Abstract
Serum amyloid A (SAA) is a small apolipoprotein that binds to high-density lipoproteins in the serum. Although SAA seems to play a role in host defense and lipid transport and metabolism, its specific functions have not been defined. Despite the growing implications that SAA plays a role in the pathology of various diseases, a high-resolution structure of SAA is lacking because of limited solubility in the high-density lipoprotein-free form. In this study, complementary methods including glutaraldehyde cross-linking, size-exclusion chromatography, and sedimentation-velocity analytical ultracentrifugation were used to show that murine SAA2.2 in aqueous solution exists in a monomer–hexamer equilibrium. Electron microscopy of hexameric SAA2.2 revealed that the subunits are arranged in a ring forming a putative central channel. Limited trypsin proteolysis and mass spectrometry analysis identified a significantly protease-resistant SAA2.2 region comprising residues 39–86. The isolated 39–86 SAA2.2 fragment did not hexamerize, suggesting that part of the N terminus is involved in SAA2.2 hexamer formation. Circular-dichroism spectrum deconvolution and secondary-structure prediction suggest that SAA2.2 contains ≈50% of its residues in α-helical conformation and <10% in β-structure. These findings are consistent with the recent discovery that human SAA1.1 forms a membrane channel and have important implications for understanding the 3D structure, multiple functions, and pathological roles of this highly conserved protein.
Serum amyloid A (SAA) proteins are a family of apolipoproteins found predominantly associated with high-density lipoprotein (HDL) in plasma (1), with different isoforms being unequally expressed constitutively and in response to inflammatory stimuli (2). Although synthesized primarily in the liver, extrahepatic tissue/cellular expression of SAA has been widely documented (3). SAA has been linked to functions related to inflammation, pathogen defense, HDL metabolism, and cholesterol transport and thereby has been implicated (3) in several pathological conditions including atherosclerosis, rheumatoid arthritis, Alzheimer's disease, and cancer.
SAA is known best for its role during the acute phase response to an inflammatory stimulus such as infection, tissue injury, and trauma (2). During active inflammation the concentration of SAA in plasma can increase up to 1,000-fold within 24 h (4). It is believed that persistently high levels of SAA during chronic inflammation may contribute to the occasional development of the potentially fatal disease reactive amyloidosis [amyloid A (AA) amyloidosis] (5). In AA amyloidosis, AA, an N-terminal (1–76) fragment of SAA (6), frequently is found to form amyloid deposits in the liver, kidney, and spleen. However, the presence, in vivo, of full-length SAA in amyloid deposits (7) and the ability of various SAA isoforms to form fibrils in vitro (8–10) suggest that proteolytic cleavage may not be a prerequisite for AA deposition but rather a postdeposition event. Of the three loci that express SAA in humans, SAA1 is the major, although not the only, precursor of AA deposits (11). Similarly, type A (i.e., BALB/c) mice contain two SAA isoforms, SAA2.1 and SAA1.1 [formerly known as SAA1 and SAA2, respectively (12)], of which only the latter deposits into amyloid after chronic inflammation induced with casein or azocasein (13). In contrast, the CE/J mouse strain produces a single SAA isoform, SAA2.2 (formerly known as SAA CE/J), which is amyloid-resistant (14, 15).
Although the exact in vivo functions of SAA are still obscure, its high conservation from fish to humans (16), wide expression profile in tissues/cells, and dramatic increase in expression levels during the acute phase response suggest a fundamental protective role for SAA. Yet, despite its small size (12 kDa) and highly significant functions, there is very limited structural information about SAA because of its inherent poor solubility in the apolipoprotein form. It is intriguing to understand how such a small protein is able to mediate or directly carry out such a wide range of functions related to inflammatory reaction and other host-defense mechanisms (3). The various functions of SAA may be modulated by factors such as conformational changes induced by ligand binding or by the ability to adopt more than one oligomeric state. Deciphering the molecular basis of the functional and potentially pathological properties of SAA will require understanding its structure under various conditions. In this study, we make a significant advancement toward understanding the structure of SAA, as we show by various methods that murine SAA2.2 can exist in aqueous solution as a hexamer containing a putative central channel.
Materials and Methods
SAA2.2 Expression and Purification.
The murine SAA2.2 cDNA was cloned into a pET21-a(+) vector between the NdeI and BamHI sites and transformed into Escherichia coli strain BL21 (DE3) pLysS-competent cells as described (17). Cells were lysed in 2-amino-2-hydroxymethyl-1,3-propanediol (Tris) buffer (20 mM, pH 8.2) by three consecutive freezing-thawing cycles, and the lysate was added gradually to buffer A (Tris buffer/6 M urea) and concentrated by ultrafiltration before loading onto a DEAE anion-exchange column (Sigma). SAA2.2 was eluted by a linear gradient of 20–100% buffer B (buffer A/0.4 M NaCl), and the fractions were pooled and desalted by several cycles of ultrafiltration using buffer A. SAA2.2 was purified further by chromatofocusing as described (8). The resulting fractions were precipitated with a 70% (final concentration) saturated ammonium sulfate solution, and the dissolved pellet was dialyzed against Tris buffer (pH 8.2) or 6 mM PBS (7.4) at 4°C.
Glutaraldehyde Cross-Linking (GCL).
SAA2.2 samples (0.1 mg/ml) in PBS were incubated at room temperature (20–22°C) for 20 min in various glutaraldehyde (Sigma) concentrations ranging from 0.006% to 0.6% (wt/vol). The cross-linking reaction was quenched by adding Tris buffer from a concentrated (1 M) stock solution to a final concentration of 0.1 M. The cross-linked SAA2.2 samples were analyzed by SDS/PAGE.
HPLC.
Reverse-phase (RP) HPLC was carried out on a Gold Noveau Beckman Coulter instrument, by using an analytic 4.6-mm-i.d. C4-RP column (Vydac, Hesperia, CA) running at 0.66 ml/min. A 5–90% linear gradient [0.1% (vol/vol) trifluoroacetic acid/90% (vol/vol) acetonitrile aqueous solution] was applied, and the column was regenerated with a 0.1% trifluoroacetic acid aqueous solution. Size-exclusion chromatography (SEC) experiments were performed on a Superdex 75 PC 3.2/30 column (Amersham Pharmacia) equilibrated with buffer B at a flow rate of 0.05 ml/min. The signal was monitored at 220 nm.
Limited Trypsin Proteolysis.
Experiments were performed at room temperature by using SAA2.2 samples (0.4 mg/ml) in Tris buffer (20 mM, pH 8.2) and a trypsin/SAA2.2 ratio of 1:120 (wt/wt). The proteolysis was quenched at various reaction times by adding 0.1% (vol/vol) trifluoroacetic acid or 1 mM phenylmethylsulfonyl fluoride. Subsequent RP-HPLC and mass spectrometry (MS) analysis of the SAA2.2 trypsin digest were performed after a 4-fold dilution in Tris buffer to generate 0.1 mg/ml SAA2.2 samples.
MS.
Liquid chromatography/electrospray MS services were provided by The W. M. Keck Biotechnology Resource Center at Yale University (New Haven, CT). Liquid chromatography/MS analysis was done in a Finnigan-MAT (San Jose, CA) LCQ quadrupole ion-trap mass spectrometer connected to an automated HPLC system with a 300-μm × 25-cm Vydac C18 column running at 3 μl/min.
Circular Dichroism (CD).
CD spectra were recorded on an OLIS (Bogart, GA) CD instrument. Data were collected in a 0.1-cm pathway quartz cuvette at 20°C. Secondary (2°) structure analysis from the CD spectra of SAA2.2 (0.1 mg/ml) and its fragment from residues 39 to 86 (SAA39–86, 0.17 mg/ml) in Tris buffer was carried out by using the programs CONTINLL, CDSSTR, and SELCON3 using a wavelength range of 190–240 nm and a reference set of 43 proteins (18).
Sedimentation-Velocity Analytical Ultracentrifugation.
Sedimentation-velocity data were collected in a Beckman Coulter XL-A centrifuge. A double-sector cell, equipped with a 12-mm Epon centerpiece and quartz windows, was loaded with ≈400 μl of SAA2.2 sample (0.27 mg/ml in 50 mM Tris, pH 8.2/150 mM NaCl). Data were collected at rotor speeds of 3,000–60,000 rpm in continuous mode at 25°C, with a step size of 0.005 cm and an average of 3 scans per point. The sedimentation-velocity absorbance profiles were analyzed to obtain the apparent distribution of sedimentation coefficients g(s*) for all the quaternary structures in solution by using the DCDT software provided by J. S. Philo (19). The partial specific-volume value of 0.71 cm3/g was determined based on the SAA2.2 amino acid sequence (20).
Electron Microscopy (EM) and Image Processing.
SAA2.2 (purified by gel filtration) was adsorbed to glow-discharged carbon-coated copper grids at 4°C. Grids were washed with two drops of deionized water and stained with two drops of freshly prepared 0.75% uranyl formate. Specimens were inspected with a Philips Tecnai 12 electron microscope operated at 120 kV, and images were taken at a nominal magnification of ×52,000 by using low-dose procedures. For image processing, 12 images were digitized with a Zeiss SCAI scanner using a pixel size of 4.04 Å at the specimen level. From the digitized images, 6,959 particles were selected and subjected to multireference alignment and image-classification procedures by using the SPIDER image-processing package (21).
Results
SAA2.2 Is a Hexamer in Solution.
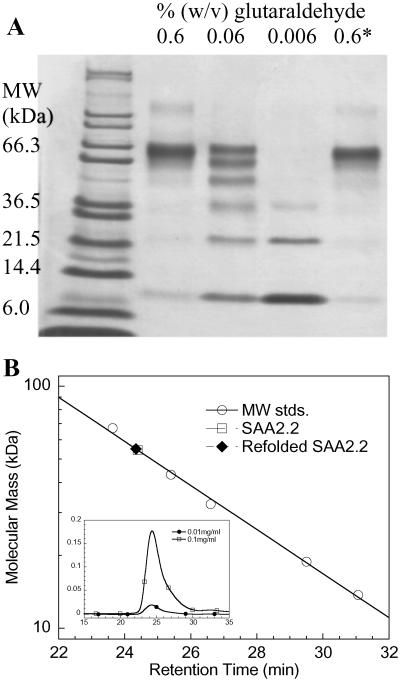
Because of its inherent tendency to self-assemble, it is not clear whether SAA carries out its various functions as a monomeric species or a higher oligomer. However, because of its multifunctional role, we hypothesized that SAA may function in more than one oligomeric state. The increased solubility of the murine SAA2.2 variant provides an opportunity to investigate the oligomeric state of the apolipoprotein in vitro. To determine the quaternary structure of SAA2.2 in solution, several complementary methods were used including GCL, SEC, sedimentation-velocity analytical ultracentrifugation, and EM. Glutaraldehyde reacts with the amino group of lysine side chains and therefore has been used extensively to cross-link oligomeric proteins (22). After cross-linking SAA2.2 with glutaraldehyde, its oligomeric state was analyzed by SDS/PAGE (Fig. 1A). The main band seen in lane 2 suggests that the oligomeric structure of SAA2.2 is consistent with a hexamer. A 10-fold decrease in glutaraldehyde concentration reveals (lane 3) all of the possible incompletely cross-linked SAA2.2 species (monomer through hexamer). Lane 5 shows that SAA2.2 unfolded in 6 M urea regains its oligomeric structure after refolding.
Fig 1.
Probing the oligomeric structure of SAA2.2. (A) GCL of SAA2.2 (0.1 mg/ml). The last lane (0.6*) shows cross-linking of a refolded SAA2.2 sample after it was unfolded in 6 M urea. (B) Size-exclusion HPLC of SAA2.2 at 0.1 and 0.01 mg/ml. Molecular mass standards (MW stds.) are BSA (67 kDa), ovalbumin (43 kDa), superoxide dismutase (32 kDa), horse myoglobin (19 kDa), and ribonuclease A (14 kDa). A 10-fold dilution of the SAA2.2 sample, from 0.10 to 0.01 mg/ml (10–1.0 μM), shows the same retention time, suggesting a submicromolar-to-nanomolar dissociation constant (Kd).
As a complement to the glutaraldehyde studies, SEC was used to probe the oligomeric state of SAA2.2. Fig. 1B shows a molecular mass calibration curve based on the retention times of six different proteins. The retention time (24.5 min) observed for SAA2.2 at ≈20°C yields a molecular mass of 55 kDa, which is lower than expected (70 kDa) but still consistent with the GCL data. The slight delay (0.75 min) in retention time is likely caused by the presence of smaller oligomeric species, as suggested by the observed asymmetric peak (Fig. 1B Inset). The presence of some monomeric SAA2.2 at 20°C is supported by temperature-induced denaturation experiments (L.W. and W.C., unpublished results). At 4°C, SAA2.2 elutes from the SEC column as a sharp symmetrical peak (Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org) with a retention time slightly faster than BSA, consistent with a homogenous hexameric structure.
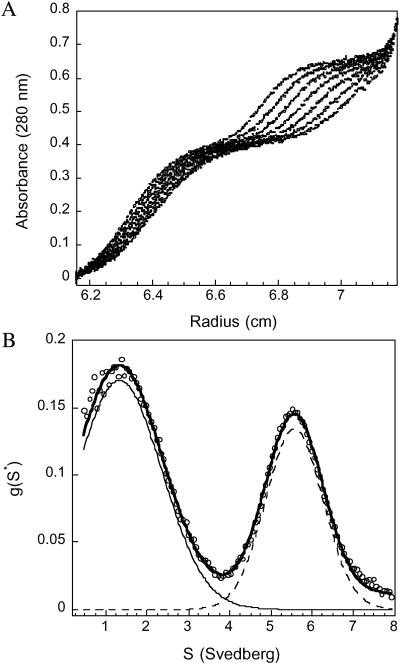
The additional bands shown in GCL experiments and the asymmetric SEC peak suggest that SAA2.2 may not exist in solution as a homogenous hexamer under our experimental conditions. Dynamic light-scattering experiments, which yielded a hydrodynamic radius for SAA2.2 consistent with a hexameric structure (3.76 nm; see Fig. 8, which is published as supporting information on the PNAS web site), also implied some quaternary structural heterogeneity. Therefore, we carried out sedimentation-velocity analytical ultracentrifugation experiments, and the sedimentation-velocity profiles of SAA2.2 at 25°C show two resolvable boundaries (Fig. 2), suggesting a mixture of at least two SAA2.2 species under these conditions. Analysis of the data reveals a sedimentation-coefficient distribution plot [g(s*)] that is consistent with two sedimenting species in solution. Fitting the g(s*) plot to a two-sedimenting-species model gave an excellent fit with sedimentation coefficients and molecular masses that correspond to a SAA2.2 monomer [s1 = 1.33 S (s20,w = 1.17 S ± 0.05), molecular mass (Mw)1 = 12 kDa, slow-moving boundary] and hexamer [s2 = 5.56 S (s20,w = 4.9 S ± 0.06), Mw2 = 73 kDa, fast-moving boundary], corresponding to ≈65% monomer and 35% hexamer based on initial absorbance values.
Fig 2.
SAA2.2 exists as a mixture of hexamer–monomer in equilibrium. (A) Sedimentation-velocity profiles of a 0.27 mg/ml sample of SAA2.2. Scans for analysis were recorded every 1 min; for clarity only eight representative scans 3 min apart are shown. The velocity data exhibit two resolvable boundaries, one corresponding to the monomer and the second boundary corresponding to hexamer. (B) Analysis of the sedimentation profiles in A using the time-derivative method (dc/dt) reveals the presence of two sedimenting species with an average sedimentation coefficient of 1.5 (±1) and 5.4 S (±1). The solid line represents a two-species fit of the data (o) using a Gaussian function.
EM Reveals a Putative Central Channel.
Electron micrographs of SAA2.2 specimens (prepared at 4°C where the hexamer is the predominant species in solution; unpublished data) showed particles with a diameter of ≈8 nm that were quite homogeneous in size and appearance (Fig. 3). Even in the raw images a stain accumulation in the center of the doughnut-shaped particles was evident, suggesting the presence of a central cavity or a putative pore. A similar staining pattern was seen with uranyl acetate (Fig. 9, which is published as supporting information on the PNAS web site), suggesting that the stain accumulation in the center of the particle is not a uranyl formate-specific staining artifact caused by decreased staining of protein components in the center of the hexamer. Approximately 7,000 of the doughnut-shaped 8-nm particles were selected and subjected to image classification specifying 50 output classes. Most of the resulting class averages showed few features other than a central stain accumulation, the putative pore, with a diameter of ≈2.5 nm (Fig. 3-1). The lack of fine structure in the averages is not surprising considering the very small size of the particles. Nevertheless, one of the class averages indeed showed indications of the ring being composed of six subunits (Fig. 3-2). This average, which contained 204 particles, was therefore sixfold-symmetrized to produce the projection map shown in Fig. 3-3. If this structure is approximated by six spheres with a diameter of 3.2 nm, and using a protein density of 1.35g/cm3 (23), the six spheres would accommodate a mass of ≈84 kDa. This value is relatively close to 70 kDa, the mass expected for a SAA2.2 hexamer.
Fig 3.
EM and projection structure of SAA2.2. Micrographs revealed doughnut-shaped particles that were homogeneous in size. (1) Representative projection average (177 particles) of the particle with a diameter of ≈8 nm and a central pore with a diameter of ≈2.5 nm. (2) Projection average (204 particles) that indicated a sixfold symmetry, which was applied to generate the sixfold-symmetrized average shown in 3. The side length of 1–3 is 20 nm. The circles in the raw image indicate bright spots that might represent monomeric SAA2.2.
Central Region of SAA2.2 Is Partially Trypsin-Resistant.
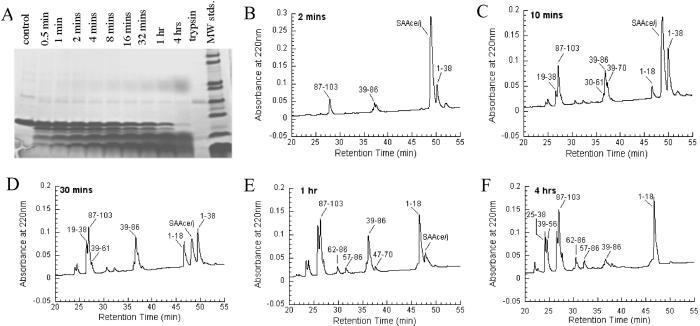
Because stably folded regions of a protein are usually more resistant to proteolysis, enhanced proteolytic susceptibility may identify protein regions that are either unfolded or highly flexible (24). Therefore, we used limited trypsin proteolysis to identify the regions of SAA2.2 that may comprise the hexamer intersubunit interface. Trypsin cleaves peptide bonds after the basic amino acids lysine and arginine, and SAA2.2 has 13 potential cleavage sites at positions 18, 24, 29, 33, 38, 46, 56, 61, 70, 83, 86, 89, and 102. Partial trypsin digestion of SAA2.2 generated a series of fragments including a 5-kDa fragment that is partially resistant to trypsin cleavage (Fig. 4A). The sequence of the various fragments (Fig. 4 B–F) was determined by liquid chromatography/MS (Fig. 5; Table 1).
Fig 4.
Time course of limited trypsin digestion of SAA2.2 (0.4 mg/ml) analyzed by SDS/PAGE (A) and RP-HPLC (B–F). The labels on RP-HPLC plots are based on the MS results (Fig. 5 and Table 1). MW stds., molecular mass standards.
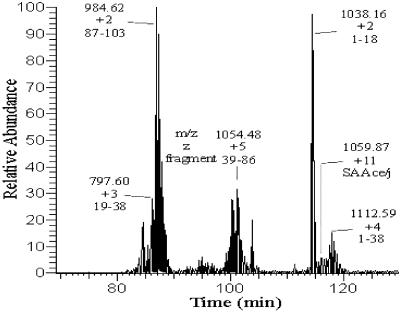
Fig 5.
Liquid chromatography/MS analysis of the SAA2.2 fragments generated after 30 min of trypsin proteolysis.
Table 1.
MS of SAA2.2 fragments generated by limited proteolysis
| Retention time, min | Assigned sequence | Expected mass, Da | LC/MS, Da |
|---|---|---|---|
| 26.68 | 19–38 | 2,389.6 | 2,391.1 |
| 26.87 | 87–103 | 1,967.2 | 1,967.9 |
| 36.93 | 39–86 | 5,293.6 | 5,295.0 |
| 46.62 | 1–18 | 2,074.3 | 2,075.3 |
| 49.27 | 1–103 | 11,670.7 | 11,647.8 |
| 50.39 | 1–38 | 4,445.9 | 4,447.4 |
Inspection of a proteolysis time-course experiment (Fig. 4 B–F) shows that residues 38/39 and 86/87 contain the two sites most susceptible to trypsin proteolysis (B), generating a 5.3-kDa fragment comprising residues 39–86. This fragment is not completely degraded after 4 h, suggesting that it is the protease-resistant fragment observed by SDS/PAGE (Fig. 4A). Therefore, it seems that arginines 38 and 86 are located in solvent-exposed and flexible regions of SAA2.2. The resulting 1–38 fragment is cleaved rapidly at arginine 18, generating the fragments 1–18 and 19–38. It is interesting that the 1–38 fragment is preferentially cleaved at position 18. It is possible that the 1–38 and 19–38 fragments may be interacting with the 39–86 fragment to form the corresponding 10- and 8-kDa bands that are seen by SDS/PAGE (Fig. 4A) but not by MS analysis. Also, the hydrophobic 1–18 fragment may be self-associating to form the diffuse growing band that runs with an apparent molecular mass of 36 kDa (Fig. 4A).
Prediction and Experimental Determination of the Secondary Structure of SAA2.2.
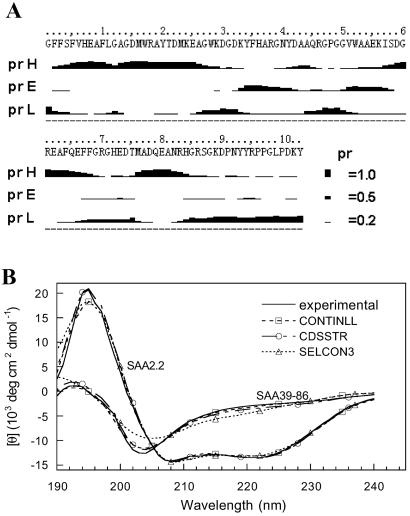
The secondary (2°) structure of SAA2.2 was predicted by using several algorithms from the ProteinPredict web server (http://cubic.bioc.columbia.edu/predictprotein). These methods are ≈76% accurate (25), and all gave similar results. In Fig. 6A we show representative results from PROF, which is an improved version of PHD, a profile-based neural network algorithm (26). The algorithm predicted (Fig. 6A) that SAA2.2 has a probability higher than 0.5 that 45% of its residues are in an α-helical structure and ≈5% in β-structure. The C terminus of SAA2.2 is predicted to lack any stable 2° structure. The values in 2° structure content are similar to those determined by CD for mice (27) and human (28, 29) SAA1, as well as structure prediction for human SAA1 (29, 30). The far-UV CD spectrum of SAA2.2 (Fig. 6B) is consistent with the predicted secondary structure, because its deconvolution using the programs CONTIN, SELCON3, and CDSSTR (18) indicate that the average content of α-helix and β-sheet structure in SAA2.2 is 54% and 7% (Fig. 6B), respectively. Reconstructions of the CD spectra based on the deconvoluted 2° structures produce model spectra that are virtually identical to the experimental data (Fig. 6B).
Fig 6.
Secondary-structure analysis of SAA2.2 and SAA39–86. (A) Structure prediction of SAA2.2 using the PROF algorithm (26) yields the probability of each residue to be in α-helix (pH), β-sheet (pE), or loop/other (pL) structure. (B) Far-UV CD spectra (solid lines) of SAA2.2 (0.1 mg/ml) and SAA39–86 (0.17 mg/ml) were deconvoluted by using the programs CONTINLL, CDSSTR, and SELCON3 (18). The average secondary structures obtained for SAA2.2 and SAA39–86 were 54% α-helix, 7% β-structure, and 39% other and 17% α-helix, 31% β-structure, and 52% other, respectively. The reconstructed CD spectra from the deconvoluted secondary structure are in excellent agreement with the experimental spectra.
Discussion
Implications of Hexameric SAA2.2 on the 3D Structure of SAA.
Our findings that lipid-free SAA2.2 in solution forms a hexamer containing a putative central channel, and knowledge of its 2° structure and solvent-exposed regions provides insight about the 3D structure of the SAA protein family. For instance, the N- and C-terminal 12 and 17 residues, respectively, are unlikely to contribute to SAA2.2 hexamerization, because the proline-rich C terminus is highly solvent-exposed and most likely unstructured (30), and the N terminus is involved in HDL/cholesterol binding (9, 31). Because residues 30–42 are also exposed in human and murine SAA (32, 33), the region available for hexamerization seems limited to residues 13–29 and 43–86, which are predicted to have a high α-helical structure content (Fig. 6A). The observation that the isolated fragment 39–86 does not hexamerize (data not shown) and exhibits a lower content of α-structure (20% α-helix) than expected (Fig. 6B) further supports our suggestion that part of the N terminus is required for hexamer formation.
The EM data (Fig. 3) show that six SAA2.2 subunits assemble into a hexameric ring (8 nm in diameter) surrounding a putative central channel (2.5 nm in diameter). Therefore, it seems that monomeric SAA2.2 interacts via two different interfaces, which may involve the helical N-terminal residues 13–29 and the central residues 56–86. Alternatively, it is conceivable that SAA2.2 may form a hexamer via domain swapping, because this is an efficient mechanism for a small protein to form higher-order oligomeric structures (34).
Implications for SAA Function.
A better understanding of the structure of SAA could help elucidate how its various functions (3) are exerted, regulated, and participating in disease. The wide range of functions suspected for SAA suggests that it may exist in more than one oligomeric form, perhaps modulated by ligand binding or the levels of SAA expression. In particular, it is conceivable that the drastic increase in SAA expression levels that occurs during inflammation could determine its oligomeric state and thereby its function. It is not clear how hexameric SAA with a central channel would function, especially concerning its apparently central role of binding HDL and transporting cholesterol. Sequence-specific antibodies raised against synthetic peptides corresponding to different amino acid regions of human SAA1 showed that the same epitopes are exposed irrespective of whether SAA1 is lipid-bound or free in solution (32), suggesting that the conformation and oligomeric state of SAA is very similar in both cases. Therefore, SAA may bind to HDL as a hexamer. A different function for SAA may involve the formation of toxic channels on the cell membrane of bacteria, thereby protecting the host against bacterial infection (29). This idea is supported by the previous observation of E. coli cell lysis caused by SAA overexpression (8) and the recent report that human SAA1 can form voltage-independent ion channels in lipid bilayers membranes (29).
Various functions of SAA are linked to regions that are shown to be flexible or solvent-exposed within the hexamer and therefore may not require hexamer formation. The C terminus of human and murine SAA shows binding affinity to heparan sulfate (35), a ubiquitous component on the cell surface and within the extracellular matrix (ECM). The observation that peptides derived from the C terminus of SAA can also bind to heparan sulfate in vitro (35) suggests that hexameric SAA is not required for this interaction. SAA also contains a highly conserved RGD-like adhesion motif (RGN, residues 39–41 in SAA2.2) that adheres to various cells including platelets (36). Peptides derived from SAA and comprising amino acids 29–42 also exhibited platelet-adhesion function, suggesting that hexameric SAA is not required for this function. This is consistent with our proteolysis studies and sequence-specific antibody experiments (32, 37), which indicate that this region is solvent-exposed.
Implications of the Hexameric Structure of SAA2.2 in Reactive Amyloidosis.
Despite three decades of research, the molecular mechanism of SAA amyloid formation in AA amyloidosis remains poorly understood. Studies with peptide fragments (38) and the full-length protein (39) suggest that the N terminus of SAA largely determines its amyloidogenicity. Very little is known about the role of the central region of SAA in AA amyloid formation, but it is likely to be relevant, because human SAA1 is found predominantly in AA deposits, even though the sequence of its N terminus is identical to SAA2 (11, 40). Furthermore, the high probability (≥0.5) of β-structure predicted (Fig. 6A) for the central residues 30–55 and the high β-structure content observed for the SAA2.2 39–86 fragment (Fig. 6B) suggest that this region may be more important in amyloid formation than previously believed.
Our discovery that SAA2.2 forms a hexamer in solution may have implications concerning the amyloidogenicity of SAA isoforms and the pathogenic mechanism of AA amyloidosis. Because amyloid fibrils are rich in β-sheet structure, the predominantly α-helical secondary structure (Fig. 6) of SAA suggests that the hexamer is not an intermediate precursor to amyloid formation. We hypothesize that a hexamer-to-monomer dissociation accompanied by a conformational change that leads to an increase in β-sheet structure may result in the formation of an amyloidogenic precursor intermediate. Thus, factors that could destabilize the hexameric structure, including ligand binding or the amino acid sequence of the SAA isoform, may play a role in SAA fibrillogenesis. Alternatively, the presence of a putative central channel in SAA may be relevant to AA amyloidosis, as suggested by the accumulating evidence that the formation of toxic channels or pores on cell membranes may be involved in the pathology of amyloid diseases (41, 42). Whether hexameric SAA or a higher oligomerized species play a similar role in the pathogenic mechanism of AA amyloidosis remains to be determined.
Supplementary Material
Acknowledgments
We are especially grateful to J. Sipe for providing the SAA2.2 construct, F. Xia and D. Moriarty for help with HPLC experiments, and Y. Cheng for taking the EM images. The molecular EM facility at Harvard Medical School was established by a donation from The Giovanni Armenise Harvard Center for Structural Biology and is maintained through National Institutes of Health Grant GM62580. This work was supported by a grant from the American Heart Association and a Camille and Henry Dreyfus New Faculty Award (to W.C.).
Abbreviations
AA, amyloid A
SAA, serum AA
HDL, high-density lipoprotein
GCL, glutaraldehyde cross-linking
SEC, size-exclusion chromatography
CD, circular dichroism
EM, electron microscopy
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Benditt E. P. & Eriksen, N. (1977) Proc. Natl. Acad. Sci. USA 74, 4025-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uhlar C. M. & Whitehead, A. S. (1999) Eur. J. Biochem. 265, 501-523. [DOI] [PubMed] [Google Scholar]
- 3.Urieli-Shoval S., Linke, R. P. & Matzner, Y. (2000) Curr. Opin. Hematol. 7, 64-69. [DOI] [PubMed] [Google Scholar]
- 4.McAdam K. P. W. J. & Sipe, J. D. (1976) J. Exp. Med. 144, 1121-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillmore J. D., Lovat, L. B., Persey, M. R., Pepys, M. B. & Hawkins, P. N. (2001) Lancet 358, 24-29. [DOI] [PubMed] [Google Scholar]
- 6.Husebekk A., Skogen, B., Husby, G. & Marhaug, G. (1985) Scand. J. Immunol. 21, 283-287. [DOI] [PubMed] [Google Scholar]
- 7.Westermark P. & Sletten, K. (1982) Clin. Exp. Immunol. 49, 725-731. [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada T., Kluve-Beckerman, B., Liepnieks, J. J. & Benson, M. D. (1994) Biochim. Biophys. Acta 1226, 323-329. [DOI] [PubMed] [Google Scholar]
- 9.Patel H., Bramall, J., Warters, H., De Beers, M. C. & Woo, P. (1996) Biochem. J. 318, 1041-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu J., Zhu, H., Guo, J. T., de Beer, F. C. & Kindy, M. S. (2000) Lab. Invest. 80, 1797-1806. [DOI] [PubMed] [Google Scholar]
- 11.Liepnieks J. J., Beckerman, B. K. & Benson, M. D. (1995) Biochim. Biophys. Acta 1270, 81-86. [DOI] [PubMed] [Google Scholar]
- 12.Sipe J. (1999) Amyloid 6, 67-70. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman J. S., Ericsson, L. H., Eriksen, N., Walsh, K. A. & Benditt, E. P. (1984) J. Exp. Med. 159, 641-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sipe J. D., Carreras, I., Gonnerman, W. A., Cathcart, E. S., de Beer, M. C. & de Beer, F. C. (1993) Am. J. Pathol. 143, 1480-1485. [PMC free article] [PubMed] [Google Scholar]
- 15.de Beer M. C., de Beer, F. C., McCubbin, W. D., Kay, C. M. & Kindy, M. S. (1993) J. Biol. Chem. 268, 20606-20612. [PubMed] [Google Scholar]
- 16.Uhlar C. M., Burgess, C. J., Sharp, P. M. & Whitehead, A. S. (1994) Genomics 19, 228-235. [DOI] [PubMed] [Google Scholar]
- 17.Liang J., Elliott-Bryant, R., Hajri, T., Sipe, J. D. & Cathcart, E. S. (1998) Biochim. Biophys. Acta 1394, 121-126. [DOI] [PubMed] [Google Scholar]
- 18.Sreerama N. & Woody, R. W. (2000) Anal. Biochem. 287, 252-260. [DOI] [PubMed] [Google Scholar]
- 19.Philo J. S. (1997) Biophys. J. 72, 435-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durchschlag H. (1986) in Thermodynamic Data for Biochemistry and Biotechnology, ed. Hinz, H.-J. (Springer, New York), pp. 45.
- 21.Frank J., Radermacher, M., Penczek, P., Zhu, J., Li, Y., Ladjadj, M. & Leith, A. (1996) J. Struct. Biol. 116, 190-199. [DOI] [PubMed] [Google Scholar]
- 22.Craig W. S. (1988) Methods Enzymol. 156, 333-345. [DOI] [PubMed] [Google Scholar]
- 23.Matthews B. W. (1968) J. Mol. Biol. 33, 491-497. [DOI] [PubMed] [Google Scholar]
- 24.Fontana A., Fassina, G., Vita, C., Dalzoppo, D., Zamai, M. & Zambonin, M. (1986) Biochemistry 25, 1847-1851. [DOI] [PubMed] [Google Scholar]
- 25.Rost B. & Eyrich, V. A., (2001) Proteins, Suppl. 5, 192–199. [DOI] [PubMed]
- 26.Rost B. (1996) Methods Enzymol. 266, 525-539. [DOI] [PubMed] [Google Scholar]
- 27.McCubbin W. D., Kay, C. M., Narindrasorasak, S. & Kisilevsky, R. (1988) Biochem. J. 256, 775-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bausserman L. L., Herbert, P. N., Forte, T., Klausner, R. D., McAdam, K. P., Osborne, J. C., Jr. & Rosseneu, M. (1983) J. Biol. Chem. 258, 10681-10688. [PubMed] [Google Scholar]
- 29.Hirakura Y., Carreras, I., Sipe, J. D. & Kagan, B. L. (2002) Amyloid 9, 13-23. [DOI] [PubMed] [Google Scholar]
- 30.Turnell W., Sarra, R., Glover, I. D., Baum, J. O., Caspi, D., Baltz, M. L. & Pepys, M. B. (1986) Mol. Biol. Med. 3, 387-407. [PubMed] [Google Scholar]
- 31.Liang J. S., Schreiber, B. M., Salmona, M., Phillip, G., Gonnerman, W. A., de Beer, F. C. & Sipe, J. D. (1996) J. Lipid Res. 37, 2109-2116. [PubMed] [Google Scholar]
- 32.Malle E., Herz, R., Artl, A., Ibovnik, A., Andreae, F. & Sattler, W. (1998) Scand. J. Immunol. 48, 557-561. [DOI] [PubMed] [Google Scholar]
- 33.Elliott-Bryant R., Liang, J. S., Sipe, J. D. & Cathcart, E. S. (1998) Scand. J. Immunol. 48, 241-247. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y. & Eisenberg, D. (2002) Protein Sci. 11, 1285-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ancsin J. B. & Kisilevsky, R. (1999) J. Biol. Chem. 274, 7172-7181. [DOI] [PubMed] [Google Scholar]
- 36.Urieli-Shoval S., Shubinsky, G., Linke, R. P., Fridkin, M., Tabi, I. & Matzner, Y. (2002) Blood 99, 1224-1229. [DOI] [PubMed] [Google Scholar]
- 37.Malle E., Munscher, G., Muller, T., Vermeer, H. & Ibovnik, A. (1995) J. Immunol. Methods 182, 131-144. [DOI] [PubMed] [Google Scholar]
- 38.Westermark G. T., Engström, U. & Westermark, P. (1992) Biochem. Biophys. Res. Commun. 182, 27-33. [DOI] [PubMed] [Google Scholar]
- 39.Kirschner D. A., Elliott-Bryant, R., Szumowski, K. E., Gonnerman, W. A., Kindy, M. S., Sipe, J. D. & Cathcart, E. S. (1998) J. Struct. Biol. 124, 88-98. [DOI] [PubMed] [Google Scholar]
- 40.Meek R. L., Hoffman, J. S. & Benditt, E. P. (1986) J. Exp. Med. 163, 499-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lashuel H. A., Hartley, D., Petre, B. M., Walz, T. & Lansbury, P. T., Jr. (2002) Nature 418, 291. [DOI] [PubMed] [Google Scholar]
- 42.Kagan B. L., Hirakura, Y., Azimov, R., Azimova, R. & Lin, M. C. (2002) Peptides (Tarrytown, NY) 23, 1311-1315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.