion. BioTechniques. 2000;28:660–662. doi: 10.2144/00284st01. [DOI] [PubMed] [Google Scholar]
- 22.Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baird G. S., Zacharias D. A., Tsien R. Y. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martincic K., Campbell R., Edwalds-Gilbert G., Souan L., Lotze M., Milcarek C. Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11095–11100. doi: 10.1073/pnas.95.19.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfeffer S. R., Stahl S. J., Chamberlin M. J. Binding of Escherichia coli RNA polymerase to T7 DNA: displacement of holoenzyme from promoter complexes by heparin. J. Biol. Chem. 1977;252:5403–5407. [PubMed] [Google Scholar]
- 26.Cooke C., Hans H., Alwine J. C. Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol. 1999;19:4971–4979. doi: 10.1128/mcb.19.7.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooke C., Alwine J. C. Characterization of specific protein–RNA complexes associated with the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol. 2002;22:4579–4586. doi: 10.1128/MCB.22.13.4579-4586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caputi M., Zahler A. M. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 2002;21:845–855. doi: 10.1093/emboj/21.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagani F., Buratti E., Stuani C., Baralle F. E. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J. Biol. Chem. 2003;278:26580–26588. doi: 10.1074/jbc.M212813200. [DOI] [PubMed] [Google Scholar]
- 30.Fogel B. L., McNally M. T. A cellular protein, hnRNP H, binds to the negative regulator of splicing element from Rous sarcoma virus. J. Biol. Chem. 2000;275:32371–32378. doi: 10.1074/jbc.M005000200. [DOI] [PubMed] [Google Scholar]
- 31.Hastings M. L., Wilson C. M., Munroe S. H. A purine-rich intronic element enhances alternative splicing of thyroid hormone receptor mRNA. RNA. 2001;7:859–874. doi: 10.1017/s1355838201002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacquenet S., Mereau A., Bilodeau P. S., Damier L., Stoltzfus C. M., Branlant C. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of tat mRNA and binds protein hnRNP H. J. Biol. Chem. 2001;276:40464–40475. doi: 10.1074/jbc.M104070200. [DOI] [PubMed] [Google Scholar]
- 33.Chen C. D., Kobayashi R., Helfman D. M. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing in the rat β-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romano M., Marcucci R., Buratti E., Ayala Y. M., Sebastio G., Baralle F. E. Regulation of 3′ splice site selection in the 844ins68 polymorphism of the cystathionine β-synthase gene. J. Biol. Chem. 2002;277:43821–43829. doi: 10.1074/jbc.M208107200. [DOI] [PubMed] [Google Scholar]
- 35.Caputi M., Zahler A. M. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J. Biol. Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- 36.Canadillas J. M. P., Varani G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 2003;22:2821–2830. doi: 10.1093/emboj/cdg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpson P. J., Monie T. P., Szendroi A., Davydova N., Tyzack J. K., Conte M. R., Read C. M., Cary P. D., Svergun D. I., Konarev P. V. Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure. 2004;12:1631–1643. doi: 10.1016/j.str.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 38.Honore B., Baandrup U., Vorum H. Heterogeneous nuclear ribonucleoproteins F and H/H′ show differential expression in normal and selected cancer tissues. Exp. Cell Res. 2004;294:199–209. doi: 10.1016/j.yexcr.2003.11.011. [DOI] [PubMed] [Google Scholar]