Abstract
Messenger RNAs that have the stability determinants, adenylate uridylate-rich elements (AREs), in their 3′ untranslated region (UTR) code for key products that regulate early and transient biological responses. We used a computational laboratory approach for amplification of large, including full-length, protein-coding regions for ARE genes. Statistical analysis of the initiation regions in the 5′ UTR of ARE-mRNAs was performed. Accordingly, several 5′ primers and a single universal 3′ primer that targeted the initiation consensuses and ARE regions, respectively, were designed. Using optimized conditions, the primers were able to enrich and amplify large protein-coding regions for the ARE gene family. The selective amplification of ARE cDNAs was verified using specific polymerase chain reactions (PCRs) to known ARE mRNA molecules and monitoring the abundance of the non-ARE β-actin signal. A mini-library from the amplified ARE products was constructed for further confirmation of ARE selection. Distinct ARE amplified cDNA pools were selectively generated by distinct 5′ primers. The biological utility of the method was shown with differential display. The up-regulation of several ARE-mRNAs, including the full-length coding region of the small inducible cytokine A4 (SCYA4) gene, was shown in endotoxin-stimulated monocytic cells. The integrated computational and laboratory approach should lead to enhanced capability for discovery and expression analysis of early and transient response genes.
A subset of the genome that is essential in cellular growth and in early and transient response to exogenous agents such as inflammatory inducers, growth stimuli, stress, and microbes is the adenylate uridylate (thymidylate)-rich element (ARE)-containing gene family. Our recent analysis showed that the ARE-gene family encodes a large number of previously unrecognized ARE-mRNAs and constitutes as much as 8% of human mRNAs (Bakheet et al. 2001). In addition, the ARE-gene family contains functionally diverse proteins that mediate different biological processes including cell growth and differentiation, transcription, innate immune response, inflammation, signal transduction, and many others (Bakheet et al. 2001). A common trait of the ARE-mRNAs is that they are expressed early and transiently. In a cDNA microarray study using B cell lymphoma and peripheral blood mononuclear cells (Lam 2001), it was concluded that our computationally extracted ARE motif (Bakheet et al. 2001) was preferentially found in the most unstable mRNA (<2 hr) and observed with decreasing frequency in stable mRNAs (>8 hr). This is opposite to that of non-ARE genes in which stable mRNA constituted the majority (60%) of mRNAs studied (Lam 2001). Stabilization of the ARE mRNAs can cause prolonged responses that may subsequently lead to diseased states. It has been shown that certain AREs act as instability determinants (Lagnado et al. 1994; Chen and Shyu 1995). For instance, the stable β-globin mRNA was rendered unstable when its 3′ untranslated region (UTR) was replaced with the GMCSF multiple ARE 3′ UTR (Shaw and Kamen 1986), whereas the unstable interleukin-1β mRNA was rendered stable when AREs were removed (Kastelic et al. 1996). Despite the accumulating evidence of the functional role of AREs in mRNA stability, the repertoire of the ARE genes and their regulatory pathways remains largely unknown.
Current approaches in gene discovery and expression profiling methods have several limitations. Many methods yield partially informative sequence data such as expressed sequence tag (EST) sequencing (Adams et al. 1991), degenerate PCR, serial analysis of gene expression (SAGE) (Velculescu et al. 1995), and conventional differential display (Liang and Pardee 1992). Also, there are methods that require previous presence of sequences such as cDNA and oligonucleotide microarrays (Duggan et al. 1999) and yield overwhelming technical and analytical tasks that arise from genome-wide analysis. In addition, these techniques are biased toward a certain threshold of mRNA abundance. Gene prediction approaches from the human genome project using computer programs have several limitations, such as the variable degree of exon locations and accuracy and the fact that cDNA clones are not available for study of the protein function. Thus, strategies are needed to address these limitations. One solution is the use of bioinformatics approaches to facilitate laboratory methods for targeting informative protein-coding regions. In this paper, we targeted the ARE gene family, a subset of the genome that is functionally and structurally related. This exercise was facilitated by different bioinformatics programs that included database sequence retrieval, UTR assembly and alignment, and statistical analysis. A system of gene discovery and expression analysis integrated with computational means leading to the amplification of the ARE-mRNA repertoire and its full-length protein-coding sequence is presented here.
RESULTS
ARE mRNA Sequence and Initiation Site Analysis
A total of 605 ARE-mRNA sequences that include 3′ UTR, full-length CDS, and at least 10 bp of 5′ UTR were obtained from the AU-rich element-containing mRNA database (ARED) (Bakheet et al. 2001) by use of the Assemble program (GCG). The initiation context sequences in the 5′ UTR, that is, those that flank the start codon, ATG, in the ARE-mRNA database were analyzed. It has been reported that the initiation regions contain conserved elements that are important in translation (Kozak 1987a,b). Thus, we chose this region to design 5′ primers for use in a PCR-based protocol. Sequences were divided into 16 subsets by using the formula, NATGN, where N = A or C or G or T. This is followed by alignment of the truncated 5′ UTR (−7 bp, ATG, +2 bp). Sixteen consensus patterns at the certainty level of 75% at each position were derived from the alignment (Table 1). The overall consensus initiation site in the ARE-mRNAs was SSMAMSATGRM at 50% certainty level at each position.
Table 1.
Consensus Sequences of Initiation Context Sequences in Human ARE mRNAs
| Aa | BHDVMMAATGAV | Ag | BVRMAATGGV |
| Ca | BSHMRVCATGAV | Cg | VVVVRSCATGGM |
| Ga | HBVVRVGATGAD | Gg | BVVSRVGATGGM |
| Ta | BDDVRHTATGAM | Tg | VDBHRBTATGGM |
| Ac | HDDVRBAATGCD | At | DRBVRMAATGTY |
| Cc | VRSVRMCATGCB | Ct | BVBMRYCATGTS |
| Gc | SSBBRMGATGCB | Gt | VDBVRRGATGTY |
| Tc | VBDWWRTATGCM | Tt | DVBVWDTATGTY |
Truncated 5′ UTRs regions of 605 ARE mRNAs (a subset of ARED; Bakheet et al. 2001) were computationally extracted as explained in the Methods second. They included at least −7 bp, ATG, +2 bp sequence and subsequently stratified into 16 subsets based on the formula NATGN (underlined). The truncated 5′ UTR regions in each subset were aligned using PileUp (GCG program), and consensus sequences were obtained using Consensus program (GCG) at 75% certainty level at each position. Letter codes follow ambiguous DNA IUB codes, for example, N = A, C, G, or T.
UTR, untranslated region; ARED, AU-rich element-containing mRNA database.
Statistical analysis of the 16 10-mer (−6 ATG, +1) consensus sequences was performed (Table 2). The most common consensus in initiation regions was Cg consensus VVVVR SCATGGM (Table 2), which occurs in 22% of all ARE-mRNA sequences analyzed. Other frequent initiation consensuses were Ca, Ag, and Gg (Tables 1 and 2); each accounts for 9% to 10% of all ARE mRNAs. Not all consensuses were unique to the initiation regions; depending on which consensus, there were varying degrees of internal sites in addition to the initiation region. The most common consensus sequence around any ATG was Aa consensus (Tables 1 and 2) that exists in 35% of the entire ARE-mRNA molecules, whereas the least occurring consensus sequences were those that were flanked by T base upstream of ATG, for example, Ta, Tc, Tg, and Tt consensuses (Table 2). The highest proportion of consensus in initiation regions in any subset was Gc consensus in which 71% of the sites (initiation plus internal) were initiation sequences. The consensus site per mRNA (number of total consensus/number of mRNAs, Table 2) ranged from 1.0 to 1.65.
Table 2.
Statistical Analysis of 10-mer-Initiation Region Consensus
| Subset | % Total ARE mRNAa | No. total consensus sitesb | Initiation sites (% of consensus sites)cd | No. ARE mRNA (% total)fe |
|---|---|---|---|---|
| Aa | 5 | 346 | 29 (8%) | 210 (35%) |
| Ca | 9 | 175 | 53 (30%) | 147 (25%) |
| Ga | 5.3 | 142 | 31 (22%) | 126 (21%) |
| Ta | 4.1 | 59 | 24 (41%) | 57 (9%) |
| Ac | 2.2 | 214 | 13 (6%) | 148 (25%) |
| Cc | 5.5 | 67 | 32 (48%) | 65 (11) |
| Gc | 6.4 | 53 | 37 (71%) | 52 (9%) |
| Tc | 2.2 | 98 | 13 (13%) | 80 (13%) |
| Ag | 9.7 | 243 | 56 (23%) | 172 (29%) |
| Cg | 22.4 | 225 | 130 (58%) | 170 (28%) |
| Gg | 10.4 | 217 | 60 (28%) | 175 (29%) |
| Tg | 3.8 | 91 | 22 (24%) | 59 (10%) |
| At | 3.4 | 64 | 20 (31%) | 66 (11%) |
| Ct | 5.5 | 153 | 32 (21%) | 134 (22%) |
| Gt | 3.3 | 177 | 19 (11%) | 128 (21%) |
| Tt | 1.4 | 43 | 8 (19%) | 43 (7%) |
Consensuses, based on the 16 divisions from Table 1, were statistically analyzed in relation to the overall 605 ARE mRNA entries. Each subset contains the ARE mRNAs in which consensuses were derived from their initiation regions; their percentages were given. The 10-mer consensus of each subset was used in the FindPattern program to search matching patterns in the overall 605 ARE mRNA database.
Total number of matching sites including both initiation and internal sites in ARE mRNA sequences.
Number of consensus sites unique to the initiation regions that also equals the ARE mRNAs in each subset.
Percentages of consensuses in the initiation sites compared with all sites (initiation and internal sites) at each subset.
Number of sequences of fpercentages of the overall 605 ARE mRNA sequences that contained the consensus pattern (both initiation and internal).
Ten nucleotides from each consensus sequence were used for incorporation in the 5′ primers. The 5′ primers were designed with two parts: one variable part of 10-mer, which incorporated consensus sequence (−6 bpATG1 + bp; Table 1), whereas the core part contained fixed sequences. Thus, each primer of the 16 primers (Tables 1 and 2) together with the single 3′ ARE primer targets a subset of ARE cDNAs in a single PCR reaction.
Taq-Mediated Amplification of ARE cDNA
The monocytic leukemia cell line, THP-1, was used as a cellular study model. It is a tumor cell line that corresponds to immature monocytic cells and constitutively produces the ARE mRNAs, interleukin-8 (IL-8) and tumor necrosis factor (TNF-α), and non-ARE mRNA, IL-8 receptor (IL-8R) (Khabar et al. 1997; Murayama et al. 1997; Al-Humidan et al. 1998). In many of the experiments, the cells were treated with both lipopolysaccharide (LPS), a potent inducer of cytokines, and cycloheximide (CHX). CHX blocks the synthesis of proteins, enhances expression of early response genes, and increases stability of the transient ARE mRNAs (Shaw and Kamen 1986; Reeves and Magnuson 1990).
By use of a universal 3′ primer that targets the ARE region and one of the 5′ primer set, selective amplification of ARE cDNA was achieved using optimized conditions of Taq-derived amplification (termed here ARE-cDNA PCR). The goal of ARE-cDNA PCR is to amplify the typical ARE-containing mRNA sequences (ARE mRNAs) with suppression of the amplification of non-ARE mRNA sequences. To verify the selective amplification of ARE cDNAs, we subsequently set up second specific PCR reactions for several examples of ARE molecules: IL-8 mRNA, TNF-α mRNA, and c-fos mRNA, each having different lengths of the pentamer repeat ATTTA. The 5′ primers for the specific PCRs were designed at or near the initiation sites so that amplification is enriched toward the large (i.e., containing the full-length regions) and not the shorter amplified ARE products, if any. The non-ARE messages of β-actin and IL-8R that contain a single ARE pentamer in a non-ARE context region in the 3′ UTR were used as controls. The choice of these rigorous controls was to monitor the stringent selective amplification of the typical ARE-cDNAs but not non-ARE-cDNAs. The second specific PCR was performed under relatively stringent conditions, for example, only 4 ng of ARE cDNA and the use of low dNTPs concentration and cycle number within the exponential phase of amplification to allow semiquantitative comparison and to eliminate or minimize amplification of original cDNA carried over from original cDNA template. Thus, the specific signals would be attributed to the amplification of the coding region containing products in the ARE-cDNA PCR, as described below.
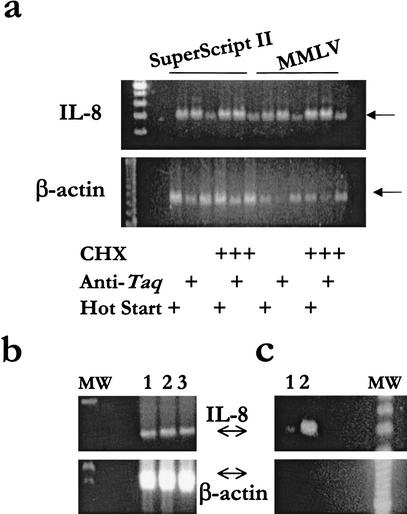
Optimum conditions in the ARE-cDNA PCR were initially fine-tuned from many trials that included amount of RNA, type of reverse transcriptase (RT), amount of input cDNA, primer concentration, type of Taq enzyme, annealing temperatures, and start conditions. We observed that the optimum selectivity for amplification of the ARE-cDNA of IL-8 was dependent on the use of CHX and start condition of PCR using either SuperScript II or Moloney murine leukemia virus (MMLV) (Fig. 1a). The optimum selectivity, that is, the specific amplification of ARE-cDNA was verified by observing the enhanced IL-8 amplified signal and the minimum or lack of β-actin cDNA signal. The results showed that CHX treatment of the cells before RNA extraction increased the ARE cDNA signal as expected (Fig. 1a). Also, ARE-cDNA PCR was optimal with the use of anti-Taq-mediated start conditions when compared with direct hot start or regular PCR. Subsequently, we used the optimum conditions of the ARE-CDNA PCR, which were the adoption of CHX treatment of the cultured cells before RNA extraction, and the use of anti-Taq-mediated PCR reaction (Fig. 1). In comparison with regular abundance of IL-8 and β-actin cDNA signals as observed by RT-PCR (Fig. 1b), an almost reversal of abundance was achieved with the optimized ARE-cDNA PCR. The specific amplification of IL-8 that resulted in a strong signal was not attributable to original cDNA carried over from the original cDNA (40 ng), used in the ARE-PCR, to the second specific PCR (Fig. 1c). The residual and minimum β-actin signal was apparently from original cDNA carried over to the second specific PCR and not from ARE-cDNA PCR (Fig. 1c). The β-actin cDNA amplification can also be eliminated by dilution, for example, 1/10, of the ARE-cDNA PCR into a second ARE-cDNA PCR (data not shown). In addition, we used PCR with specific primers to the non-ARE IL-8 receptor mRNA, which resulted in no IL-8 receptor specific signals (data not shown).
Figure 1.
Selectivity of the ARE mRNA, IL-8, and suppression of the abundant β-actin mRNA. Total RNA samples (1 μg) from THP-1 or from LPS/cycloheximide- (CHX) treated THP-1 cell line were extracted and subjected to reverse transcriptase (RT) using either Moloney murine leukemia virus (MMLV) or SuperScriptII. Forty ng cDNA was used for the ARE-cDNA PCR at conditions of 10 μM dNTP, 1 Taq U enzyme, and 1 μM of primers. One of the 5′ primers, Ca (Table 1), which contains computationally derived consensus that targets the initiation sites of a subset of ARE-mRNAs that includes IL-8 mRNA, was used along with the 3′ ARE primer that targets the 15-mer ARE region. (a) Aliquots (2 μL) of the amplified ARE products were subjected to relatively stringent polymerase chain reaction (PCR) specific to IL-8 and β-actin (50 μM dNTPs and 25 cycles). (b) Specific PCR to either IL-8 or β-actin was performed to view the relative abundance of the ARE mRNA IL-8 versus the housekeeping β-actin mRNA. Total RNA was used from three different cell lines (1, THP-1; 2, Hela; and 3, WISH). (c) An amount of 4 ng, which mimics the amount of cDNA carried over from the original ARE-cDNA tubes to the second specific PCR tubes, was used as a template. This template was used for second specific PCR for IL-8 and β-actin without the use of ARE-cDNA PCR (lane 1) or with the use of ARE-cDNA PCR (control, lane 2). Arrows indicate the predicted size of IL-8 (289 bp) and β-actin (642 bp). MW: 100-bp size marker.
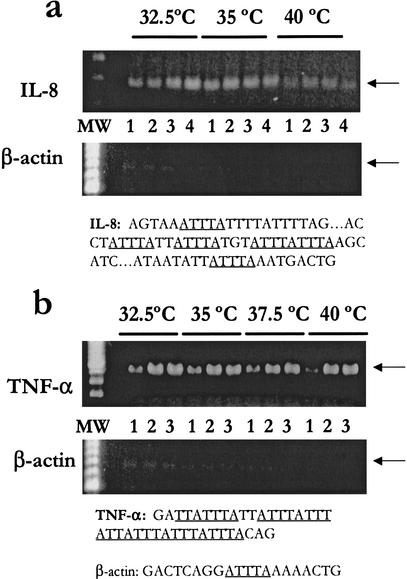
The effect of the initial annealing temperatures on the amplification of cDNAs with different ARE structures was studied (Fig. 2). The results showed that small differences in ARE annealing temperatures, that is, during the first four cycles, had significant effects in the case of IL-8, which has discontinuous multiple nonamers, TTATTTAWW (Fig. 2a) but not with TNF-α, which has continuous overlapping multiple nonamers (Fig. 2b). In other words, IL-8 has two overlapping pentameric repeats (ATTTATTTA), whereas TNF-α has five overlapping pentameric repeats. The normally abundant β-actin signal was largely suppressed in all lanes because the optimized conditions adopted from previous experiments were used. The optimum ARE annealing temperature for ARE-cDNA PCR in regard to selectivity of IL-8 amplified products was 35°C when compared with ARE annealing temperatures of 40°C and 32.5°C (Fig. 2a). In contrast, there were no significant effects of variations in the initial annealing temperature (32.5–40°C) of the ARE-cDNA PCR on amplification of continuous multiple ARE-cDNA as in the case of TNF-α (Fig. 2b). Thus, the 15-mer two overlapping nonamers in the 3′ ARE primer appeared to anneal to multiple targets of two overlapping nonamers (more than three pentameric repeats), leading to enhanced amplification of TNF-α. Unlike the case with IL-8 cDNA, amplification at lower initial temperature still led to significant TNF-α signals as a result of the presence of multiple (more than six) partial continuous and overlapping repeats (Fig. 2b). Temperatures as high as 45°C were also slightly tolerated in the case of TNF-α cDNA amplification, and amplification was decreased dramatically at 50°C, whereas no specific amplification was seen at 55°C (data not shown). The minimum number of cycles required for optimum PCR signal for TNF-α and IL-8 was 25; the increasing number of cycles did not result in further improvement of signals, indicating that the amplification was above exponential linearity at a cycle number higher than 25 (Fig. 2, lanes 1–3). In all of the experiments, DNA contamination was monitored by lack of larger PCR products because primers for the specific PCRs were designed to span more than one exon.
Figure 2.
Effect of initial annealing temperature and number of cycles on selectivity of the discontinuous and continuous ARE amplified products. Total RNA samples (1 μg) from LPS and CHX-treated THP-1 cell line were extracted and subjected to RT using MMLV. Forty ng cDNA was used for the ARE-cDNA PCR using the 5′ primer Ca (Table 1) and the 3′ ARE primer using different initial annealing temperatures (first four cycles) followed by a different number of cycles (lane 1, 20 cycles; lane 2, 25 cycles; lane 3, 30 cycles; lane 4, 35 cycles) at high annealing temperature (60°C). Aliquots of the amplified ARE products were subjected to PCR at stringent conditions specific to IL-8 (a) or TNF-α (b) in addition to β-actin to monitor selectivity of ARE-mRNA amplification. The specific amplified product of IL-8 is not attributable to cDNA carryover from original cDNA because PCR from the amount of carryover cDNA (4 ng) failed to show detectable IL-8 and TNF-α messages at the same PCR conditions (data not shown). Arrows indicate the predicted size of IL-8 (289 bp), TNF-α (548 bp), and β-actin (642 bp). MW: 100-bp size marker.
Mini-Libraries and Random Cloning and Sequencing
To show the utility of the computationally facilitated ARE-cDNA PCR, and to analyze and confirm the amplified sequences, we adopted a random cloning and sequencing approach. The random cloning and sequencing approach was performed by construction of a mini-library in which the amplified ARE products from ARE-cDNA PCR were cloned into pUC19 or pCR2.1 vectors, and clones were randomly picked for sequencing. We also used the gel-format differential display with modifications to display the amplified ARE cDNAs (details below). Sequence data confirmed the presence of AREs with different lengths that were dependent on the initial annealing temperature. Most of these partial cDNA fragments (57%) are novel, that is, 23 of the 30 cDNAs are uncharacterized, as determined by BLAST search against GenBank and EST databases (Table 3). Also, this indicates that enrichment of transient rare ARE-mRNAs were made possible by this method. Among the previously known cDNA fragments, several sequences corresponded to known cDNAs (Table 3) having a 15-bp ARE pattern consisting of one to four overlapping nonamers. Among them, several were noted to be typical ARE-mRNAs, interleukin-1β, c-fos, and plasminogen activator inhibitor protein (PIA2). In addition, there were several matches to the human dbEST databases.
Table 3.
Summary of Identity of ARE-Expressed Sequencesa
| Total: 30 distinct clones |
| cDNA/mRNA with known function (7) |
| Interleukin-1β (4 AREs) |
| c-fos (3 AREs) |
| Hypoxia-induced factor α (3 AREs) |
| Plasminogen activator inhibitor protein (2 AREs) |
| Diacylglycerol kinase delta (1 ARE) |
| Amyloid beta (A4) precursor (1 ARE) |
| Small inducible cytokine A4 (1 ARE) |
| Matched EST hits (9) |
| Matched EST + cDNA (16) |
| No significant matches (14) |
ARE-PCR products generated from random cloning of pUC19 ARE mini-library or excised from ARE-cDNA differential display gel were sequenced. Sequence identities were generated using BLAST search of GenBank and hEST databases. ARE refers to the 13-bp computationally derived ARE motif consensus, that is, WW WTATTTATWWW Bakheet et al. 2001).
The accession numbers BG604230 to BG604251 were obtained from dbEST for the sequence data in this article.
ARE, adenylate uridylate-rich element; EST, expressed sequence tag; PCR, polymerase chain reaction.
Although the mini-library from the ARE-cDNA PCR was constructed for the purpose of validating the ARE selectivity, we assessed the size distribution in a sample of 10 randomly picked clones. Despite the fact that insert size can be influenced by library construction methods and size exclusion procedures, the mini-library produced an average of 0.9 kb and a range of 800–1100 bp among nine clones; one clone had a size of 400 bp. These mini-libraries are within the size range and average insert size that are comparable or superior to those reported by others in constructing PCR-based mini-libraries (Bertiol et al. 1994; Peterson et al. 1998).
Primer-Target Annealing Characteristics and Size Distribution of Amplified ARE-cDNA Products
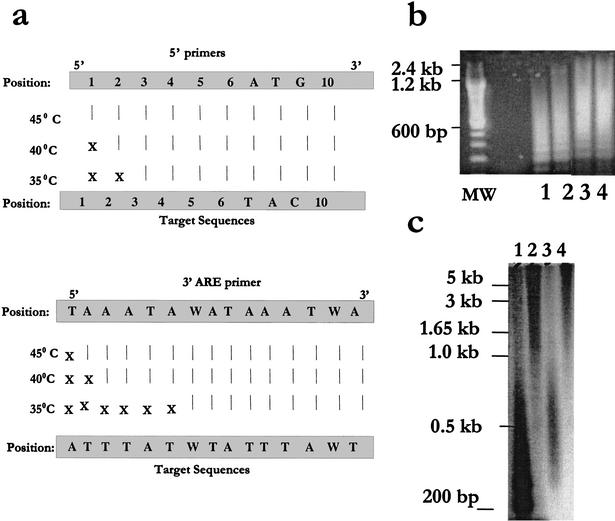
The annealing specificity of our primers under different annealing temperatures (35°C, 40°C, and 45°C) was assessed using the optimized ARE-cDNA PCR conditions, including the use of anti-Taq start conditions. The use of anti-Taq start conditions minimized primer-target mispriming when compared with regular or hot start conditions (data not shown). We analyzed 26 target sites for the 3′ and 5′ primers (from 13 ARE-PCR products) that were retrieved by BLAST search and found to be portions of seven known genes, six EST records, and seven hits in the human genome project database with no characterized cDNA/EST among the 30 sequences in Table 3. The ARE 3′ primer annealed longer than the 5′ primer; approximately two to three AT bases for each one G/C base in the 5′ primer. There was complete homology between at least eight bases at the 3′ end of both the 5′ and the ARE 3′ primers and the target region, whereas mispriming occurred only toward the 5′ end (Fig. 3a). In addition, mispriming was reduced with higher annealing temperatures (Fig. 3a). Figure 3a also shows and confirms the findings of the specific PCRs performed with IL-8, TNF-α, and c-fos: The higher the annealing temperatures, the higher proportion of cDNAs with longer ARE stretches were amplified, whereas those cDNAs with shorter stretches were not efficiently amplified. Thus, we have chosen to perform most of our subsequent ARE-cDNA PCRs with a temperature of 40°C to 42.5°C, which allowed the amplification of a 13-bp ARE pattern with at least two overlapping ATTTA repeats.
Figure 3.
Characteristics of primer-target annealing and size distribution. (a) The annealing specificities of our primers under different annealing temperatures (35°C, 40°C, and 45°C) at the optimized ARE-cDNA PCR conditions (40 ng cDNA template, anti-Taq start conditions, 1 U Taq, 50 μM dNTP, and 30 cycles of amplification) were assessed. Twenty-six target sites for the 3′ and 5′ primers from 13 ARE-PCR products that were found to be portions of several known genes and genome/EST hits retrieved by BLAST search (Table 3) were evaluated. Vertical lines designate perfect matches (all at the 3′ end of the primer), and X designates mismatches. (b) Size distribution of amplified ARE-cDNA products. To visualize ethidium bromide smear for size distribution in agarose gels, amplified ARE-cDNA products (using an extension time of 3 min) were subject to a second ARE-cDNA PCR (Fig. 1) using 40μM dNTP. Samples were from THP-1 (lanes 1 and 3) and THP-1 treated with LPS + CHX (lanes 2 and 4) using two different 5′ primers (Ac and Gg; Table 1). MW: size markers. (c) Size distribution of the amplified radio labeled ARE-cDNA products. ARE-cDNA PCR from THP-1 samples, labeled with (α-32P)-dCTP, was performed at the optimized conditions (above) in the absence of (lanes 1 and 3) or presence of 0.1 U of pfu (lanes 2 and 4). Southern blotting was performed and autoradiogram was subsequently developed. Column purification was applied (lanes 3 and 4) to remove access dNTPs, primers, and short PCR products.
In addition to the fact that the ARE amplified products containing the full-length coding regions (1.0, 1.2, and 1.8 kb) that belong to the IL-8, TNF-α, and c-fos cDNAs, respectively, were efficiently amplified as described previously, the size distribution of the ARE-PCR products was assessed. The ARE-cDNA products were visualized by ethidium bromide smear of agarose gel showing a range of up to 3.0 kb when the extension time was increased to 3 min (Fig. 3b). The agarose gel smear was further evaluated by using radioactively labeled ARE-PCR products (Fig. 3c). The size distribution of the amplified ARE-cDNA products ranged from 200 bp to more than 1.65 kb using the standard ARE-cDNA PCR (Fig. 3c, lane 1). The proofreading polymerase, pfu, an enzyme that is suitable for generating long PCR products, was also added in small amounts (0.1 U) to the ARE-cDNA PCR to improve the size distribution; the extension time was increased to 3.5 min. As a result, the size distribution favored longer ARE-PCR products, ranging from 0.5 kb to more than 5 kb (Fig. 3c, lane 2). Column purification, which was applied before gel electrophoresis to remove access dNTPs, primers, and short PCR products, resulted in size distribution ranging from ∼300 bp to 1.5 kb and 0.8 kb to >5 kb in the absence or presence of pfu, respectively (Fig. 3c, lanes 3 and 4). It should be noted that the agarose gel smear is useful for size distribution but not for visualization of discrete PCR products as in the case of polyacrylamide gel-based electrophoresis (PAGE), which are limited to shorter DNA sizes; the PAGE differential display was used in the next experiment.
Specificity of ARE-PCR Subsets and Biological Utility
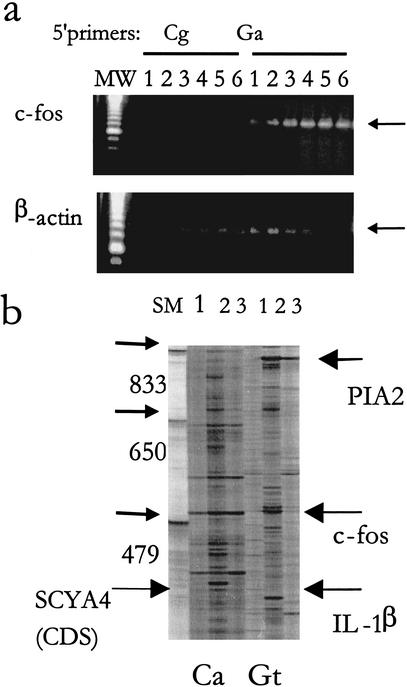
The specificity of 5′ primers that target the distinct subset of the 16 amplified ARE pools was verified by using two 5′ primers that amplify distinct subsets of the ARE-cDNAs. This was shown with c-fos amplification (Fig. 4a) by the 5′ primer Ga but not the 5′ primer Cg (Table 1). The optimum initial annealing temperature for the two overlapping nonamers containing c-fos was 40°C (Fig. 4a). The specificity of the 5′ primers was also verified in the case of specific TNF-α amplification by the primer Ca but not the primer Ag (data not shown).
Figure 4.
Specificity of subset-specific 5′ primers. (a) Forty ng cDNA was used for the ARE-cDNA PCR with different initial annealing temperatures in the first four cycles (1, 32.5°C; 2, 35°C; 3, 37.5°C; 4, 40°C; 5, 42.5′°C; and 6, 45°C) followed by a consistent cycle of high annealing temperature (60°C). Two different 5' primers were used: Ga 5′ primer (which includes 10-mer sequence that targets a subset containing c-fos mRNA) and Cg 5′ primer as a control. Aliquots of the amplified ARE products were subjected to PCR specific to c-fos and β-actin. (b) Long Range display using two different primers showing two different patterns of display using 5′ primer Ca and Gt. cDNA samples (40 ng) from THP-1 cells (lane 1), from THP-1 cells that were treated with LPS+CHX (lane 2), or THP-1 cells that were treated with PMA (lane 3), were subjected to ARE-cDNA PCR at 42.5°C initial annealing temperature as described above. Samples were loaded on 4.5% urea-denaturing polyacrylamide gel and electrophoresed in the Genomyx LR sequencing apparatus for 16 hr. Arrows indicate differentially expressed bands that were upregulated by treatment of the LPS and CHX and with known identity. The full-length coding region of SCYA4 (CDS) is shown with large arrow. MW: molecular weight markers using amplified products of known sizes.
The long-range differential display (Fig. 4b) also showed that different amplified ARE-cDNA patterns were distinctly displayed using the 5′ primers Ca and Gt (Table 1). In this particular experiment, we used RNA from untreated, LPS + CHX treated, and phorbol myristate acetate-treated cells. Many bands were up-regulated in response to LPS and CHX. Several bands with known sequence identity were overexpressed as a result of LPS + CHX, namely, IL-1β, c-fos, plasminogen activator inhibitor protein (PIA), and small inducible cytokine A4 (SCYA4); each has significant sequence information. In particular, the full-length CDS of SCYA4 (278 bp CDS, which is a part of the 450-bp band in addition to a portion of the 3′ UTR) was displayed on the gel, indicating the feasibility of targeting full-length coding regions for small mRNA molecules in the long-range differential display gel. Although we have not confirmed the differential expression of these ARE genes, data in the literature using the same cell line and inducers as in our study (Collart et al. 1987; Schwartz and Bradshaw 1992; Kastelic et al. 1996) support the differential expression of these genes.
DISCUSSION
The ARE-gene family comprises a large component of the human transcriptome, 8% of the total mRNA population (Bakheet et al. 2001), and encodes proteins that are involved in transient biological processes and are important in several disease states. In this study, we devised an integrated computational laboratory approach not only to selectively amplify the ARE-cDNAs but also to target a large proportion of the protein-coding region of the ARE-cDNA family. The ARE-cDNA PCR was used for several pilot applications to show its utility, namely, the mini-library construction, EST generation, and differential display.
The computational approach in targeting large, including full-length, protein-coding regions of the ARE-cDNA family relies on designing one universal primer that targets ARE regions (the ARE 3′ primer) and several 5′ primers that target consensus sequences largely unique to regions near to the 5′ UTR, including the initiation sites. The ARE primer is based on the computationally derived 13-bp pattern that is specific to the 3′ UTR and not to coding regions (Bakheet et al. 2001) or 5′ UTR (our unpublished observations), thus allowing priming downstream of the coding region. It should be noted that we used a simple computational method for the derivation of ARE consensuses (Bakheet et al. 2001) that may not permit the subtlety of other computational models such as those using a matrix, profile, or Markov models. Our consensuses in the initiation regions contained the conserved pattern CACCATGG in 30% of total ARE mRNAs similar to the Kozak sequence: CRCCATG (Kozak 1987a,b). It is also similar to the pattern of a larger list available in the TransTerm database: CAMCATGGC (Dalphin et al. 1999). The overall consensus initiation site in the ARE-mRNAs was SSMAMSATGRM with 50% certainty level at each position. In comparison, the initiation consensus of nonclustered random human sequences was SSSRMSATGRM (Dalphin et al. 1999). We noted that the initiation context consensus occurs also inside the coding regions. The internal sites predominantly occur toward the initiation codon and the 5′ UTR. The presence of more than one ATG codon that includes the Kozak sequence in the 5′ UTR has been recently noted (Suzuki et al. 2000). The presence of internal initiation consensus may indicate the possibility of alternative translation sites leading to different protein isoforms. Alternative translation initiation sites have been experimentally determined in many mRNA species (van der Velden and Thomas 1999).
Previous attempts of arbitrary PCR/RNA fingerprinting protocols that target gene families, including zinc fingers, 3′-ARE-containing UTR regions, and MADS-box coding sequences (Asson-Batres et al. 1994; Fischer et al. 1995; Johnson et al. 1996; Gonsky et al. 1997; Dominguez et al. 1998), were tried but had limitations that our ARE-cDNA procedure avoids. For example, in a study that targeted G-protein coupled receptors (Lopez-Nieto and Nigam 1996; Consalez et al. 1999), a statistically designed primer set targeted at the protein-coding regions of mammalian G-protein coupled receptors needed 496 pair of primer sets to span 77.7%. The detection rate of the two subsets of ARE-mRNAs, those containing two and three ARE pentamers, using the computational approach exceeds 80% with only a definitive set of the primers that equals 16 primers. Procedures by others targeted the short regions between the 3′ ends and the AREs (Asson-Batres et al. 1994; Gonsky et al. 1997; Dominguez et al. 1998) that use 3′ primers to the polyA tail and 5′ primers to ARE regions, resulting in largely AT-rich regions that are characteristically homologous, redundant, and in which size distribution is restricted. Also, none of these approaches by others address the computational targeting of large or full-length protein-coding regions as described in this paper.
Although most of the ARE-cDNA PCR reactions in our approach yielded a mixture of two size populations of the coding regions (full and truncated), the amplified ARE products primed with the 5′ primer at internal sites yielded a significant proportion of the coding region. The large fragments find their use with construction of ARE-cDNA libraries, microarray expression approaches, and isolation of full-length cDNA using 5′ RACE or from the human genome project data. The significant coding information generated from our ARE-cDNA PCR protocol is an improvement in contrast with other approaches such as restriction fragment length polymorphism- (RFLP) PCR (Fischer et al. 1995), SAGE, (Velculescu et al. 1995), and restriction enzyme-digested cDNA amplification (READS), (Prashar and Weissman 1996), which yield products with much less sequence information. The informative protein-coding regions can be compared for homology with known genes in the databases. The amplified fragments shorter than 1.5 kb can be used with long-range differential display techniques. Use of conventional differential display is limited to ∼600 bp. Primarily, the conventional differential display is intended for smaller ARE products (<600 bases); thus, shortening with restriction enzymes can also be used with a protocol such as RFLP differential display (Fischer et al. 1995; Kato 1995; Ivashuta et al. 1999). Unlike specific domain-targeting PCR approaches such as those used in conjunction with differential display (Fischer et al. 1995; Johnson et al. 1996), focusing on fewer genes, this method targets a broader family of genes because of the presence of ARE-elements, yet it is not as complex as the overall expressed genome repertoire.
The ARE-cDNA approach was largely successful in the selective amplification of ARE-cDNA as verified by (1) selective amplification of IL-8, TNF-α, and c-fos cDNA and (2) by the presence of AREs in the amplified ARE-cDNA fragments, as revealed by sequencing, which were either excised from differential display gels or randomly picked from pUC19 mini-libraries. The thermoprofiling control of the initial annealing temperatures allows extra flexibility in targeting subsets of ARE-cDNAs on the basis of the number of AREs. None of the cDNA fragments lack AREs, indicating the specificity of the ARE-cDNA PCR. This is probably because the mispriming that is frequently encountered with arbitrary PCR and mRNA fingerprinting has been significantly minimized with the use of anti-Taq and temperatures at or higher than 40°C. Anti-Taq has been successfully used for its outcome in enhanced specificity of primer annealing in PCR (Morrison et al. 1998). A limitation that is commonly inherited in PCR protocols is that longer cDNAs are amplified less efficiently than shorter cDNAs. Our approach showed that IL-8, TNF-α, and c-fos cDNAs, in which their respective predicted size of the products that were generated from ARE-cDNA PCR (i.e., 1.0, 1.2, and 1.8 kb, respectively) including the full-length coding regions, were efficiently and selectively amplified, as verified by use of specific primers in which the 5′ primer was designed at or near initiation sites. In addition, assessment of size distribution of electrophoresed amplified ARE-cDNA products showed a size distribution of up to 3.0 kb when the PCR extension step was increased to 3 min; longer PCR products (>4 kb) were also generated when pfu polymerase was used.
An additional advantage of the ARE-cDNA PCR is the ability of detecting rare genes. About half of the sequence information obtained from the pUC19/pCR2.1 mini-libraries and bands excised from differential display gels did not match any mRNA/cDNA or EST database entries. This indicates that the ARE-cDNA PCR targets rare messages that are otherwise masked by overexpressed genes in many techniques or not normally represented in conventional cDNA libraries. Among the previously known cDNAs are interleukin-1β, c-fos, plasminogen activator inhibitor protein (PIA2), small inducible cytokine A4 (SCYA4), hypoxia-induced factor alpha, amyloid A4 protein, and diacylglycerol delta kinase (all have typical ARE stretches), whereas IL-1β, c-fos, and PIA2 belong to previously characterized ARE-mRNAs as such (Chen et al. 1994; Kastelic et al. 1996; Maurer et al. 1999). Differential display results indicated the strong up-regulation of several typical ARE mRNAs including IL-1β because of the treatment of the potent cytokine inducer LPS and the protein synthesis inhibitor CHX. IL-1β is a typical ARE mRNA with three ARE clusters that is known to be up-regulated by LPS (Kastelic et al. 1996). The presence of the ARE-cDNA bands, despite the treatment with CHX as a protein synthesis inhibitor, indicates the expected biology of ARE-mRNAs, in which many of them encode transient and early response proteins that are independent of protein synthesis of other gene products. The full-length CDS of SCYA4 (278-bp CDS contained in a 450-bp band that also includes a portion of the 3′ UTR) was displayed on the gel, indicating the feasibility of targeting full-length coding regions by the computational/laboratory strategy. Although we have not confirmed the differential expression of these ARE genes, the data in the literature support differential expression of the genes (IL-1β, c-fos, PIA2) in monocytic cells (Collart et al. 1987; Schwartz and Bradshaw 1992; Kastelic et al. 1996). PMA, which is known to differentiate THP-1 cells to monocytes (Tsuchiya et al. 1982), also induced differential expression when compared with control cells, although fewer bands were observed than with LPS and CHX treatment. PIA2 was also expressed in response to PMA in accord with PMA induction of PIA2 in monocytic cell lines (Gyetko et al. 1988).
The ARE-cDNA PCR method can be used as both a discovery and expression tool. There are several advantages of the ARE-cDNA PCR as a discovery tool for ARE genes when compared with gene prediction from the human genome: (1) The amplified ARE products are the result of modulated transcripts and tissue specificity, whereas predicted genes are not; (2) the sequences are likely more accurate than the predicted genes; and (3) there is no need for downstream laboratory verification for exon accuracy and expression as with the predicted genes. Unlike those expression-profiling approaches such as those that depend on existing clones of known sequences for use with cDNA microarrays, the ARE-cDNA PCR when coupled with cDNA microarray yields information on both known and novel genes. In contrast with techniques for discovery of modulated transcripts such as subtractive hybridization and differential screening of cDNA libraries that cannot be used for mRNA profiling, ARE-cDNA PCR can be used for mRNA profiling when coupled with differential display, RLFP-differential display, and microarray. Many techniques may be biased toward a certain threshold of mRNA abundance and require largely high RNA input samples (Duggan et al. 1999). In the ARE-cDNA PCR, as little as 40 ng of total RNA can be used for one single PCR reaction; this allows its utility with tissues in which the amount of RNA is of great concern. A combined discovery and expression profile tool is SAGE (Velculescu et al. 1995), which has the limitation of exact identification of genes by the short sequence tags and lack of physical clones. Thus, the described method circumvents many of these limitations in addition to the multiple other potential uses of the ARE-cDNA PCR method.
In brief, the described computationally facilitated PCR approach and its putative applications in gene discovery and expression analysis reduces the limitations of other technologies and contains novel and significant improvements. The broader aspect of the ARE-cDNA PCR is obvious because of the diversity of the ARE-mRNA repertoire; this diversity spans many biological processes that are not limited to cellular growth, differentiation, immune response, inflammation, and cardiovascular toning. Thus, the method should constitute a valuable tool in discovering novel genes and pinpointing suspicious genes that are dysregulated, for example, in cancer but not in normal states. In addition, as more sequences are discovered, the method will ultimately help to understand the diversity, complexity, and potential involvement of expressed ARE genes in human disease.
METHODS
Sequence Retrieval and Analysis
Sequence retrieval and analysis was performed using the GCG Wisconsin Package (Genetics Computer Group [GCG]/Oxford Molecular Co.) and source codes written in PERL (Practical Extraction and Report Language). A human minimally redundant AU-rich element (ARE)-containing mRNA/cDNA database (ARED) was previously constructed using GCG and written PERL codes using GenBank Release 113 (National Center for Biotechnology Information, NCBI) and contains 895 sequences (Bakheet et al. 2001).
A 12-bp region comprising the 7 bp before (−7 bp) the start of the coding region domain sequence (CDS) in the 5′ UTR, ATG, and 2 bp after the start codon (+2 bp) from each sequence that belongs to ARED was constructed using the Assemble (GCG package). The truncated 5′ UTR list was divided into 16 subsets according to the formula NATGN, using the FindPattern program (GCG). The Pileup program (GCG), progressive pairwise algorithm, was performed to align the truncated 5′ UTR sequences in each of the 16 subsets; its output was written in multiple sequence format file (MSF). The MSF file was edited to be used as input to the Consensus program (GCG) to calculate nucleotide frequencies in positions flanking ATGN or NATGN; consensus patterns were generated for use in the design of 5′ primers. All of the 16 consensuses were subsequently used as patterns in the FindPattern program (GCG) to search for hits in the overall ARE mRNA/cDNA database, which comprised the 16 subsets. ARE motif positions were deduced using FindPattern output (GCG); calculation of the fragment lengths (from beginning of the CDS to the AREs) and statistical analysis was performed using the Excel program (Microsoft).
Primer Design
Primers for PCR were synthesized by the genomics facility at King Faisal Specialist Hospital and Research Center or GIBCO-BRL. The 3′ ARE primer was designed to incorporate sequences that target the 15-mer ARE target. The ARE primer sequence is 5′-GGCGGATCCGGGCTAAATAWATAAATWA-3′. The primer contains a BamHI site, GGATCC. Sixteen 5′ primers were designed to incorporate 10-mer initiation context consensus generated from the above analysis. All primers, in addition to the desired 5′-end sequences, were elongated with GC-rich sequence incorporating restriction sites to facilitate cloning and in vitro transcription. The upstream primers contain a common 5′-end sequence in addition to the variable 10-mer consensus patterns: 5′ACGACTCACTATAGGAA CAGA + 10-mer 3′. The computer program, Oligo 6.0 (Molecular Biology Insights, Inc.) was used to verify the physiochemical properties of the oligonucleotides for suitability in PCR reaction.
Cells and Reagents
The monocytic cell line THP-1 was obtained from the American Type Culture Collection (ATCC) and grown in RPMI 1640 supplemented with 10% FBS (low endotoxin-FBS; HyClone, UT) and antibiotics (Sigma Chemical Corp.). The cells (1 × 106 cells/mL in 25-cm2 flasks) were treated with 10 μg/mL CHX and 10 μg/mL of lipopolysaccharides (Sigma).
RNA and cDNA Synthesis
Total RNA was extracted from cells by the guanidine isothiocyanate method using Tri Reagent (Molecular Research Center). In many of the experiments, RNA was subject to DNAse I treatment (10 U per reaction, Promega) and followed by chloroform extraction, precipitation, and resuspension in diethyl pyrocarbonate (DEPC)-treated water.
The RT reaction was performed using 1 μg total RNA and 2 μM cocktail of anchored primers: oligo(dT)12A, oligo(dT)12C, and oligo(dT)12G primers, 50 μM each dNTP (Perkin Elmer), 40 U RNAsin (Promega), and 200 U of MMLV RT or SuperScript II (GIBCO BRL). The samples were heated to inactivate RT.
ARE-cDNA PCR
The cDNA (40 ng) was amplified using 1 U of Taq DNA polymerase (Amplitaq, Perkin Elmer) per reaction (20μL of total volume) that was treated with anti-Taq antibody (Clonetech). In certain experiments, direct hot-start PCR was used instead. The cDNA was subject to PCR using the single 3′ universal primer (the ARE primer) and each of the 5′ primers that was specific to initiation context consensus sequences as explained above. PCR conditions were performed with 1 μM final concentration of the primers, 10 μM each dNTP, 1.5 mM MgCl2, and 1X PCR buffer (Perkin Elmer). Thermal cycling using Gene Amp PCR System 9600 (Perkin Elmer) was first performed at 95°C for 2 min to inactivate the anti-Taq. This was followed by four cycles with denaturation of 94°C for 1 min, variable annealing temperature “controlled stringency” for 2 min, and an extension step at 72°C for 2–3 min. The variable annealing temperature in the controlled stringency step is any temperature between 35°C and 45°C depending on the ARE motif repeats targeted in ARE cDNA. The four initial cycle steps were then followed by 30 high stringency cycles of the following protocol: 94° C for 1 min, a fixed annealing temperature of 60 °C for 2 min, and an extension step at 72°C for 2–3 min.
In some experiments, a two-step PCR protocol was used to increase specificity and amount of ARE-cDNA PCR products for subsequent applications. Briefly, 1/100 of the PCR reaction was subjected to a second round of high stringency PCR using the following protocol: 95°C for 1 min, followed by a fixed annealing temperature of 60°C for 1 min, and an extension step at 72 °C for 2–3 min. Final concentrations of 1 μM of the same primers were used, 50 μM of dNTPs, 1 U Taq, and 1X PCR buffer.
Size Distribution of the Amplified ARE-cDNA Products
Two approaches were performed to assess the size distribution of the amplified ARE-cDNA products. One approach was performing the two-step PCR protocol (above) to visualize the ethidium bromide staining of the PCR products in agarose gels. The second approach was performing ARE-cDNA PCR in the presence of 0.1 μL (α-32P) dCTP (3000 Ci/mmole) per 20 μL reaction. In some of the experiments, pfu polymerase (Stratagene, Inc.) at 0.1 U per reaction was used in mixture with Taq (1 U). Southern blotting was performed using Zeta Probe nitrocellulose membranes. The blots were exposed to x-ray films (30 min) and the autoradiograms were subsequently developed.
Second Specific PCR
PCRs specific for ARE cDNAs, IL-8, TNF-α, c-fos, and the non-ARE cDNAs were performed. β-Actin and IL-8 receptor PCRs were performed under relatively stringent conditions to eliminate or minimize amplification of cDNA carryover from original PCR (ARE-cDNA PCR) tubes so that (enhanced) signals of specific targets would be attributed to the ARE-cDNA PCR. Specifically, 2 μL from the ARE-cDNA PCR reaction was amplified in the presence of 0.4 μM primers, 50 μM each dNTP, 1X PCR buffer, and with 1 U of Taq DNA polymerase per reaction.
The PCR products for IL-8, TNF-alpha, c-fos, and beta-actin have the sizes of 289 bp, 548 bp, 624 bp, and 642 bp, respectively. To monitor genomic DNA contamination, the primers were designed to span intronic sequences so that the genomic PCR products of 1216, 1450, 2130, and 1320 bp for the respective molecules would indicate the contamination. The sequences of the primer pairs used were as follows: IL-8 sense: ATG ACT TCC AAG CTG GCC GTG GCT; IL-8 antisense: T CTC AGC CCT CTT CAA AAA CTT CTC; TNF-alpha sense: CTT CTG CCT GCT GCA CTT TGG A; TNF-α antisense: TCC CAA AGT AGA CCT GCC CAG A; c-fos sense: GGG GAT AGC CTC TCT TAC TAC CAC; c-fos antisense: GCT GCA TAG AAG GAC CCA GAT AG; β-actin sense: ATC TGG CAC CAC ACC TTC TAC AAT GAG CTG CG; and β-actin antisense: CGT CAT ACT CCT GCT TGC TGA TCC ACA. The 5′ primers were designed at or near the initiation sites so that amplification is enriched toward the larger and not shorter amplified ARE products. The cycling profile was as follows: 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min for 25 cycles; the total reaction volume was 20 μL. The PCR products were resolved on 2% agarose gel and visualized with ethidium bromide. Size markers (100 bp) were obtained from GIBCO and used to verify the size of PCR products.
Construction of Mini-Libraries, Random Cloning, and Sequencing
ARE-cDNA PCR products were cloned in either pCR2.1 vector (TA cloning kit; Invitrogen) or the pUC19 vector (T-7 blue blunt-end cloning system; Novagen) according to the manufacturer's instructions. Positive colonies were randomly picked and further propagated in LB medium. Plasmids were extracted by Qiagen Miniprep kit. Sequencing was initially performed with Taq dye terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, ABI) in automated fluorescent DNA sequencer 373A (ABI) according to the manufacturer's instructions. Sequencing was performed with vector-specific primers flanking the PCR products such as M13 forward and reverse primers. PCR products, 2 μL, were treated with 0.2 U exonuclease I (New England Biolabs, ) and 0.2 U sheep alkaline phosphatase in a 10-μL final volume to degrade excess primers and dNTPs, respectively. These reactions were performed for 1 hr at 37°C and terminated with heat (85°C, 15 min). Templates (2.5 μL each for M13R and M13F reaction) were mixed with the sequencing DYEnamic ET reagent premix (Pharmacia), along with 10 pM of the sequencing primers (M13). Cycling was performed according to the manufacturer's instructions. Sequencing reactions were precipitated, washed, air-dried, and resuspended in loading buffer. Parameters for injecting and running the samples were performed according to the manufacturer's instructions (Amersham-Pharmacia).
Differential Display, Long Range Differential Display, and Cloning of Differential Display Products
The cDNAs (40 ng/mL) were amplified as explained above in the ARE-cDNA PCR but with the use of 0.5 μM (35S)dATP (1200 Ci/mmole;). Aliquots of 4 μL from the PCR reactions were loaded on a 6% urea-denaturing polyacrylamide gel and electrophoresed for 2.5 hr in TBE buffer. Unless otherwise indicated, the long-range sequencing gel apparatus was used (Genomyx LR, Beckman) using 4.5% urea-denaturing polyacrylamide gel with 16 hr electrophoresis for resolution of long PCR fragments. Gels were fixed and dried; x-ray autoradiograms were developed for 48 hr. Bands were excised from the gel and rehydrated in 100 μL 10 mM Tris-HCl at pH 8.0 and 1 mM EDTA for 15 min at 25°C followed by elution at 95°C for 15 min. The PCR products were precipitated with sodium acetate/ethanol, centrifuged, and resuspended in 10 μL water. An aliquot of 5 μL was used for reamplification using the same 5′ primer and 3′ primer set that was originally used in ARE-cDNA PCR and final concentration of dNTPs, 100 μM each, and 30 cycles of 94°C for 45 sec, 60°C for 1 min, 72°C for 2 min, and final extension cycle of 72°C for 7 min. Aliquots of the PCR products were visualized on 2% agarose or low-melting temperature gel to confirm reamplification and PCR product size. The PCR products were cloned in pCR2.1 or in pUC19 as described above.
Acknowledgments
We thank Dr. Brian Meyer and his team at the Genomics Service Facility for their helpful assistance. We also thank Dr. Meyer for critical review of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL khabar@kfshrc.edu.sa; FAX 966-1-442-7858.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.204902. Article published online before print in May 2002.
REFERENCES
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Al-Humidan A, Edwards CK, Al-Sofi A, Dzimiri M, Al-Sedairy ST, Khabar KSA. A carbocyclic nucleoside analogue is a TNF-alpha inhibitor with immunosuppressive action: Role of prostaglandin E2 and protein kinase C and comparison with pentoxifylline. Cell Immunol. 1998;188:12–18. doi: 10.1006/cimm.1998.1324. [DOI] [PubMed] [Google Scholar]
- Asson-Batres MA, Spurgeon SL, Bagby GC., Jr Identification of AU-rich 3′ untranslated regions in mRNA from sea urchin coelomocytes. Ann NY Acad Sci. 1994;712:34–41. doi: 10.1111/j.1749-6632.1994.tb33560.x. [DOI] [PubMed] [Google Scholar]
- Bakheet T, Frevel M, Williams BRG, Greer W, Khabar KSA. ARED: Human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertiol DJ, Smoker M, Brown AC, Jones MG, Burrows PR. A method based on PCR for the construction of cDNA libraries and probes from small amounts of tissue. Biotechniques. 1994;16:1054–1058. [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Chen CY, Chen TM, Shyu AB. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol Cell Biol. 1994;14:416–426. doi: 10.1128/mcb.14.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Belin D, Vassalli JD, Vassalli P. Modulations of functional activity in differentiated macrophages are accompanied by early and transient increase or decrease in c-fos gene transcription. J Immunol. 1987;139:949–955. [PubMed] [Google Scholar]
- Consalez GG, Cabibbo A, Corradi A, Alli C, Sardella M, Sitia R, Fesce R. A computer-driven approach to PCR-based differential screening, alternative to differential display. Bioinformatics. 1999;15:93–105. doi: 10.1093/bioinformatics/15.2.93. [DOI] [PubMed] [Google Scholar]
- Dalphin ME, Stockwell PA, Tate WP, Brown CM. TransTerm, the translational signal database, extended to include full coding sequences and untranslated regions. Nucleic Acids Res. 1999;27:293–294. doi: 10.1093/nar/27.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez O, Ashhab Y, Sabater L, Belloso E, Caro P, Pujol-Borrell R. Cloning of ARE-containing genes by AU-motif-directed display. Genomics. 1998;54:278–286. doi: 10.1006/geno.1998.5548. [DOI] [PubMed] [Google Scholar]
- Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet. 1999;21:10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- Fischer A, Saedler H, Theissen G. Restriction fragment length polymorphism-coupled domain-directed differential display: A highly efficient technique for expression analysis of multigene families. Proc Natl Acad Sci. 1995;92:5331–5335. doi: 10.1073/pnas.92.12.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsky R, Knauf JA, Elisei R, Wang JW, Su S, Fagin JA. Identification of rapid turnover transcripts overexpressed in thyroid tumors and thyroid cancer cell lines: Use of a targeted differential RNA display method to select for mRNA subsets. Nucleic Acids Res. 1997;25:3823–3831. doi: 10.1093/nar/25.19.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyetko MR, Webb AC, Sitrin RG. Modulation of urokinase-type plasminogen activator and plasminogen activator inhibitor-2 expression by U-937 mononuclear phagocytes. Effects of 1 alpha, 25-dihydroxyvitamin D3 and phorbol ester. J Immunol. 1988;141:2693–2698. [PubMed] [Google Scholar]
- Ivashuta S, Imai R, Uchiyama K, Gau M. The coupling of differential display and AFLP approaches for nonradioactive mRNA fingerprinting. Mol Biotechnol. 1999;12:137–141. doi: 10.1385/MB:12:2:137. [DOI] [PubMed] [Google Scholar]
- Johnson SW, Lissy NA, Miller PD, Testa JR, Ozols RF, Hamilton TC. Identification of zinc finger mRNAs using domain-specific differential display. Anal Biochem. 1996;236:348–352. doi: 10.1006/abio.1996.0178. [DOI] [PubMed] [Google Scholar]
- Kastelic T, Schnyder J, Leutwiler A, Traber R, Streit B, Niggli H, MacKenzie A, Cheneval D. Induction of rapid IL-1 beta mRNA degradation in THP-1 cells mediated through the AU-rich region in the 3′ UTR by a radicicol analogue. Cytokine. 1996;8:751–761. doi: 10.1006/cyto.1996.0100. [DOI] [PubMed] [Google Scholar]
- Kato K. Description of the entire mRNA population by a 3′ end cDNA fragment generated by class IIS restriction enzymes. Nucleic Acids Res. 1995;23:3685–3690. doi: 10.1093/nar/23.18.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabar KS, Al-Zoghaibi F, Al-Ahdal MN, Murayama T, Dhalla M, Mukaida N, Taha M, Al-Sedairy ST, Siddiqui Y, Kessie G, et al. The alpha chemokine, interleukin 8, inhibits the antiviral action of interferon alpha. J Exp Med. 1997;186:1077–1085. doi: 10.1084/jem.186.7.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987a;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 1987b;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- Lagnado CA, Brown CY, Goodall GJ. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: The functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LT. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2001;2:0041.0041–0041.0011. doi: 10.1186/gb-2001-2-10-research0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction [see comments] Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Lopez-Nieto CE, Nigam SK. Selective amplification of protein-coding regions of large sets of genes using statistically designed primer sets [see comments] Nat Biotechnol. 1996;14:857–861. doi: 10.1038/nbt0796-857. [DOI] [PubMed] [Google Scholar]
- Maurer F, Tierney M, Medcalf RL. An AU-rich sequence in the 3′-UTR of plasminogen activator inhibitor type 2 (PAI-2) mRNA promotes PAI-2 mRNA decay and provides a binding site for nuclear HuR. Nucleic Acids Res. 1999;27:1664–1673. doi: 10.1093/nar/27.7.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TB, Weis JJ, Wittwer CT. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques. 1998;24:954–958. , 960, 962. [PubMed] [Google Scholar]
- Murayama T, Ohara Y, Obuchi M, Khabar KS, Higashi H, Mukaida N, K. Matsushima K. Human cytomegalovirus induces interleukin-8 production by a human monocytic cell line, THP-1, through acting concurrently on AP-1- and NF- kappaB-binding sites of the interleukin-8 gene. J Virol. 1997;71:5692–5695. doi: 10.1128/jvi.71.7.5692-5695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson LA, Brown MR, Carlisle AJ, Kohn EC, Liotta LA, Emmert-Buck MR, Krizman DB. An improved method for construction of directionally cloned cDNA libraries from microdissected cells. Cancer Res. 1998;58:5326–5328. [PubMed] [Google Scholar]
- Prashar Y, Weissman SM. Analysis of differential gene expression by display of 3′ end restriction fragments of cDNAs. Proc Natl Acad Sci. 1996;93:659–663. doi: 10.1073/pnas.93.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R, Magnuson NS. Mechanisms regulating transient expression of mammalian cytokine genes and cellular oncogenes. Prog Nucleic Acid Res Mol Biol. 1990;38:241–282. doi: 10.1016/s0079-6603(08)60713-8. [DOI] [PubMed] [Google Scholar]
- Schwartz BS, Bradshaw JD. Regulation of plasminogen activator inhibitor mRNA levels in lipopolysaccharide-stimulated human monocytes. Correlation with production of the protein. J Biol Chem. 1992;267:7089–7094. [PubMed] [Google Scholar]
- Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ishihara D, Sasaki M, Nakagawa H, Hata H, Tsunoda T, Watanabe M, Komatsu T, Ota T, Isogai T, et al. Statistical analysis of the 5′ untranslated region of human mRNA using “Oligo-Capped” cDNA libraries. Genomics. 2000;64:286–297. doi: 10.1006/geno.2000.6076. [DOI] [PubMed] [Google Scholar]
- Tsuchiya S, Kobayashi Y, Goto Y, Okumura H, Nakae S, Konno T, Tada K. Induction of maturation in cultured human monocytic leukemia cells by a phorbol diester. Cancer Res. 1982;42:1530–1536. [PubMed] [Google Scholar]
- van der Velden AW, Thomas AA. The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int J Biochem Cell Biol. 1999;31:87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression [see comments] Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]