Abstract
Identification of therapeutic strategies to prevent or cure diseases associated with amyloid fibril deposition in tissue (Alzheimer's disease, spongiform encephalopathies, etc.) requires a rational understanding of the driving forces involved in the formation of these organized assemblies rich in β-sheet structure. To this end, we used a computer-designed algorithm to search for hexapeptide sequences with a high propensity to form homopolymeric β-sheets. Sequences predicted to be highly favorable on this basis were found experimentally to self-associate efficiently into β-sheets, whereas point mutations predicted to be unfavorable for this structure inhibited polymerization. However, the property to form polymeric β-sheets is not a sufficient requirement for fibril formation because, under the conditions used here, preformed β-sheets from these peptides with charged residues form well defined fibrils only if the total net charge of the molecule is ±1. This finding illustrates the delicate balance of interactions involved in the formation of fibrils relative to more disordered aggregates. The present results, in conjunction with x-ray fiber diffraction, electron microscopy, and Fourier transform infrared measurements, have allowed us to propose a detailed structural model of the fibrils.
The ability of soluble proteins or protein fragments to convert spontaneously into amyloid fibrils under some circumstances is a very important biological phenomenon, because this process is related to a range of human disorders such as spongiform encephalopathies, Alzheimer's disease, etc. (1, 2). Although soluble precursors of amyloidogenic proteins do not have any obvious sequence homology or common folding patterns, x-ray fiber diffraction data indicate that all amyloid fibrils share a characteristic cross-β-structure (3, 4). This finding suggests that the key elements of the fibril formation process may be common to all proteins and that, therefore, a highly simplified system that is able to polymerize into β-sheets can offer additional insights into the molecular details of amyloid fibril formation. Up till now, the search for such simple model systems has been based mainly on empirical approaches such as, for example, the identification of fragments of amyloidogenic proteins that can self-assemble into amyloid fibrils (5–8). Some rational designs based on alternating hydrophobic and hydrophilic residues have been found to have some success in identifying the interactions behind β-sheet polymerization (9). Although such systems have helped to clarify many aspects of this process, our detailed understanding of the factors promoting amyloid formation remains limited.
To probe the interactions driving β-sheet aggregation, we have investigated the effect of specific residues on the propensity of a given sequence to form amyloid fibrils. To this end, we have used a computer-based method (10–12) to design a series of self-associating hexapeptides able to form polymeric β-sheet structures arranged in a similar fashion to that assumed to be present in amyloid fibrils. This rational approach, together with the simplicity of the designed model system, has served to dissect the intermolecular forces driving amyloid fibril formation and to propose a structural model of the fibrils that is consistent with the organization of the protofilament core of amyloid fibrils.
Methods
Peptide Design.
A detailed description of the algorithm PERLA is available elsewhere (10–12). In this work, the molecular mechanics force-field energies and entropy changes were scaled by a factor of 0.5, and the relative dielectric constant used was 16.0.
Peptide Synthesis and Purification.
Peptides were synthesized by using the standard Fmoc solid-phase chemistry. Peptide homogeneity and composition were analyzed by analytical HPLC and mass spectrometry (95% purity).
Fibril Sample Preparation.
Stock peptide solutions were prepared by dissolving a weighed amount of peptide into buffered solution (20 mM glycine/HCl, pH 2.6/20 mM Na2HPO4/NaH2PO4, pH 6.0/20 mM Na2HPO4/NaH2PO4, pH 7.4/20 mM KCl/NaOH, pH 12.5), water, or NaCl aqueous solution (0.05, 0.1, 0.25, 0.5, and 1.0 M) whose pH had been adjusted previously to 2.6. Samples were immediately sonicated (10 min) to dissemble preformed nuclei and centrifuged (5 min at 16,100 × g) to deposit insoluble material. The concentration of the stock solutions was determined by measuring the absorbance at 280 or 220 nm for peptides without aromatic residues. Solutions of concentrations of 50, 200, 500, and 1,000 μM were prepared by diluting a known volume of stock solution into buffer. Then, peptide solutions were incubated at room temperature and checked by CD and EM at different incubation times (t = 0, 7 days, and 1 month).
Far-UV CD.
CD spectra were acquired on a Jasco-710 instrument calibrated with (1S)-(+)-10-camphorsulphonic acid. Usually, 20 scans were acquired in the range 190–250 nm at a temperature of 25°C by taking points every 0.2 nm, with a scan rate of 100 nm per min, an integration time of 1 s, and a band width of 1 nm. Time-scan experiments were carried out recording ellipticity at 218.5 nm, with a sampling period of 10–20 s, response times of 2 s, and a band width of 1 nm.
Electron Microscopy.
Aliquots (5 μl) of the fibril preparation were adsorbed to glow-discharged carbon-coated collodion film on 400-mesh copper grids for 1 min. The grids were blotted, washed twice in droplets of MilliQ water, and stained with 1% (wt/vol) uranyl acetate. For cryo-EM studies, fibrils were adsorbed onto carbon-coated holey films (Qantifoil, Jena, Germany) with a hole diameter of 3.5 μm and quick-frozen in liquid ethane. Electron microscopy was carried out in a Philips CM200FEG by using a GATAN-626 cryo-holder. Images were recorded on Kodak SO163 film at a nominal magnification of ×38,000 and digitized at 21 μm stepsize, corresponding to 0.55 nm actual dimension on the specimen. Suitable areas were analyzed by using the image processing software SUPRIM (13).
Unidirectional Metal Shadowing.
Amyloid fibrils were adsorbed on carbon-coated EM grids. The samples were quick-frozen in LN2 and freeze dried at 5 × 10−7 mbar and 180 K for 2 h. The dried specimens were then unidirectionally shadowed with a thin layer (≈0.5 nm) of tantalum/tungsten at an elevation angle of 45°. Freeze-drying and metal shadowing took place in a specially designed vacuum-transfer system that is directly connected to a CM12 electron microscope (Midilab, Eidgenössische Technische Hochschule, Switzerland).¶ This system allows for a direct specimen transfer into the microscope onto a modified GATAN-626 cryo-holder.¶ Images were recorded with a GATAN-794 1 K charge-coupled device at a nominal magnification of ×35,000 on the microscope, which corresponds to a pixel size of 0.53 nm.
Fourier-Transform Infrared Spectroscopy.
FTIR spectra were recorded by using an FTS 175C spectrometer (Bio-Rad, Cambridge, MA) equipped with a liquid N2-cooled mercury cadmium telluride detector and purged with a continuous flow of N2 gas. Solutions of ≈1 mg/ml peptide were prepared and incubated in buffered solution (20 mM glycine-d5/DCl, pD 2.6). The samples then were concentrated to ≈10 mg/ml by ultracentrifugation at 300,000 × g for 1 h, and the resulting pellet was resuspended in buffer. This procedure was repeated twice to remove any remaining soluble peptide that could contribute to the spectrum, as reported (14). EM examination of the samples indicated no morphological changes as a result of the concentration procedure. Peptide solutions were placed between a pair of CaF2 windows separated by a 30-μm Mylar spacer, and spectra were collected at 25°C. For each sample, 256 interferograms were accumulated at a spectral resolution of 2 cm−1.
X-Ray Fiber Diffraction.
An aliquot of 15 μl of fibrils prepared as described above for FTIR spectroscopy (concentration of ≈10 mg/ml) was placed between two capillaries with waxed ends and allowed to dry to promote alignment of the fibrils. The resulting stalk was placed in the x-ray beam, and diffraction patterns were collected in a Cu Kα rotating anode equipped with a 345 MAR-Research image plate (MarResearch, Hamburg, Germany) for 20–30 min. Images were analyzed by using MARVIEW software.
Results
Design Procedure.
By using a computer-designed algorithm (10–12), we searched for peptide sequences optimized to self-associate into polymeric β-sheet structures. We assumed that propagation and stacking of the preformed β-sheets would result in the final assembly of amyloid fibrils. On that assumption, the design procedure was carried out by using a polymeric six-stranded antiparallel β-sheet with six residues per strand as a structural template for calculations. The three-dimensional model for the backbone was constructed by taking the six central strands of the large single-layer β-sheet exhibited by the outer surface protein A (OspA; ref. 15). The algorithm used for the design enabled identification and sorting of amino acid sequences with a high propensity to fold into the desired three-dimensional structure. To reduce the sequence space to be explored, we designed homopolymeric β-sheets (i.e., having the same sequence for all of the strands) and selected a simplified amino acid repertoire containing both polar and hydrophobic residues (Table 1). The criterion for selection of a subset from the output sequences was that the set of peptides should be as similar as possible while containing sequences both energetically favorable and unfavorable to adopt a polymeric β-sheet conformation (Table 1).
Table 1.
Designed sequences and self-assembly behavior as a function of the molecule net charge
| Sequence
|
Net charge of the molecule | |||
|---|---|---|---|---|
| +1 | Neutral | −1 | >1 | |
| No. 1 KTVIIE | Ac—K+TVIIE—Am | Ac—K+TVIIE−—am | Ac—KTVIIE−—Am | N+—K+TVIIE—C |
| pH 2.6, 98%; β | pH 7.4, 100%; β | pH 12.5, 100%; rc + β | pH 2.6, 92%, +2; rc | |
| No. 3 STVIIE | N+—STVIIE—C | Ac—STVIIE—Am | Ac—STVIIE−—am | |
| pH 2.6, 92%; β | pH 2.6, 98%; rc | pH 7.4, 100%; rc + β | ||
| No. 4 KTVIIT | Ac—K+TVIIT—Am | Ac—KTVIIT—Am | N—KTVIIT—C− | N+—K+TVIIT—C |
| pH 2.6, 100%; rc | pH 12.5, 100%; rc | pH 12.5, 100%; rc | pH 2.6, 97%, +2; rc | |
| No. 5 STVIIT | N+—STVIIT—C | Ac—STVIIT—Am | Ac—STVIIT—C− | |
| pH 2.6, 97%; β | pH 2.6, 100%; β | pH 7.4, 100%; rc + β | ||
| N+—STVIIT—C− | ||||
| pH 6.0, 96%; β | ||||
| No. 6 KTVLIE | Ac—K+TVLIE—Am | Ac—K+TVLIE−—Am | Ac—KTVLIE−—Am | N+—K+TVLIE—C |
| pH 2.6, 98%; rc | pH 7.4, 100%; β | pH 12.5, 100%; rc | pH 2.6, 92%, +2; rc | |
| No. 7 KTVIVE | Ac—K+TVIVE—Am | Ac—K+TVIVE−—Am | Ac—KTVIVE−—Am | N+—K+TVIVE—C |
| pH 2.6, 98%; rc | pH 7.4, 100%; β | pH 12.5, 100%; rc | pH 2.6, 92%, +2; rc | |
| No. 8 KTVIYE | Ac—K+TVIYE—Am | Ac—K+TVIYE−—Am | Ac—KTVIYE−—Am | N+—K+TVIYE−—C |
| pH 2.6, 98%; rc | pH 7.4, 100%; rc | pH 12.5, 100%; rc | pH 2.6, 92%, +2; rc | |
| No. 9 STVIYE | N+—STVIYE—C | Ac—STVIYE—Am | Ac—STVIYE−—Am | |
| pH 2.6, 92%; rc + β | pH 2.6, 98%; rc | pH 7.4, 100%; rc + β | ||
rc and β annotations refer to the CD signature of the peptide at the pH given. “rc” indicates random coil CD spectrum, “β” stands for β-sheet CD spectrum, and “rc + β” indicates the presence of both β-structure and unfolded conformations in solution. Bold annotations correspond to sequences and conditions that showed amyloid fibril formation as indicated by EM and x-ray diffraction data.
Sequences are given using the one-letter code for amino acid residues. Amino acids modelled for the six-residue β-strands of the six-stranded polymeric β-sheet are: at positions 1 and 6, K, E, T, and S; at positions 2 and 5, K, E, T, S, V, I, L, Y and W; and at positions 3 and 4, V, I, L, Y and W. The predicted sequence stability is No. 1 > 3 > 6 > 7 > 8 > 9 > 5 > 4.
Superscripts “+” and “−” stand for a formal positive and negative charge, respectively. “Ac” stands for an N terminus protected with an acetyl group, “Am” for a C terminus protected with an amide group, “N” for a free N terminus, and “C” for a free C terminus. pH and percentage of molecules with a given net charge at that pH are given below each peptide sequence. This percentage has been calculated by using the Arrhenius equation (pH = pK + log([acceptor]/[donor])) and the following pKa values (31): free N terminus, pKa(S) = 7.34; pKa(K) = 7.10. Free C terminus, pKa(T) = 4.07; pKa(E) = 3.65. Charged side chains, pKa(K) = 10.60; pKa(E) = 4.25.
Effective Net Charge on the Designed Peptide Sequences.
Amyloid fibril formation and amorphous aggregation are in many cases highly sensitive to solution conditions, including pH and the presence of salts (16–21). This fact indicates that the charged state, as well as the conformational state of the polypeptide chain, plays an important role in the formation of amyloid fibrils in vitro. This evidence led us to propose that, at least in some cases, the presence of noncompensated charges might act as a predominant force in the supramolecular organization of proteins into protofilaments and/or amyloid fibrils rather than into amorphous aggregates. To check this hypothesis, we studied the aggregation behavior of the selected sequences at different total net charges (0, ±1, and ±2). These charge variations were achieved by synthesizing versions of the same sequences with either both termini protected (acetylated at the N terminus and amidated at the C terminus), only one terminus protected, or with both termini free, as well as by varying the pH of the solution (pH 2.6, 6.0, 7.4, and 12.5; Table 1).
Sequences That Form Polymeric β-Sheets.
The ability of the selected sequences to form the target polymeric β-sheet was assessed by recording far-UV CD spectra under different experimental conditions. In general, all those sequences that were predicted to be energetically highly favorable to form β-sheet (nos. 1, 3, 6, and 7) displayed a β-sheet-like spectrum at concentrations equal to or higher than 0.5 mM (Table 1). Those sequences predicted to be less favorable for this polymeric topology (e.g., sequence no. 4) displayed typical random coil CD spectra under all experimental conditions examined here (Table 1). These results support the usefulness of the computational method for designing single mutations (e.g., Glu-6 → Thr from nos. 1 to 4) that disfavor β-sheet polymerization. The disagreements observed in some cases between predicted and experimental propensities to polymerize (e.g., sequence no. 5 STVIIT) might be caused by a bias of the backbone template used for the modeling toward twisted β-sheets. It is possible, however, that polymeric β-sheets are flatter than the β-sheets found in globular proteins, as suggested by some structural models of amyloid fibrils (20, 22).
From Polymeric β-Sheet to Amyloid Fibril.
A CD β-sheet signature is not unequivocally linked to the formation of amyloid fibrils, because it can also be displayed by other kinds of aggregates (16–18). To identify the factors that are important for the conversion of the polymeric β-sheets detected by CD spectroscopy into fibrils, we analyzed the samples by EM. Peptides that did not polymerize into β-sheets did not form amyloid fibrils under any conditions that we tested. Kinetic analysis of fibril formation monitored by CD and EM showed that β-sheet polymerization precedes fibril formation (Fig. 1). Nevertheless, although β-sheet formation appears to be necessary for fibril formation, it is not a sufficient criterion, as some samples that displayed a β-sheet CD spectrum did not proceed to form fibrils. Only samples from peptides 1, 3, 5, and 9 revealed the presence of fibrils. Moreover, certain point mutations involving substitutions of the Ile residues (e.g., Ile-4 → Leu from no. 1 to 6; Ile-5 → Val from no. 1 to 7) completely prevent fibril growth from preformed β-sheets. For amyloidogenic peptides with charged groups (nos. 1, 3, and 9), amyloid fibrils were only observed in our experiments when the molecule carried a net charge of ±1 (Table 1). Peptide solutions that displayed a β-sheet spectrum when the peptide net charge was zero resulted in the formation of amorphous aggregates, or the peptides simply remained soluble. All solutions where the peptide carried a net charge higher than 1 displayed a typical random coil CD spectrum and did not form fibrils under any of the conditions tested.
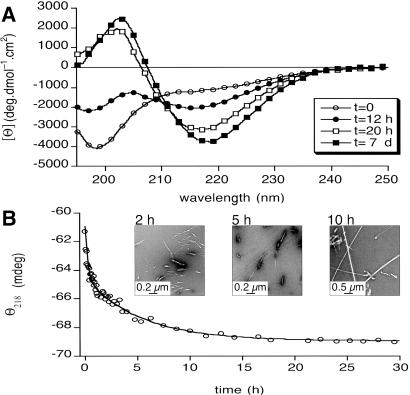
Fig 1.
Self-association kinetics of peptide no. 3. (A) Far-UV CD spectra showing the random coil → β-sheet transition (c = 0.8 mM, pH = 2.6) experienced on time (t = 0, 12 h, 20 h, and 7 days). (B) Plot of the ellipticity change at 218 nm (Θ218; β-sheet minimum) as a function of the time (c = 1.2 mM, pH = 2.6). Electron micrographs of aliquots taken at t = 2, 5, and 10 h indicate that although Θ218 reaches a plateau at t ≈ 4–5 h, the degree of maturation and number of fibrils still increases with time. Θ218 at t = 2 h indicates a high percentage of β-sheet structure, whereas EM shows the presence of amorphous species and very short filaments.
The importance of the charged state of the precursor in the present series of experiments was checked further by incubation of peptides with charged residues in the presence of different concentrations of salt (0–1 M NaCl). Up to 0.1 M NaCl, the rate of the fibrillation process increased with ionic strength, as a result attributable to a reduction of charge-repulsion effects. At higher ionic strength, however, only very short filaments and amorphous aggregates could be observed (data not shown), which is consistent with the screening of charges under such conditions.
Peptide STVIIT, having no charged amino acids but several residues whose side chains contain hydrogen-bond donor and acceptor groups, formed fibrils at an effective charge of zero if both termini were protected. However, it formed amorphous aggregates if the net charge of zero resulted from compensation of a negative and a positive charge (free N and C termini at pH 6.0), illustrating again the importance of charged groups in driving fibril formation.
Fibril Morphology as a Function of the Net Charge on the Molecule.
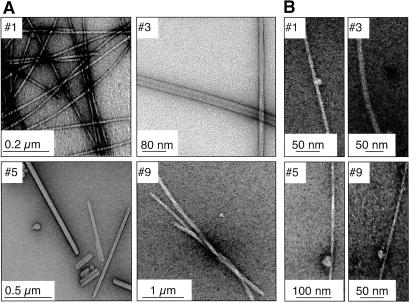
At acidic pH, fibrils grown from peptides carrying a net charge of +1 exhibited variable morphology (Fig. 2A). In contrast, the same sequences with a total net charge of −1 led to fibrils with a lower morphological variability, although differences in fibril diameter can be observed (Fig. 2B). Under these conditions, the rate of formation of fibrils was also slower. This result suggests that, under conditions where polymerization is slow, the formation of fibrils is under thermodynamic control, and all molecules are able to achieve the most favorable fibril architecture. This finding agrees well with previous evidence suggesting that amyloid fibril formation has many similarities to crystal growth, and that slow polymerization under conditions close to equilibrium allows specific interactions to occur, enabling the polypeptide chains to reach lower energy states (19, 23).
Fig 2.
Fibril morphology as a function of the net molecular charge. (A) Electron micrographs of negatively stained fibrils (c = 1 mM, t = 1 month) from samples where the net charge on the molecule is +1. (B) Electron micrographs of negatively stained fibrils (c = 1 mM, t = 1 month) from samples where the net charge on the molecule is −1.
Structural Characterization.
To carry out a more detailed characterization of the structures formed by these peptides, we selected one of the sequences (no. 3 STVIIE) that forms very regular and homogeneous fibrils (Fig. 2A).
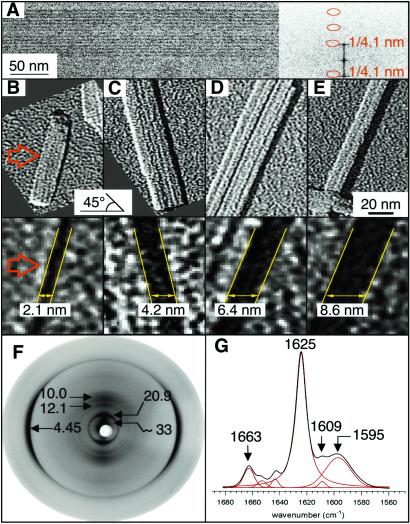
Analysis of these fibrils by cryo-EM revealed ribbon-like structures apparently formed by lateral association of protofilaments with a diameter of ≈41 Å (Fig. 3A). Metal shadowing experiments performed on the fibrils showed that although the height of the ribbons is variable, it is always a multiple of ≈21 Å. This result suggests that “mono-layers” of protofilaments are stacked one on top of another (two, three, and four) in some of the fibrils present in the preparation (Fig. 3 B–E).
Fig 3.
Structural characterization of the fibrils formed by peptide no. 3 (STVIIE) at pH 2.6 by cryo-EM, unidirectional metal shadowing, x-ray fiber diffraction, and FTIR measurements. (A) The center-to-center spacing of the protofilament was determined by cryo-EM. (B–E) The height of flattened amyloid fibrils was determined by high resolution surface shadowing. The elevation angle of the evaporation is 45°, and the azimuthal angle is horizontal (arrow in B). Accordingly, the length of the shadow in the azimuthal direction correlates directly with the height of the fibrils. The smallest (mono-) layers have been found to be in the order of 21 Å (B). In addition, these layers or “ribbons” can stack one on top of another to give multiples of 21 Å such as double (C), triple (D), or even quadruple (E) layers. (F) X-ray diffraction pattern from oriented fibers showing the reflections that define the organization of the protofilaments. One more reflection on the equator can be observed at 5.65 ± 0.03 Å that could arise from a harmonic of either 10.0- or 12.1-Å diffractions (or a combination of both). (G) FTIR amide I band of the fibrils indicating the frequencies of its main components.
X-ray fiber diffraction data from oriented fibrils reveals an anisotropic x-ray diffraction pattern with cross-β reflections, characteristic of amyloid structure, at 4.45 ± 0.05, 10.0 ± 0.5, and 12.1 ± 0.5 Å (Fig. 3F). The reflection at 4.45 Å on the meridian corresponds to the distance between chains in the H-bonding direction and indicates that the β-strands are perpendicular to the fibril axis. This reflection corresponds to a slightly smaller distance than that typically found in amyloid structures (≈4.7 Å; refs. 3 and 4). This reduction in distance might result from the closer interaction of β-strands within the fibrils formed by these small peptides relative to those formed from large peptides or proteins. The pair of reflections of similar intensity at 10.0 and 12.1 Å on the equator arise from the face-to-face separation of the β-sheets and are determined by the side-chain interactions between β-sheets (4). The existence of two inter-sheet distances, as indicated by these reflections, would be consistent with the pairing of “β-sandwiches,” because β-sheet pairing takes place through either face of the β-sheet within this motif. The high degree of order in the structure of the fibrils allowed detection of other equatorial reflections at 20.9 ± 1.3 Å and 33.2 ± 2.3 Å. The first distance coincides well with the height of an individual protofilament, as revealed by unidirectional metal shadowing (Fig. 3 B–E), and with the length of the hexapeptide chain in an extended β-conformation (21 Å, assuming an axial distance of 3.5 Å between adjacent residues). The second reflection (33.2 Å) would then correspond to the distance between the first and the fourth β-sheet within an individual protofilament (10.0 + 12.1 + 10.0 Å). Interestingly, the center-to-center spacing of an individual protofilament as estimated from cryo-EM data is ≈41 Å (Fig. 3A), which could accommodate four β-sheets and fits all of the x-ray diffraction data remarkably well, if we take into account the outer side chains of the edge β-sheets.
Second derivative analysis of the amide I band in the FTIR spectra of these fibrils showed two major components at 1,609 and 1,625 cm−1, characteristic of β-sheet structure (Fig. 3G). Among the other secondary components, a prominent band at 1,663 cm−1 exhibited the same half-width (9 cm−1) as the component at 1,625 cm−1, and it is likely to correspond to a splitting of the main β-sheet component, indicative of an antiparallel arrangement of the β-strands. The splitting band was also observed for peptides 1, 5, and 9, with a net charge of +1 (data not shown). The broad band at 1,595 cm−1 likely arose from additional interactions that occurred in the fibrils, such as protofilament packing.
The data obtained in this study do not provide direct evidence to distinguish whether or not the β-sheets that make up the protofilaments are twisted or flat. However, Fourier transform analysis of cryo-EM pictures along the protofilament length is not consistent with highly twisted β-sheets. Furthermore, if protofilaments consisted of highly twisted β-sheets, there would be a helical twist to the stack of β-sheets around a common axis, coincident with the axis of the protofilament. This twist would determine protofilaments of the same height and width, which is not consistent with our EM measurements. Indeed, very flat β-sheets have been proposed for the amyloid fibrils of the SH3 domain of phosphatidylinositol-3′-kinase and for fibrils formed by insulin (20, 22). β-Sheets without a significant twist are also observed in a number of globular proteins (24).
Structural Model.
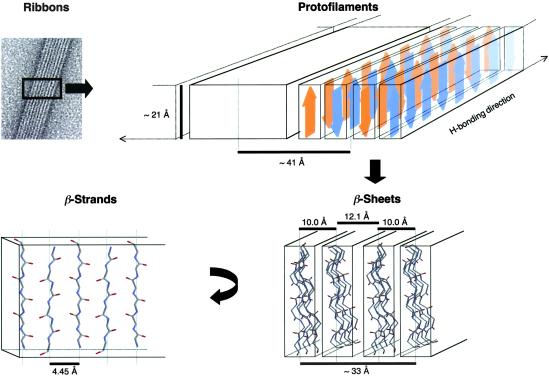
Taken together, these results are consistent with a model of the fibrils in which the protofilaments consist of four flat antiparallel β-sheets of indefinite length running parallel to the axis of the protofilaments (Fig. 4). The strands of the β-sheets are continuously H-bonded and oriented perpendicularly to the plane formed by the fibrillar ribbon. The β-sheets are then organized into two β-sandwiches, where the face-to-face distance between polymeric β-sheets is 10 Å, and the distance between the faces that pair the β-sandwiches is 12.1 Å (Fig. 4). Interestingly, this protofilament organization is identical to that proposed for the protofilament core of amyloid fibrils formed from transthyretin (TTR; ref. 3). The TTR protofilaments in this model also consist of four β-sheets organized into pairs spaced by 10.1 and 12.6 Å. Similar doublets have also been reported for betabellin, a β-sandwich protein designed de novo and for the Aβ (1–40) peptide (25, 26). Differences such as β-sheet spacing (≈12.1 Å for no. 3 vs. ≈12.6 Å for TTR) and protofilament center-to-center spacing (≈41 Å for no. 3 vs. ≈64 Å for TTR) are attributable to differences in amino acid composition and to the part of the TTR molecule that does not participate in the core of the amyloid fibril, respectively. Therefore, this designed peptide reproduces the organization of the protofilament core of naturally occurring amyloid fibrils, which validates its use as model system to study amyloid fibril formation and structure.
Fig 4.
Schematic illustration showing how the peptides might associate to form the ribbons observed by EM. Protofilaments ≈41 Å wide and ≈21 Å high consist of four antiparallel β-sheets of indefinite length running parallel to the axis of the protofilament. β-sheets are organized into two β-sandwiches, where the face-to-face distance between β-sheets is 10.0 and the distance between the faces that pair β-sandwiches is 12.1 Å. The strands of the β-sheets are continuously H-bonded in the direction of the long axis of the fibril, nontwisted with respect to each other and oriented perpendicularly to the plane formed by the fibrillar ribbon. The termini of the β-strands are solvent-exposed.
Discussion
Model Systems for Amyloidogenesis.
The difficulty in studying amyloid deposits formed by natural proteins has prompted the scientific community to devise in vitro models of the complex in vivo aggregation process (5–9, 19). Ideally, a model system for amyloidogenesis must recapitulate what is known about the in vivo process, it must possess suitable physical properties such as reversibility, good solubility, etc., to allow biophysical characterization, and it must also be small enough to permit detailed analysis of the aggregation process. Therefore, approaches such as the one presented here are an important step in developing our understanding of the more general phenomenon of amyloid deposition.
The rational design and the simplicity of the model system developed in this study serve to highlight the delicate balance that must exist between specific side-chain interactions, as some single-point mutations have dramatic effects on the aggregation of the peptide, coulombic effects (noncompensated charges), and main-chain H-bonds involved in the formation of fibrils. Fibril formation will occur only if these interactions outweigh similar ones formed in amorphous aggregates or in the solution state. In larger polypeptides some of these requirements may be overridden by the multiplicity of existing interactions. This complication emphasizes the advantage and usefulness of small systems to dissect the contribution of individual interactions to the complex process of amyloid fibril formation.
Small model systems also offer important advantages for structural analysis. They usually form more homogeneous and regular fibrils than do proteins, which increases enormously the quality of the accessible experimental data. On the other hand, the smaller length of peptides compared with amyloidogenic proteins, allows a more accurate fitting of the molecular structure within the fibrils, once the dimensions of the protofilament are known.
Interactions Driving Amyloid Fibril Formation.
Hydrophobic interactions.
We have shown that single-point mutations of the type Ile->Leu completely prevent amyloid fibril growth under the conditions that we have explored here. Therefore, a simple hydrophobic collapse cannot account for the formation of fibrils and specific interactions are crucial in the stabilization of these fibrillar aggregates relative to the soluble forms of these peptides. This result agrees with findings for proteins linked to familial neurodegenerative diseases, where just a simple point mutation can dramatically promote or accelerate the development of amyloid disease (27).
Coulombic interactions.
We and others have already carried out very exhaustive studies on the role of electrostatics in peptide and protein self-assembly (16–21, 28). From the peptide model system presented here, we have extracted some complementary information on how coulombic interactions modulate the supramolecular organization of peptides having some propensity to polymerize into β-sheets. Under our experimental conditions, designed amyloidogenic peptides with charged residues polymerize into fibrils only when the net charge is ±1. Neutral or higher effective charge on the molecules prevent fibril formation, either because of amorphous aggregation or high-charge repulsion caused by an excess of uncompensated charges, respectively. These results can be explained on the assumption that nonspecific aggregation and fibril formation are competing phenomena (19). Under conditions where the effective charge on the molecule is zero, e.g., it results from the presence of a positive and negative charge in the peptide, any charge would be compensated for by a charge of opposite sign in a hypothetical polymeric antiparallel β-sheet. Because there are no uncompensated charges at the β-sheet level, β-sheets could pack against each other in many ways, leading to amorphous aggregates. In contrast, a net positive or negative charge on the peptide would favor the organization of supramolecular structures in which the distances between charges of the same sign are maximized, thus favoring ordered polymerization. With a large excess of uncompensated charges present, however, it would be energetically highly unfavorable for the molecules to aggregate, and the peptides would remain in solution. Because coulombic interactions are absent in uncharged peptides, self-association processes will be driven exclusively by backbone–backbone H-bonds and side chain–side chain interactions. In this case, such interactions can result in the formation of highly ordered polymers, such as those observed for peptide 5.
In larger proteins, where the number of charged residues may be large and their distribution more complex, the charge balance that promotes fibril formation must be more complicated. What, then, is the relevance of these findings for amyloidogenic proteins in general? Most importantly, they highlight the relevant role of the charges in the supramolecular organization of proteins into fibrils, suggesting that amyloid fibril formation in proteins might take place preferentially for particular charge states of the molecule. This interpretation of the results is also in agreement with experimental data obtained both from peptides and proteins, where the propensity to form fibrils is highly dependent on the pH and ionic strength of the solutions from which aggregation occurs (16–21, 29).
Interactions that mediate protofilament packing.
Because our data suggest that the β-strands in these fibrils are not twisted with respect to their immediate neighbors and are oriented perpendicularly to the plane formed by the fibrillar ribbon, the peptides should expose their charged ends to the solvent. Solvation of non-compensated charges will, in this case, stabilize the system. Because the full-length molecule is essentially the entire protofilament core, interactions between protofilaments can only take place either through the side chains of the fourth and first β-sheet and/or through interactions mediated by water molecules, which would explain the lack of optical density between protofilaments. However, in amyloid fibrils formed by larger peptides or proteins, protofilament interactions are likely to be determined by the regions of the protein which are not integrated within the protofilament core (20).
The present results show that the rational design of a peptide-based model system of amyloid fibril formation can greatly facilitate the identification and rationalization of those elements in sequences that favor the spontaneous conversion of natural proteins into amyloid fibrils. Given that it is likely that only segments of the sequence of large proteins are involved in the β-sheet structure of amyloid fibrils, our results suggest that, in addition, computational methods should be powerful tools for solving the challenging problems associated with the identification of highly amyloidogenic regions of native proteins. Such methods should also be important in the design of molecules capable of inhibiting polymerization or of disrupting preformed amyloid structures, effects that are crucial to many of the therapeutic strategies preventing or treating amyloid diseases that are currently under scrutiny (29, 30). More generally, these methods might also be a very promising approach to solving one of the major challenges in polymer chemistry, namely, rational control over macromolecular architecture by controlling variables such as molecular length, sequence, and stereochemistry.
Note Added in Proof.
The importance of electrostatic interactions on muscle acylphosphatase aggregation has recently been published in the early edition of PNAS (32). The results presented by Chiti et al. strengthen our investigation of the net molecular charge in aggregation and amyloid fibril formation.
Acknowledgments
We thank Dr. Pérez-Paya at the Universidad de Valencia for the synthesis of the peptides in his laboratory and Dr. Heinz Gross at the Eidgenössische Technische Hochschule (Switzerland) for allowing us to use his Midilab setup to perform the unidirectional shadowing experiments. We acknowledge the support of a European Union biotechnology grant (to L.S.) and a European Union Marie Curie Fellowship (to M.L.P.). We also acknowledge support from the Wellcome Trust (to J.Z. and C.M.D.).
Abbreviations
TTR, transthyretine
EM, electron microscopy
This paper was submitted directly (Track II) to the PNAS office.
Gross, H., Krusche, K. & Tittman, P., Twelfth International Congress of Electron Microscopy, 1990, Seattle, WA, abstr. 510–511.
References
- 1.Rochet J.-C. & Landsbury, P. T., Jr. (2000) Curr. Opin. Struct. Biol. 10, 60-68. [DOI] [PubMed] [Google Scholar]
- 2.Dobson C. M. (1999) Trends Biochem. Sci. 24, 329-332. [DOI] [PubMed] [Google Scholar]
- 3.Blake C. & Serpell, L. (1996) Structure (London) 4, 989-998. [DOI] [PubMed] [Google Scholar]
- 4.Sunde M., Serpell, L., Bartlam, M., Fraser, P. E., Pepys, M. B. & Blake, C. C. (1997) J. Mol. Biol. 273, 729-739. [DOI] [PubMed] [Google Scholar]
- 5.Halverson K., Fraser, P. E., Kirschner, D. A. & Lansbury, P. T., Jr. (1990) Biochemistry 29, 2639-2644. [DOI] [PubMed] [Google Scholar]
- 6.von Bergen M., Friedhoff, P., Biernat, J., Heberle, J., Mandelkow, E. M. & Mandelkow, E. (2000) Proc. Natl. Acad. Sci. USA 97, 5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giasson B. I., Murray, I. V. J., Trojanowsky, J. Q. & Lee, V. M.-Y. (2001) J. Biol. Chem. 276, 2380-2386. [DOI] [PubMed] [Google Scholar]
- 8.Balbirnie M., Grothe, R. & Eisenberg, D. S. (2001) Proc. Natl. Acad. Sci. USA 98, 2375-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West M. W., Wang, W., Patterson, J., Mancias, J. D., Beasley, J. R. & Hecht, M. H. (1999) Proc. Natl. Acad. Sci. USA 96, 11211-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lacroix E., (1999) Ph.D. thesis (Universite Libre de Bruxelles).
- 11.Finsinger S., Serrano, L. & Lacroix, E. (2001) Protein Sci. 10, 809-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.López de la Paz M., Lacroix, E., Ramírez-Alvarado, M. & Serrano, L. (2001) J. Mol. Biol. 312, 229-246. [DOI] [PubMed] [Google Scholar]
- 13.Schroeter J. P. & Bretaudiere, J. P. (1996) J. Struct. Biol. 116, 131-137. [DOI] [PubMed] [Google Scholar]
- 14.Zurdo J., Guijarro, J. I. & Dobson, C. M. (2001) J. Am. Chem. Soc. 123, 8141-8142. [DOI] [PubMed] [Google Scholar]
- 15.Pham T. N., Koide, A. & Koide, S. (1998) Nat. Struct. Biol. 5, 115-119. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S., Holmes, T., Lockshin, C. & Rich, A. (1993) Proc. Natl. Acad. Sci. USA 90, 3334-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S. & Rich, A. (1997) Proc. Natl. Acad. Sci. USA 94, 23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caplan M. R., Schwartzfarb, E. M., Zhang, S., Kamm, R. D. & Lauffenburger, D. A. (2002) Biomaterials 23, 219-227. [DOI] [PubMed] [Google Scholar]
- 19.Zurdo J., Guijarro, J. I., Jiménez, J. L., Saibil, H. R. & Dobson, C. M. (2001) J. Mol. Biol. 311, 325-340. [DOI] [PubMed] [Google Scholar]
- 20.Jiménez J. L., Guijarro, J. I., Orlova, E., Zurdo, J., Dobson, C. M., Sunde, M. & Saibil, H. R. (1999) EMBO J. 18, 815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aggeli A., Nyrkova, I. A., Bell, M., Harding, R., Carrick, L., McLeish, T. C. B., Semenov, A. N. & Boden, N. (2001) Proc. Natl. Acad. Sci. USA 98, 11857-11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiménez J. L., Nettleton, E. J., Bouchard, M., Robinson, C. V., Dobson, C. M. & Saibil, H. R. (2002) Proc. Natl. Acad. Sci. USA 99, 9196-9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harper J. D. & Lansbury, P. T. (1997) Annu. Rev. Biochem. 66, 385-407. [DOI] [PubMed] [Google Scholar]
- 24.Liou Y. C., Tocilj, A., Davies, P. L. & Jia, Z. (2000) Nature 406, 322-324. [DOI] [PubMed] [Google Scholar]
- 25.Lim A., Makhov, A. M., Bond, J., Inouye, H., Connors, L. H., Griffith, J. D., Erickson, B. W., Kirschner, D. A. & Costello, C. E. (2000) J. Struct. Biol. 130, 363-370. [DOI] [PubMed] [Google Scholar]
- 26.Malinchik S. B., Inouye, H., Szumowski, K. E. & Kirschner, D. A. (1998) Biophys J. 74, 537-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conway K., Harper, J. & Landsbury, P. T., Jr. (1998) Nat. Med. 4, 951-959. [DOI] [PubMed] [Google Scholar]
- 28.Otzen D. E., Kristensen, O. & Oliveberg, M. (2000) Proc. Natl. Acad. Sci. USA 97, 9907-9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soto C., Sigurdsson, E. M., Morelli, L., Kumar, R. A., Castano, E. M. & Frangione, B. (1998) Nat. Med. 4, 822-826. [DOI] [PubMed] [Google Scholar]
- 30.Soto C., Kascsak, R. J., Saborio, G. P., Aucouturier, P., Wisniewski, T., Prelli, F., Kascsak, R., Mendez, E., Harris, D. A., Ironside, J., et al. (2000) Lancet 355, 192-197. [DOI] [PubMed] [Google Scholar]
- 31.Creighton T. E., (1993) Proteins: Structures and Molecular Properties (Freeman, New York).
- 32.Chiti F., Calamai, M., Taddei, N., Stefani, M., Ramponi, G. & Dobson, C. M. (2002) Proc. Natl. Acad. Sci. USA 99, Suppl. 4, 16419-16426. [DOI] [PMC free article] [PubMed] [Google Scholar]