Abstract
P58IPK is an Hsp40 family member known to inhibit the interferon (IFN)-induced, double-stranded RNA-activated, eukaryotic initiation factor 2α (eIF2α) protein kinase R (PKR) by binding to its kinase domain. We find that the stress of unfolded proteins in the endoplasmic reticulum (ER) activates P58IPK gene transcription through an ER stress-response element in its promoter region. P58IPK interacts with and inhibits the PKR-like ER-localized eIF2α kinase PERK, which is normally activated during the ER-stress response to protect cells from ER stress by attenuating protein synthesis and reducing ER client protein load. Levels of phosphorylated eIF2α were lower in ER-stressed P58IPK-overexpressing cells and were enhanced in P58IPK mutant cells. In the ER-stress response, PKR-like ER kinase (PERK)-mediated translational repression is transient and is followed by translational recovery and enhanced expression of genes that increase the capacity of the ER to process client proteins. The absence of P58IPK resulted in increased expression levels of two ER stress-inducible genes, BiP and Chop, consistent with the enhanced eIF2α phosphorylation in the P58IPK deletion cells. Our studies suggest that P58IPK induction during the ER-stress response represses PERK activity and plays a functional role in the expression of downstream markers of PERK activity in the later phase of the ER-stress response.
Cells respond to environmental stress stimuli by regulating mRNA translation. A key step in this regulation occurs at the level of initiation through modification of the phosphorylation of eukaryotic initiation factor (eIF)2α (1). In higher eukaryotic cells, several serine/threonine eIF2α kinases that respond to different stress signals have been identified. These include protein kinase R (PKR), an IFN-induced, double-stranded RNA-activated kinase that is activated during virus infection, and PKR-like endoplasmic reticulum (ER) kinase (PERK), an eIF2α kinase that is activated during the unfolded protein response (UPR), a cellular response to the accumulation of malfolded proteins in the ER. After virus infection, as part of the antiviral mechanism host cells stimulate PKR-mediated eIF2α phosphorylation, thus shutting off global protein synthesis including the synthesis of viral proteins. During the UPR, PERK phosphorylates eIF2α to attenuate mRNA translation, thus reducing the burden of protein substrate for the ER-folding and -degradation machinery. Because PKR and PERK share a common substrate, they contain similar kinase domains (40% identity). However, their stress signal-sensor domains are different, and the kinases are localized to different compartments (the cytosol or ER, respectively) (2).
Previously we identified a cellular inhibitor of PKR, P58IPK, which is activated after influenza virus infection (3–5). The activation of P58IPK inhibits the PKR-mediated translational arrest by binding to and inactivating the kinase domain of PKR, thereby ensuring that the cellular protein-synthesis machinery remains available to synthesize viral proteins. The influenza virus has therefore found a way to co-opt a cellular activity to its purpose. It seems unlikely, however, that P58IPK evolved as a cellular gene to aid in viral replication. Rather, the activation of P58IPK in response to both influenza virus infection and heat shock suggests that P58IPK might play a role in multiple stress responses (6).
In the present study we sought to gain insight into additional functions of P58IPK by analyzing the gene's expression, which led to the identification of an ER stress-response element (ERSE) in the P58IPK promoter. We also noticed that the kinase domain of PKR, with which P58IPK interacts, is very similar to the kinase domain of PERK. In addition, both PERK and P58IPK are highly expressed in pancreatic cells (7, 8). Therefore, we investigated whether P58IPK is also involved in the regulation of PERK activity. Here we describe our results whereby P58IPK is induced during the UPR, interacts with PERK, attenuates PERK-mediated eIF2α phosphorylation during ER stress, and negatively regulates selective translation of UPR target proteins BiP and Chop. Thus, P58IPK is among a group of genes encoding molecular chaperones, protein-folding enzymes, and transcription factors that are induced after ER stress and function to restore homeostasis to stressed cells.
Materials and Methods
Plasmid Constructs.
A 1.7-kb fragment from a screen of a mouse C57BL/6 genomic DNA library (9), which contained the 5′-flanking region of the P58IPK gene, was cloned into the pGL3-Basic luciferase reporter vector (Promega). This fragment was subsequently shortened to 0.58 kb by deleting the NheI–AatII fragment. The mutant P58IPK-luciferase reporter construct was made by PCR by replacing the sequence of the ERSE to CGCGT(N9)CTAGT. The mouse pGRP78/BiP-luciferase reporter plasmid was provided by Stephen Spindler (University of California, Riverside) (10).
The His-P58IPK used in the pull-down assay (11), GST-P58IPK and GST-PERK in the in vitro kinase assay (11, 12), and PERK-myc in the microscopy experiment (12) have been described. To construct the hemagglutinin (HA) tagged HA-PERK (full-length) and HA-PERKΔC (C-term truncation) used in the pull-down assay, EcoRI–XhoI PCR fragments made from the PERK-pcDNA1 or PERKΔC-pcDNA1 (12) were cloned into the same sites of the pCMV-HA vector (BD/CLONTECH). To express the HA-P58IPK used in the microscopy experiment, an EcoRI–BglII PCR fragment of bovine P58IPK was cloned into the same sites of pCMV-HA. To create a P58IPK-inducible cell line, a BamHI–EcoRI (blunted) fragment from P58IPK-pGEX2T (13) was cloned into the BamHI and PvuII sites of pTRE2Hyg (BD/CLONTECH) to make the P58IPK-pTRE2Hyg construct.
Cell Culture, Drug Treatment, and Transfection.
All cells were maintained in DMEM supplemented with 10% FBS. The Tet-Off P58IPK-inducible cell line was constructed by transfecting P58IPK-pTRE2Hyg into mouse embryonic fibroblasts (MEF/3T3, BD/CLONTECH) following manufacturer instructions. To induce the UPR, cells were treated with tunicamycin (2 μg/ml) for 16 h or thapsigargin (1 μM) for 30 min before analysis unless otherwise indicated. Transfection was performed by using Superfect reagent (Qiagen, Valencia, CA) following manufacturer protocol.
Luciferase Assay.
NIH 3T3 cells grown on six-well plates were transfected with 1 μg of plasmid DNA per well. At 24 h posttransfection point, equally transfected cells were untreated or treated with tunicamycin (2 μg/ml) for 16 h before being lysed in 400 μl of 1× cell lysis buffer (BD/PharMingen). An aliquot of each lysate (10 μl) was mixed with 100 μl of luciferase assay reagent (Promega), and luciferase activity was measured in a Beckman Coulter LS6500 scintillation system in the single-photon mode.
Northern and Western Blot Analysis.
Northern blot analysis was performed as described (14). For Western blot analysis, cells were lysed in buffer (20 mM Hepes, pH 7.5/150 mM NaCl/1 mM EDTA/10% glycerol/1% Triton X-100) supplemented with 1× complete protease-inhibitor mixture (Roche Diagnostics) and subjected to SDS/PAGE. Antibodies against PERK (12, 15), P58IPK (16), Chop (17), and eIF2α (18) have been described. The antiactin (ICN), anti-BiP (StressGen Biotechnologies, Victoria, Canada), anti-HA (3F10, Roche Molecular Biochemicals), and antiphosphor-eIF2α (Research Genetics, Huntsville, AL) antibodies were purchased from manufacturers.
Protein-Interaction Assay.
Protein complex formation was determined by pull-down assays and a coimmunoprecipitation assay. In the pull-down assays, cell lysates containing transfected His-P58IPK or vector (pEF4/HisA) control were incubated with Ni2+-nitrilotriacetic acid beads (Qiagen) overnight and used as bait to incubate with AR42J cell lysates for 2 h. The AR42J cells (mock-treated or treated with tunicamycin or thapsigargin) were lysed and precleared with Ni2+-nitrilotriacetic acid beads before incubation with the bait. The final bead-bound fractions were subjected to Western blotting. To pull down P58IPK using PERK as bait, COS1 cells, transfected with HA-PERK or HA-PERKΔC, were mock-treated or treated with tunicamycin before being lysed. Cell lysates were incubated with the anti-HA affinity matrix (Roche Molecular Biochemicals) for 2 h. The bound fractions were incubated with purified with GST-P58IPK and subjected to Western blotting. In the coimmunoprecipitation assay, P58IPK was cotransfected with either PERK-K618A or PERKΔC into COS1 cells. Cell lysates were immunoprecipitated with anti-PERK antibody in buffer [20 mM Hepes, pH 7.5/50 mM KCl/1 mM MgCl2/0.2% Triton X-100/10% glycerol/1× protease inhibitor mixture (Roche Diagnostics)] and subjected to Western blot analysis.
In Vitro Kinase Assay.
The kinase assay was performed at 30°C for 30 min in 40 μl of kinase buffer (20 mM Hepes, pH 7.5/50 mM KCl/1.5 mM DTT/2 mM MgCl2/0.1 mM ATP) containing 2 μg of purified WT eIF2α (gift from Scott Kimball, Pennsylvania State University, Hershey, PA) or S51A mutant eIF2α (gift from Ron Wek, Indiana University, Indianapolis), 6 μCi of [γ-32P]ATP (1 Ci = 37 GBq), bacterially expressed GST-PERK (4 μg), and bacterially expressed GST-P58IPK (2, 4, or 8 μg) or GST alone as control (10, 20, or 40 μg). Hsp40 (4 μg, StressGen Biotechnologies) was included in one reaction as indicated. Reaction mixtures were subjected to 12% SDS/PAGE and visualized by autoradiography with a phosphorimager.
Immunofluorescence Microscopy.
COS1 cells transfected with HA-P58IPK and/or PERK-myc were fixed in 4% paraformaldehyde for 30 min at room temperature. Immunostaining was performed by using antibodies against HA (rat, clone 3F10, Roche Molecular Biochemicals), myc (mouse, clone 9E10, BD/PharMingen), and calnexin (mouse, clone 37, BD/Transduction Laboratories) followed by secondary antibodies conjugated with FITC (anti-rat) or Texas red (anti-mouse). Samples were examined by using an Eclipse E600 microscope (Nikon).
Results
P58IPK Is Induced During the UPR.
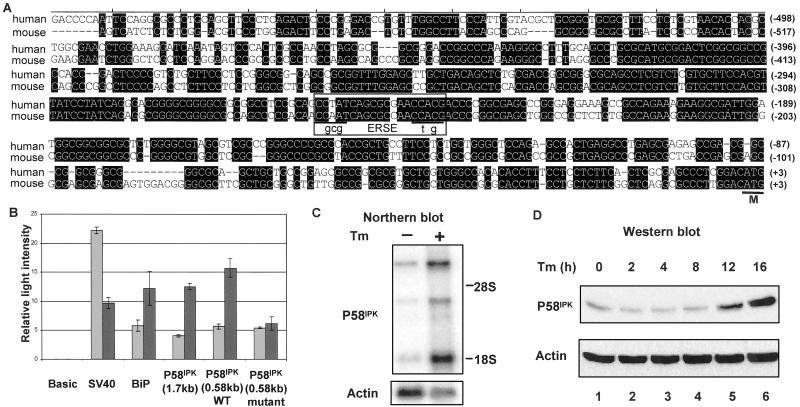
To gain insight into cellular pathways in which P58IPK might function, we cloned the murine P58IPK promoter and examined this genomic region for potential regulatory elements. A 1.7-kb DNA fragment from a P58IPK mouse genomic clone, containing the region immediately 5′ of the translational start site, was inserted upstream of a promoterless luciferase reporter gene. This DNA fragment was able to drive expression of the luciferase reporter when inserted in the forward but not in the reverse orientation (Fig. 1B and data not shown), suggesting that it functions as a P58IPK promoter. The nucleotide sequence of this region (GenBank accession no. AF495532) revealed a potential heat-shock response element (1 base mismatch from an HSF-2 binding site) located at position −1,076 and a single copy of an ERSE located at position −268 (Fig. 1A). The ERSE is a 19-nt motif with a consensus sequence of CCAAT(N9)CCACG that is commonly found in the promoter region of genes induced during the UPR (19, 20). An ERSE is found also in a similar location in the human P58IPK gene (Fig. 1A), suggesting a conserved function for this promoter region.
Fig 1.
P58IPK contains an ERSE and is induced during the UPR. (A) Sequence alignment of the 5′-flanking region of human (GenBank accession no. NT009952) and mouse (GenBank accession no. AF495532) P58IPK. Conserved nucleotides are shaded. The ERSE is boxed, and the nucleotide changes introduced into the ERSE are shown in lowercase letters. The A of the ATG initiation codon (marked as M) is designated +1. (B) NIH 3T3 cells, transfected with the indicated constructs, were either untreated or treated with tunicamycin (2 μg/ml) for 16 h and subjected to a luciferase assay. Luciferase activity is depicted as relative light intensity. Gray bars, untreated cells; black bars, tunicamycin-treated cells; SV40, simian virus 40. (C) Northern blot analysis of poly(A)+ RNA (2 μg per lane) isolated from NIH 3T3 cells grown in the presence or absence of tunicamycin (Tm). (Upper) Probed with a 1,070-bp mouse P58IPK fragment. (Lower) Probed with actin-specific probe. (D) NIH 3T3 cells were treated with tunicamycin for the indicated hours. An equal amount of cell lysate (100 μg) was loaded in each lane and subjected to immunoblot analysis by using indicated antibodies.
We next examined the responsiveness of the P58IPK promoter to ER stress. A variety of stimuli including calcium depletion from the ER lumen (triggered by thapsigargin treatment), inhibition of protein N-glycosylation (triggered by tunicamycin treatment), or the reduction of disulfide bonds (triggered by DTT treatment), are capable of disrupting ER function and inducing intracellular signaling pathways collectively referred to as the UPR. To induce the UPR experimentally, NIH 3T3 cells, transfected with various reporter gene constructs, were treated with tunicamycin and lysed for the luciferase assay. We observed that the P58IPK promoter exhibited a 3-fold increase in expression in response to tunicamycin treatment (Fig. 1B). An ER Hsp70 chaperone GRP78/BiP-luciferase construct used as a positive control exhibited a comparable level of induction. In contrast, activity of the simian virus 40-luciferase control was reduced by tunicamycin treatment. The introduction of point mutations [CGCGT(N9)CTAGT in place of CCAAT(N9)CCACG] into the ERSE abolished activation of the P58IPK promoter by tunicamycin. Thus, expression from the P58IPK promoter is induced in an ERSE-dependent manner by a cellular treatment that triggers the UPR.
Similar changes were observed in the expression of the endogenous P58IPK mRNA and protein in tunicamycin-treated cells. Steady-state levels of P58IPK mRNA increased 12-fold compared with the level present in untreated cells (specifically the 1.7-kb transcript; Fig. 1C). We also observed a similar increase in the P58IPK protein level in response to tunicamycin treatment (Fig. 1D). These results confirm the luciferase reporter assays and are consistent with a recent microarray analysis that identified the P58IPK gene among those induced by the UPR (21).
P58IPK Interacts with PERK.
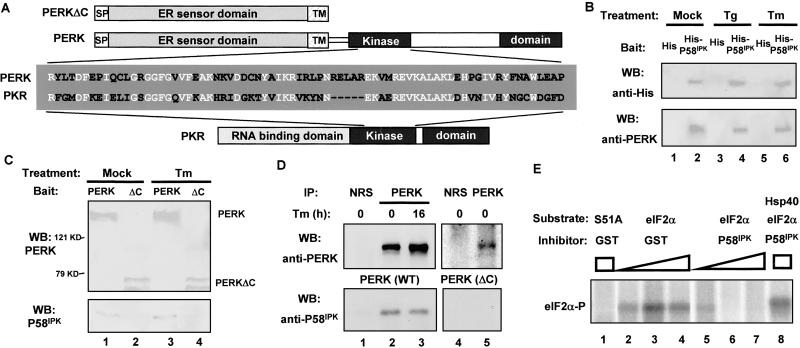
Because P58IPK is regulated during the UPR and the P58IPK-interacting region of PKR is conserved in PERK (Fig. 2A), we explored the possibility that P58IPK may interact with PERK and function as a PERK regulator. We used His-tagged P58IPK, isolated from NIH 3T3 cell lysates and bound to Ni2+ beads, as a bait to pull down PERK from rat pancreatic AR42J cell lysates. As shown in Fig. 2B, PERK was retained on beads containing His-P58IPK but not on the control beads. We also performed a reciprocal pull-down assay using HA-tagged PERK (isolated from COS1 cells and bound to the anti-HA affinity matrix) to analyze PERK-P58IPK interaction. We observed that full-length PERK (HA-PERK), but not the C-terminal truncation of PERK (HA-PERKΔC) that lacks the cytosolic kinase domain that contains a potential P58IPK-binding region (Fig. 2A), was able to pull down GST-P58IPK directly (Fig. 2C).
Fig 2.
P58IPK interacts with and inhibits PERK. (A) Schematic diagram for PERK, PERKΔC, and PKR. The P58IPK-interacting region of PKR, located in the kinase domain, was aligned with the corresponding region of PERK. Identical sequences are indicated in white. (B) His-P58IPK pulls down PERK. His-tagged P58IPK (lanes 2, 4, and 6) or His-tag alone (lanes 1, 3, and 5), bound on beads, was incubated with lysates from AR42J cells grown in the absence or presence of thapsigargin (Tg) or tunicamycin (Tm). The bead-bound protein complex was subjected to Western blotting (WB) by using the indicated antibodies. (C) HA-PERK pulls down P58IPK. HA-PERK (lanes 1 and 3) or HA-PERKΔC (lanes 2 and 4) from mock-treated or tunicamycin-treated COS1 cells was used as bait to pull down purified GST-P58IPK. The bead-bound protein complex was subjected to Western blotting by using indicated antibodies. (D) PERK coimmunoprecipitates with P58IPK. COS1 cells cotransfected with P58IPK and PERK (lanes 1–3) or PERKΔC (lanes 4 and 5), either treated with tunicamycin or untreated, were lysed and immunoprecipitated (IP) with anti-PERK antibody (lanes 2, 3, and 5) or normal rabbit serum (NRS, lanes 1 and 4). The precipitates then were subjected to Western blotting by using indicated antibodies. (E) P58IPK inhibits PERK kinase activity. Purified GST-PERK was incubated with purified WT eIF2α (lanes 2–8) or S51A mutant eIF2α (lane 1) and [γ-32P]ATP in the presence of purified GST-P58IPK (lanes 5–8) or GST control (lanes 2–4). Hsp40 was included as a specific inhibitor to P58IPK (lane 8). Reaction mixtures were subjected to SDS/PAGE and visualized by autoradiography.
To examine whether this P58IPK–PERK interaction occurs in vivo, we performed a coimmunoprecipitation assay in COS1 cells cotransfected with P58IPK and full-length PERK or C-terminal truncation of PERK (PERKΔC). Because overexpression of WT PERK caused translational inhibition and cell-growth arrest, we used a PERK mutant (PERK-K618A) of inactive kinase activity as the full-length PERK. When the full-length PERK was immunoprecipitated, we found that P58IPK was in the same immunocomplex with PERK but not in the complex precipitated by normal rabbit serum (Fig. 2D). In addition, P58IPK only coimmunoprecipitated with the full-length PERK, not with the C-terminal truncated form of PERK. Therefore P58IPK seems to be complexed with PERK in vivo, which requires the kinase domain of PERK. These in vivo coimmunoprecipitation data, together with the in vitro pull-down results of P58IPK binding to PERK, suggest that P58IPK interacts with PERK, probably with the kinase domain of PERK.
P58IPK Inhibits PERK Activity.
PERK phosphorylates eIF2α in vitro (12, 22). Therefore, to examine the functional consequences of the interaction described above, we performed an in vitro kinase assay using purified GST-PERK in the presence of either GST-P58IPK or GST alone. GST-PERK phosphorylated eIF2α (Fig. 2E, lanes 2–4) but not the S51A mutant of eIF2α (lane 1). Adding increasing amounts of purified P58IPK attenuated PERK-mediated eIF2α phosphorylation (lanes 5–7). Interestingly, adding purified Hsp40, a cellular repressor of P58IPK (6, 23), blocked the repressive effect of P58IPK (lane 8). These results indicate that P58IPK is able to repress PERK activity in vitro.
P58IPK Is Associated with the ER.
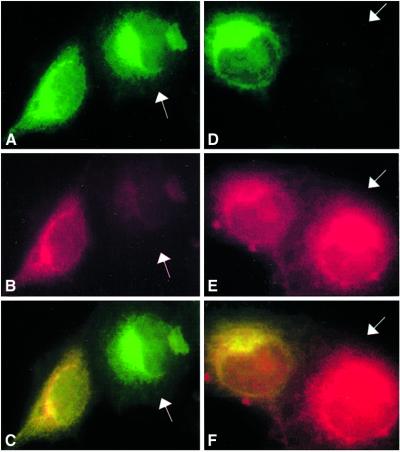
Given that P58IPK interacted with PERK and inhibited PERK kinase activity, we examined whether P58IPK and PERK are localized in the same cellular compartment. HA-tagged P58IPK and myc-tagged PERK were cotransfected into COS1 cells, and monoclonal antibodies directed to the HA and myc epitopes were used to determine P58IPK and PERK subcellular localization by immunofluorescence microscopy in fixed cells. PERK is located on ER membranes (12). As shown in Fig. 3, anti-HA staining of transfected cells produced a lacy reticular staining pattern that colocalized with PERK (A–C). In addition to PERK, P58IPK also colocalized with the endogenous ER marker calnexin, detected with the anticalnexin serum (D–F). There were no differences in P58IPK localization between nonstressed and ER-stressed COS1 cells, and similar results were obtained by using NIH 3T3 cells (data not shown). These results suggest that P58IPK is associated with the ER, the compartment in which PERK is localized.
Fig 3.
P58IPK and PERK associate with the ER. HA-P58IPK with (A–C) or without (D–F) PERK-myc was transfected into COS1 cells. Cells were fixed and costained with rat anti-HA antibody, followed by FITC-conjugated anti-rat serum and mouse anti-myc (mouse) (A–C) or mouse anticalnexin (D–F) antibodies, followed by Texas red-conjugated anti-mouse serum. The white arrows in A (P58IPK, green), B (PERK, red), and C (merger of A and B) point to the PERK nontransfected cells. The white arrows in D (P58IPK, green), E (calnexin, red), and F (merger of D and E) point to the P58IPK nontransfected cells.
Overexpression of P58IPK Reduces PERK-Mediated eIF2α Phosphorylation in Vivo.
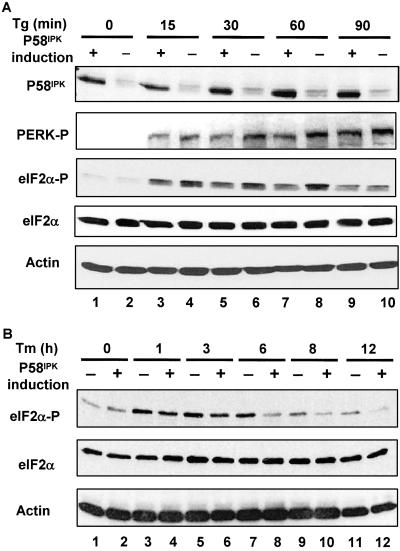
To investigate the role of P58IPK in regulating PERK in vivo, we constructed a cell line in which P58IPK expression was regulated by tetracycline (Fig. 4A). We then measured eIF2α phosphorylation and PERK activation in these cells at multiple time points during the UPR. Immunoblotting with specific antibodies revealed lower levels of phosphorylated eIF2α in P58IPK-overexpressing ER-stressed cells compared with the nonoverexpressing stressed cells. These differences in phosphorylated eIF2α levels were observed after the induction of the UPR by either thapsigargin or tunicamycin. The ability of P58IPK overexpression to reduce levels of phosphorylated eIF2α correlated with its ability to inhibit PERK activation as measured by immunoblotting with an antiserum that detects the phosphorylated, activated form of the PERK kinase (Fig. 4).
Fig 4.
Overexpression of P58IPK attenuates PERK-mediated eIF2α phosphorylation. (A) P58IPK Tet-Off-inducible cells were grown in the presence or absence of tetracycline (1 μg/ml) for 2 days. Then cells were treated with thapsigargin (Tg, 1 μM) for the indicated hours. Equal amounts of cell lysates from these cells were subjected to Western blotting by using the indicated antibodies. (B) Similar to A except that cells were treated with tunicamycin (Tm, 2 μg/ml) for the indicated period.
Deletion of P58IPK in Mouse Embryonic Stem (ES) Cells Increases eIF2α Phosphorylation and Induction of UPR Target Proteins.
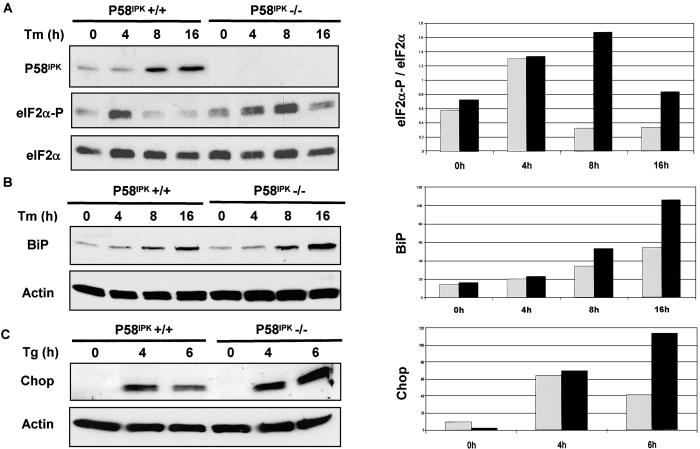
As an alternative approach to study the role of P58IPK as an inhibitor of PERK in vivo, we compared the levels of phosphorylated eIF2α in WT and P58IPK −/− ES cells. The mutant cells exhibited higher levels of eIF2α phosphorylation than the WT cells, a difference that was particularly obvious at later time points of the ER-stress response when P58IPK is induced in the WT cells (Fig. 5A). These data, together with the above observation that overexpression of P58IPK attenuated PERK-mediated eIF2α phosphorylation, suggest that P58IPK represses eIF2α phosphorylation during the UPR.
Fig 5.
Deletion of P58IPK enhances UPR-mediated eIF2α phosphorylation and BiP/Chop induction. (A) P58IPK+/+ (BL/6) and P58IPK−/− mouse ES cells, treated with tunicamycin (Tm, 2 μg/ml) for the indicated hours, were lysed and subjected to Western blotting by using the indicated antibodies. Densitometry analysis was performed on the Western data. The ratio of the phosphor-eIF2α signal versus the total eIF2α signal is shown on the y axes by values in arbitrary units. Gray bars, P58IPK+/+ cells; black bars, P58IPK−/− cells. (B) Tunicamycin-treated cells (for the indicated hours) were immunoblotted with anti-BiP antibody and analyzed by densitometry. (C) Cells were treated with thapsigargin (Tg) for the indicated hours, immunoblotted with anti-Chop antibody, and analyzed by densitometry.
During the UPR, PERK-mediated eIF2α phosphorylation attenuates protein synthesis to reduce ER client-protein load while selectively promoting expression of certain UPR target genes such as BiP and Chop. It has been shown that BiP and Chop induction in the UPR depends highly on PERK-mediated eIF2α phosphorylation (21, 24, 25); in other words, the levels of Chop and BiP proteins serve as markers of PERK activity in the UPR. Our finding that expression of both BiP and Chop was enhanced in the P58IPK −/− ES cells, particularly at a later phase of the UPR (Fig. 5 B and C), indicates that P58IPK modulation of PERK activity impacts on the intensity of signaling in the UPR.
Discussion
The mammalian ER-stress response (the UPR) consists of an early phase in which protein synthesis is inhibited by eIF2α phosphorylation and a later phase in which genes that promote increased ER capacity are induced. The early, PERK-dependent phase plays an important role in acutely reducing the load of client proteins that the ER must handle and is strongly protective against ER stress (2). However, realization of the later, synthetic phase of the UPR requires new protein synthesis, particularly the UPR target proteins. Therefore, cells must be able to terminate PERK signaling and promote eIF2α dephosphorylation in the later stages of the UPR.
We report here on a role for P58IPK in terminating PERK activation in this later phase of the UPR. We find that the P58IPK gene is transcriptionally induced in the UPR through an ERSE in its promoter, and its encoded protein accumulates in ER-stressed cells. P58IPK binds to the kinase domain of PERK and inactivates it. P58IPK overexpression attenuates PERK activation by ER stress, whereas P58IPK −/− cells have higher persistent PERK activation during the ER-stress response.
P58IPK is a member of the Hsp40 family of chaperones, and its proposed role in modulating PERK signaling has interesting parallels with another chaperone, the ER luminal Hsp70 family member BiP/GRP78. BiP binding to the luminal domain of PERK blocks PERK oligomerization and activation. As the load of unfolded client proteins in the lumen of the organelle increases during the UPR, BiP is engaged progressively in their folding, and BiP–PERK complexes dissociate. Unbound PERK oligomerizes and is activated by trans-autophosphorylation, initiating signaling in the PERK pathway. BiP is also a UPR-induced gene (21, 24–26). As ER client-protein synthesis is inhibited and BiP protein levels rise, the equilibrium in the ER is shifted once again toward the reformation of PERK–BiP complexes, restoring the kinase to its inactive monomeric form (15, 27, 28). Our results suggest that P58IPK may play a similar role in PERK inactivation by binding to its kinase domain from the cytoplasmic side. Thus BiP and P58IPK may cooperate to terminate PERK signaling by independently targeting its luminal and cytoplasmic domains.
BiP and P58IPK-mediated PERK inactivation is not the only means by which signaling in the translational arm of the UPR is terminated. A third stress-inducible gene, GADD34, helps restore translation in the late, synthetic phase of the UPR. GADD34 recruits the catalytic subunit of a protein phosphatase, PP1c, to eIF2α, promoting its dephosphorylation. Expression of GADD34 strongly depends on eIF2α phosphorylation and is mediated in part by the transcription factor ATF4, the translation of which is induced under conditions of eIF2α phosphorylation. Thus GADD34 is part of a simple negative-feedback loop for terminating eIF2α phosphorylation and translational recovery (25). Induction of P58IPK is mediated by an ERSE in its promoter region, and therefore it is likely that it responds to signaling in the ATF6- and IRE1 → XBP-1-mediated arms of the UPR (29–31). These observations suggest that unlike GADD34, P58IPK affects integration of signals from UPR pathways that do not depend on eIF2α phosphorylation into promoting translational recovery. The modulatory role of P58IPK in PERK activity is likely to be significant even at physiological levels of ER stress, which is reflected in the high levels of P58IPK in the pancreas, an organ in which PERK activity is known to play an important role under basal conditions (32).
P58IPK was identified originally as a cellular repressor of PKR that is activated posttranscriptionally by the stress of viral infection. This study reveals an important role for transcriptional activation of P58IPK in the context of the UPR. However, it does not exclude a role for posttranscriptional activation of P58IPK in this setting, too. It is tempting to speculate that in the context of viral infection, as viral glycoproteins fill the ER causing ER stress, the posttranscriptional activation of P58IPK may act synergistically with the UPR-mediated transcriptional activation to afford influenza virus a measure of relief from both PKR- and PERK-mediated translational repression. It therefore seems that with respect to P58IPK, influenza virus has found a way to co-opt a gene that normally coordinates translational recovery in the UPR to serve its selfish needs.
Acknowledgments
We thank Eunice Badiable for technical support and the construction of the P58IPK-inducible cell line, John Morton and Warren Ladiges for providing the mouse ES cells, and Ted Holzman for bioinformatics support. We are also grateful to Dr. Stephen Spindler for providing the mouse GRP78-luciferase plasmid and Drs. Ron Wek and Scott Kimball for providing purified eIF2α proteins. This investigation was supported by National Institutes of Health Grants AI22646 (to M.G.K.) and ES08681 and DK47119 (to D.R.). C.L.F. is supported by Howard Hughes and Mary Gates undergraduate research fellowships.
Abbreviations
eIF2α, eukaryotic initiation factor 2α
PKR, protein kinase R
ER, endoplasmic reticulum
PERK, PKR-like ER kinase
UPR, unfolded protein response
ERSE, ER stress-response element
HA, hemagglutinin
ES, embryonic stem
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF495532).
References
- 1.Kaufman R. J. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Mathews, M. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 503–528.
- 2.Ron D. & Harding, H. P. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Mathews, M. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 547–560.
- 3.Lee T. G., Tomita, J., Hovanessian, A. G. & Katze, M. G. (1990) Proc. Natl. Acad. Sci. USA 87, 6208-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee T. G., Tomita, J., Hovanessian, A. G. & Katze, M. G. (1992) J. Biol. Chem. 267, 14238-14243. [PubMed] [Google Scholar]
- 5.Lee T. G., Tang, N., Thompson, S., Miller, J. & Katze, M. G. (1994) Mol. Cell. Biol. 14, 2331-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melville M. W., Tan, S. L., Wambach, M., Song, J., Morimoto, R. I. & Katze, M. G. (1999) J. Biol. Chem. 274, 3797-3803. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y., An, J., Liang, J., Hayes, S. E., Sandusky, G. E., Stramm, L. E. & Yang, N. N. (1999) J. Biol. Chem. 274, 5723-5730. [DOI] [PubMed] [Google Scholar]
- 8.Ladiges W., Morton, J., Blakely, C. & Gale, M. (2000) Mech. Ageing Dev. 114, 123-132. [DOI] [PubMed] [Google Scholar]
- 9.Korth M. J., Edelhoff, S., Disteche, C. M. & Katze, M. G. (1996) Genomics 31, 238-239. [DOI] [PubMed] [Google Scholar]
- 10.Tillman J. B., Mote, P. L., Walford, R. L. & Spindler, S. R. (1995) Gene 158, 225-229. [DOI] [PubMed] [Google Scholar]
- 11.Yan W., Gale, M. J., Jr., Tan, S. L. & Katze, M. G. (2002) Biochemistry 41, 4938-4945. [DOI] [PubMed] [Google Scholar]
- 12.Harding H. P., Zhang, Y. & Ron, D. (1999) Nature 397, 271-274. [DOI] [PubMed] [Google Scholar]
- 13.Tang N. M., Ho, C. Y. & Katze, M. G. (1996) J. Biol. Chem. 271, 28660-28666. [DOI] [PubMed] [Google Scholar]
- 14.Korth M. J., Lyons, C. N., Wambach, M. & Katze, M. G. (1996) Gene 170, 181-188. [DOI] [PubMed] [Google Scholar]
- 15.Bertolotti A., Zhang, Y., Hendershot, L. M., Harding, H. P. & Ron, D. (2000) Nat. Cell Biol. 2, 326-332. [DOI] [PubMed] [Google Scholar]
- 16.Barber G. N., Thompson, S., Lee, T. G., Strom, T., Jagus, R., Darveau, A. & Katze, M. G. (1994) Proc. Natl. Acad. Sci. USA 91, 4278-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X. Z., Lawson, B., Brewer, J. W., Zinszner, H., Sanjay, A., Mi, L. J., Boorstein, R., Kreibich, G., Hendershot, L. M. & Ron, D. (1996) Mol. Cell. Biol. 16, 4273-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scorsone K. A., Panniers, R., Rowlands, A. G. & Henshaw, E. C. (1987) J. Biol. Chem. 262, 14538-14543. [PubMed] [Google Scholar]
- 19.Roy B. & Lee, A. S. (1999) Nucleic Acids Res. 27, 1437-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida H., Haze, K., Yanagi, H., Yura, T. & Mori, K. (1998) J. Biol. Chem. 273, 33741-33749. [DOI] [PubMed] [Google Scholar]
- 21.Scheuner D., Song, B., McEwen, E., Liu, C., Laybutt, R., Gillespie, P., Saunders, T., Bonner-Weir, S. & Kaufman, R. J. (2001) Mol. Cell 7, 1165-1176. [DOI] [PubMed] [Google Scholar]
- 22.Shi Y., Vattem, K. M., Sood, R., An, J., Liang, J., Stramm, L. & Wek, R. C. (1998) Mol. Cell. Biol. 18, 7499-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melville M. W., Hansen, W. J., Freeman, B. C., Welch, W. J. & Katze, M. G. (1997) Proc. Natl. Acad. Sci. USA 94, 97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harding H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M. & Ron, D. (2000) Mol. Cell 6, 1099-1108. [DOI] [PubMed] [Google Scholar]
- 25.Novoa I., Zeng, H., Harding, H. P. & Ron, D. (2001) J. Cell Biol. 153, 1011-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozutsumi Y., Segal, M., Normington, K., Gething, M. J. & Sambrook, J. (1988) Nature 332, 462-464. [DOI] [PubMed] [Google Scholar]
- 27.Liu C. Y., Wong, H. N., Schauerte, J. A. & Kaufman, R. J. (2002) J. Biol. Chem. 277, 18346-18356. [DOI] [PubMed] [Google Scholar]
- 28.Ma K., Vattem, K. M. & Wek, R. C. (2002) J. Biol. Chem. 277, 18728-18735. [DOI] [PubMed] [Google Scholar]
- 29.Calfon M., Zeng, H., Urano, F., Till, J. H., Hubbard, S. R., Harding, H. P., Clark, S. G. & Ron, D. (2002) Nature 415, 92-96. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida H., Matsui, T., Yamamoto, A., Okada, T. & Mori, K. (2001) Cell 107, 881-891. [DOI] [PubMed] [Google Scholar]
- 31.Lee K., Tirasophon, W., Shen, X., Michalak, M., Prywes, R., Okada, T., Yoshida, H., Mori, K. & Kaufman, R. J. (2002) Genes Dev. 16, 452-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harding H. P., Zeng, H., Zhang, Y., Jungries, R., Chung, P., Plesken, H., Sabatini, D. D. & Ron, D. (2001) Mol. Cell 7, 1153-1163. [DOI] [PubMed] [Google Scholar]